Abstract
The family Picornaviridae consists of a large group of plus-strand RNA viruses that share a similar genome organization. The nomenclature of the picornavirus proteins is based on their position in the viral RNA genome but does not necessarily imply a conserved function of proteins of different genera. The enterovirus 2B protein is a small hydrophobic protein that, upon individual expression, is localized to the endoplasmic reticulum (ER) and the Golgi complex, reduces ER and Golgi complex Ca2+ levels, most likely by forming transmembrane pores, and inhibits protein trafficking through the Golgi complex. At present, little is known about the function of the other picornavirus 2B proteins. Here we show that rhinovirus 2B, which is phylogenetically closely related to enterovirus 2B, shows a similar subcellular localization and function to those of enterovirus 2B. In contrast, 2B proteins of hepatitis A virus, foot-and-mouth disease virus, and encephalomyocarditis virus, all of which are more distantly related to enteroviruses, show a different localization and have little, if any, effects on Ca2+ homeostasis and intracellular protein trafficking. Our data suggest that the 2B proteins of enterovirus and rhinovirus share the same function in virus replication, while the other picornavirus 2B proteins support the viral life cycle in a different manner. Moreover, we show that an enterovirus 2B protein that is retained in the ER is unable to modify Ca2+ homeostasis and inhibit protein trafficking, demonstrating the importance of Golgi complex localization for its functioning.
The family Picornaviridae is a group of small, nonenveloped cytolytic viruses that include a number of important human and animal pathogens. The picornavirus family consists of nine genera, including enterovirus (e.g., coxsackievirus [CBV] and poliovirus [PV]), rhinovirus (e.g., human rhinovirus [HRV]), cardiovirus (e.g., encephalomyocarditis virus [EMCV]), aphthovirus (e.g., foot-and-mouth disease virus [FMDV]), hepatovirus (hepatitis A virus [HAV]), teschovirus (e.g., porcine teschovirus), erbovirus (e.g., equine rhinitis B virus), parechovirus (e.g., parechovirus 2), and kobuvirus (e.g., aichivirus). In addition, the picornavirus family contains a number of unassigned viruses. All picornaviruses have a similar genome organization. The viral genome typically consists of a positive-stranded RNA molecule of approximately 7,500 to 8,000 nucleotides that contains one single large open reading frame preceded by a long 5′-untranslated region and followed by a much smaller 3′-untranslated region and a genetically encoded poly(A) tail. A small viral protein, VPg, is covalently linked to the 5′ end of the viral genome. Translation of the RNA genome yields a polyprotein of approximately 2,200 amino acids (aa) that is divided into the P1, P2, and P3 regions. The polyprotein is processed by virus-encoded proteases to generate the individual structural and nonstructural proteins. Processing of the P1 region yields the structural capsid proteins 1A (VP4), 1B (VP2), 1C (VP3), and 1D (VP1), whereas processing of the P2 and P3 regions yields the nonstructural replication proteins 2A, 2B, 2C, 3A, 3B (VPg), 3C, and 3D as well as cleavage intermediates (2BC, 3AB, and 3CD) that are relatively stable and may serve other functions from those of their individual constituents. It should be emphasized that the nomenclature of the picornavirus proteins is based on their position in the viral RNA genome (1A to 3D from the 5′ to the 3′ end) and does not necessarily imply a conservation of function between the different genera. The functions of the nonstructural picornavirus proteins have been investigated by analysis of well-defined mutants, by expression in bacteria and eukaryotic cells, and by enzymatic assays in vitro. Multiple functions have been attributed to the mature viral proteins and the cleavage intermediates, but their exact role in the picornaviral replication cycle is still not fully understood (reviewed in references 22, 28, and 42).
Little is known about the function of the picornavirus 2B proteins. Most of our current understanding of 2B stems from studies of enteroviruses. In enterovirus-infected cells, 2B is present both as a mature protein and as part of the 2BC protein, a relatively stable precursor protein that is involved in cytosolic accumulation of the secretory pathway-derived membrane vesicles, where viral replication takes place (4, 31, 35). Studies of both PV and CBV indicate that 2B plays an important role in the modification of intracellular membrane structures and functions. The 2B protein is a small hydrophobic membrane protein that localizes at endoplasmic reticulum (ER) and Golgi complex membranes (4, 15, 33). Increasing evidence indicates that 2B forms homomultimers that build pores in ER and Golgi complex membranes (1, 11, 12-14, 41), thereby reducing the levels of Ca2+ and H+ in the lumens of these organelles in infected cells (8, 14, 39). Individual expression of 2B furthermore results in inhibition of protein trafficking through the Golgi complex (14, 17). It is unknown whether these activities represent different functions of 2B or whether the one activity is the consequence of the other. The observation that 2B mutants that are impaired in increasing the efflux of ions from the ER and Golgi complex are also impaired in inhibiting protein trafficking suggests that these activities are somehow connected (8, 14). The relevance of these 2B activities for the viral life cycle is still poorly understood. Mutations that interfere with the ability of 2B to disturb ER and Golgi complex ion homeostasis and/or to inhibit membrane trafficking cause early defects in viral RNA replication (9, 40). These 2B functions may be required for the activity of the precursor 2BC to accumulate membranous replication vesicles, but other possibilities cannot be excluded.
In this study, we investigated whether the structural and functional aspects of the enterovirus 2B protein are conserved among other members of the picornavirus family. To this end, the phylogenetic relationships and degree of conservation between the 2B proteins of CBV, PV, rhinovirus, hepatovirus, aphthovirus, and cardiovirus were defined. In addition, subcellular localization and possible effects on organelle Ca2+ homeostasis and intracellular protein trafficking were investigated. Finally, the importance of the subcellular localization of enterovirus 2B for its functions was studied.
MATERIALS AND METHODS
Cells and medium.
Buffalo green monkey (BGM) cells were grown in minimal essential medium (Gibco) supplemented with 10% fetal bovine serum, 100 units penicillin per ml, and 25 μg streptomycin per ml. Cells were grown at 37°C in a 5% CO2 incubator.
Antisera.
Rabbit polyclonal anti-enhanced green fluorescent protein (anti-EGFP) was described previously (15). Mouse monoclonal anti-c-myc (clone 9E10) and anti-Flag M2 antisera were obtained from Sigma-Aldrich. Rabbit polyclonal anti-calreticulin was obtained from Affinity Bioreagents, Inc. Fluorescein isothiocyanate-conjugated goat anti-rabbit polyclonal antibody, Texas Red-conjugated goat anti-mouse polyclonal antibody, and Texas Red-conjugated goat anti-rabbit polyclonal antibody were obtained from Jackson ImmunoResearch Laboratories.
Plasmids.
pVSV-G(ts045)-GFP (37) was a kind gift from P. Keller and K. Simons, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany. The expression constructs pER-AEQ (30), pGolgi-AEQ (27), p2B-EGFP, 2B-EGFP-AAAA, 2B-EGFP-KKAA, and p2B-myc p2B-I64S/V66S-EGFP for CBV3 have been described previously (12, 15). For the construction of p2B-EGFP and p2B-myc plasmids for the other picornavirus 2B proteins, the 2B coding sequences were amplified and cloned into the p2B-EGFP and p2B-myc constructs, from which the CBV3 2B region was removed using SalI and BamHI restriction sites. The forward primers contained the SalI restriction site (italics) and a start codon preceded by a Kozak sequence (underlined). The reverse primers contained a BamHI restriction site (italics). The PV1 2B protein was amplified using the cDNA clone pXpA (29) and primers p374-11 (5′-GGGGGGGTCGACGCCACCATGGGCATCACCAATTACATAGAG-3′; nucleotide [nt] 3833) and p374-12 (5′-CCCCCCGGATCCTGCTTGATGACATAAGGTAT-3′; nt 4123). The HRV14 2B protein was amplified using the cDNA clone pWR3.26 (21) and primers p374-1 (5′-TGGGACAGTGTCGACGCCACCATGGGGCTGAGTGATTACATCACAGGT-3′; nt 3635) and p374-2 (5′-GATCCAGCTGGATCCTTGTCTTTCAATGTAAGGCAC-3′; nt 3925). The HAV 2B protein was amplified using the cDNA clone pHAV/7 (a plasmid containing the cDNA of the cell culture-adapted HAV175 virus) (10) and primers p374-13 (5′-TGGGACAGTGTCGACGCCACCATGGCTAAAATTTCTCTTTTTTATACTGAG-3′; nt 3243) and p374-4 (5′-GATCCAGCTGGATCCCTGAGTCCTTAACTCCATCTTCTGGA-3′; nt 3995). The FMDV 2B protein was amplified using the cDNA clone pMR15 (FMDV type 01K) (32) and primers p374-7 (5′-TGGGACAGTGTCGACGCCACCATGGGGCCCTTCTTTTTCTCCGACGTTAGG-3′; nt 3883) and p374-8 (5′-GATCCAGCTGGATCCCTGTTTCTCTGCTCTCTCAAGGTCTTCGGG-3′; nt 4344). The 2B protein of EMCV (strain mengovirus) was amplified using the cDNA clone pM16.1 (18) and primers p374-5 (5′-TGGGACAGTGTCGACGCCACCATGCCTTTCACGTTTAAACCAAGACAACGG-3′; nt 3966) and p374-6 (5′-GATCCAGCTGGATCCCTGCTGTTGGAAAAGTGAGATCACAGG-3′; nt 4415).
Phylogenetic analysis.
Alignments were made using the Blosum62 similarity matrix (20). A phylogenetic tree was constructed using Clustal W (1.81) multiple sequence alignment software (36).
Subcellular localization.
Immunofluorescence was performed as described previously (15). Briefly, BGM cells were grown on coverslips and transfected with 1 μg of picornavirus p2B-myc or with 0.5 μg p2B-myc and 0.5 μg pEGFP-Golgi per well, using the FuGENE transfection reagent (Roche) as described previously (13). Cells were fixed, permeabilized, and stained at 24 h posttransfection. Primary antibodies were diluted 1:200 (anti-c-Myc) or 1:150 (anti-calreticulin). Conjugates were diluted 1:200. Cells were analyzed by confocal laser scanning microscopy (CLSM) (Leica TCS NT; Leica Lasertechnik GmbH, Heidelberg, Germany).
Intraorganelle calcium homeostasis.
ER and Golgi complex Ca2+ concentrations were determined using the calcium-sensitive photoprotein aequorin in a bioluminescence assay as described previously (8). Briefly, BGM cells were grown on coverslips and transfected with 0.2 μg of picornavirus p2B-myc and 0.2 μg of either pCI-neo, pCI-ER-AEQ, or pCI-Golgi-AEQ per well, using the FuGENE transfection reagent (Roche) as described previously (13). At 24 h posttransfection, aequorins were reconstituted with coelenterazine-N. To efficiently reconstitute the aequorin chimeras and to measure [Ca2+] reliably, the luminal [Ca2+] must first be reduced. This was achieved by incubating the cells in HEPES-Tris medium supplemented with coelenterazine (i.e., the prosthetic group of aequorin) and ionomycin (a Ca2+ ionophore) in the absence of Ca2+. Cells were then transferred to a luminometer and superfused with HEPES-Tris medium without Ca2+ (132 mM NaCl, 4.2 mM KCl, 1 mM MgCl2, 5.5 mM d-glucose, 10 mM HEPES, 500 μM EGTA) at 2 ml per min. Cells were subsequently superfused with HEPES-Tris buffer lacking EGTA and containing 1 mM CaCl2. The recorded aequorin luminescence data were calibrated offline into [Ca2+] values by using a computer algorithm based on the [Ca2+] response curves of mutant aequorins (2).
Inhibition of protein trafficking.
Immunofluorescence was performed as described above (see “Subcellular localization”). BGM cells were transfected with 0.5 μg p2B-myc and 0.5 μg pVSV-G-GFP(ts045), incubated at 40°C for 18 h, and additionally incubated at 32°C for 2 h. Cells were fixed, permeabilized, and stained. Cells were analyzed by CLSM (Leica TCS NT; Leica Lasertechnik GmbH, Heidelberg, Germany).
Coimmunoprecipitation.
BGM cells were cotransfected with EGFP-tagged and myc-tagged or Flag-tagged 2B proteins. At 24 h posttransfection, cells were lysed and EGFP-tagged proteins were precipitated using an anti-EGFP antibody, as described previously (15). Samples were run in sodium dodecyl sulfate-polyacrylamide gels, transferred to nitrocellulose membranes, and immunodetected with antisera against both EGFP and either c-myc or Flag. Proteins were visualized using a chemiluminescence detection system (Amersham Pharmacia Biotech).
Statistical analysis.
Data are presented as mean values ± standard deviations. Differences were tested for significance by means of Student's t test.
RESULTS
Phylogenetic analysis of picornavirus 2B proteins.
A phylogenetic analysis of the 2B proteins of enterovirus, rhinovirus, cardiovirus, aphthovirus, and hepatovirus was performed. The enterovirus genus can be divided into two subgroups (on the basis of differences in amino acid sequence), the CBV-like subgroup and the PV-like subgroup. Both CBV3 2B and PV1 2B were included for a direct comparison of their effects in the same cell type, using the same expression constructs. The rhinovirus genus is divided into groups A and B (34). Sequence comparison of the 2B proteins of HRV group A and HRV group B showed that they were very similar (data not shown), and therefore only HRV14 (HRV group B) was used as a representative of the rhinovirus genus. EMCV was used as a representative of the cardiovirus genus, FMDV was used as a representative of the aphthovirus genus, and HAV was used as a representative of the hepatovirus genus.
Phylogenetic analysis showed that the 2B proteins of CBV3 (99 aa), PV1, and HRV14 (both 97 aa) are closely related (Fig. 1A). The 2B proteins of both PV1 and HRV14 share 51% and 55% identity (71% and 74% similarity), respectively, with CBV3 2B and 49% identity (71% similarity) with each other (Fig. 1B). Like CBV3 2B, both PV1 2B and HRV14 2B contain two hydrophobic regions (referred to as HR1 and HR2) (Fig. 1C), of which the first is predicted to form a cationic amphipathic α-helix, typical of membrane-lytic polypeptides (Fig. 1D), separated by a short hydrophilic sequence (38).
FIG. 1.
Genetic analysis of picornavirus 2B proteins. (A) Phylogenetic tree of the 2B proteins constructed with Clustal W software. The Blosum62 similarity matrix was used to perform sequence alignment analysis. The phylogenetic tree shows that the 2B proteins of CBV3, PV1, and HRV14 are grouped together, whereas the 2B proteins of viruses from other genera are more distantly related. (B) Identity matrix for 2B proteins. Identity was calculated using the Blosum62 similarity matrix. (C) Pairwise alignment of CBV3, PV1, and HRV14 2B proteins. All three proteins contain two hydrophobic regions (HR1 and HR2) spaced by a 5-aa hydrophilic sequence. Dots represent residues that are identical to those in CBV3 2B. Dashes indicate gaps in the alignment. (D) Top view of the amphipathic α-helices of the first hydrophobic region of CBV3, PV1, and HRV14 2B proteins. Note that all three proteins contain a hydrophobic backbone and a hydrophilic face that contains three cationic residues. Hydrophobic residues are boxed. CBV3, coxsackievirus B3; HRV14, human rhinovirus 14; PV1, poliovirus 1; FMDV, foot-and-mouth disease virus; EMCV, encephalomyocarditis virus; HpaV2, human parechovirus 2; PTV, porcine teschovirus; ERBV, equine rhinitis B virus; HAV, hepatitis A virus.
The 2B proteins of EMCV, FMDV, and HAV share little, if any, relationship (<20% sequence identity) to the enterovirus and rhinovirus 2B proteins (Fig. 1A and B). They also share little sequence identity with each other or with the 2B proteins of aichivirus (kobuvirus), human parechovirus 2 (parechovirus), porcine teschovirus (teschovirus), and equine rhinitis B virus (erbovirus) (Fig. 1A and B). The EMCV, FMDV, and HAV 2B proteins are larger than the enterovirus and rhinovirus 2B proteins (EMCV 2B, 151 aa; FMDV 2B, 155 aa; and HAV 2B, 251 aa). They contain one or more hydrophobic regions, but none of them contains a cationic amphipathic α-helix typical of the group of membrane-lytic polypeptides, as can be found in all enterovirus and rhinovirus 2B proteins.
Subcellular localization of picornavirus 2B proteins.
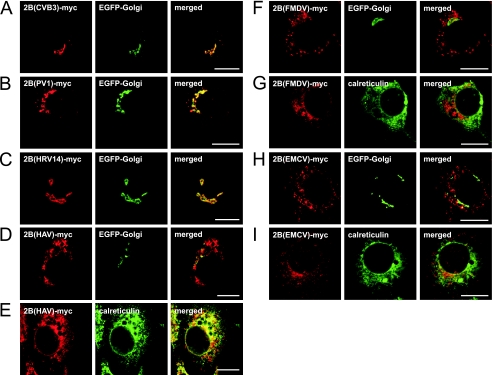
CBV3 2B is present predominantly in the Golgi complex upon individual expression (15). Here we determined the subcellular localization of the other picornavirus 2B proteins. To this end, the coding regions of the picornavirus 2B proteins were amplified and cloned into a eukaryotic expression vector that drives the expression of proteins carrying a c-myc tag at the C terminus. The experiments were performed with BGM kidney cells because all viruses used in this study are able to replicate in kidney cell lines, indicating that the 2B proteins have the ability to fulfill their function(s) in these cells (5, 7, 25, 26). BGM cells were transfected with the expression constructs, and the subcellular distribution of the 2B proteins was compared with that of markers for the Golgi complex (EGFP-Golgi) and the ER (calreticulin), as described previously (15).
Like CBV3 2B, PV1 2B colocalized predominantly with the EGFP-Golgi protein (Fig. 2A and B). The closely related HRV14 2B showed a subcellular distribution that was similar to that of enterovirus 2B and also predominantly colocalized with the Golgi marker (Fig. 2C). The more distantly related HAV, EMCV, and FMDV 2B proteins showed different subcellular localizations. HAV 2B stained in a reticular pattern that included the nuclear envelope and was largely identical to staining of calreticulin (Fig. 2E), indicating that HAV 2B is localized to the ER. HAV 2B was virtually absent from the Golgi complex (Fig. 2D), but in some highly expressing cells, some colocalization of HAV 2B with EGFP-Golgi was observed (not shown). In lowly and moderately expressing cells, FMDV and EMCV 2B proteins showed no colocalization with either calreticulin or EGFP-Golgi (Fig. 2F to I), indicating that these proteins are not present in the ER or the Golgi complex. In cells expressing high levels of FMDV 2B, staining and morphology of both the ER and the Golgi complex were often affected, yet there was still no obvious colocalization of 2B and the ER or Golgi marker (not shown).
FIG. 2.
Subcellular localization of picornavirus 2B proteins. (A to D, F, and H) BGM cells were cotransfected with EGFP-Golgi and the indicated 2B-myc constructs, fixed at 24 h posttransfection, and stained with anti-c-myc antiserum. (E, G, and I) BGM cells were transfected with the indicated 2B-myc constructs, fixed at 24 h posttransfection, and stained with anti-c-myc and anti-calreticulin antisera. Cells were analyzed by CLSM. The 2B-myc proteins of CBV3 (A), PV1 (B), and HRV14 (C) colocalized with the Golgi marker. HAV 2B did not colocalize with the Golgi marker (D) but was colocalized predominantly with the ER marker (E). FMDV 2B (F and G) and EMCV 2B (H and I) did not colocalize with the Golgi and ER markers. Bars = 10 μm.
Intraorganelle calcium concentrations in 2B-expressing cells.
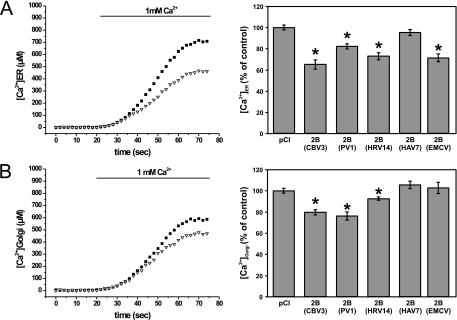
We previously demonstrated that expression of CBV3 2B reduced ER and Golgi complex Ca2+ levels (8, 14). Here we analyzed whether the other picornavirus 2B proteins also have the ability to modify ER and Golgi complex Ca2+ homeostasis, using the Ca2+-sensitive photoprotein aequorin as described previously (8). To this end, BGM cells were cotransfected with the different 2B-myc constructs and either pCI-ER-AEQ, pCI-Golgi-AEQ, or the empty pCI expression vector. Ca2+ measurements were performed at 24 h posttransfection. Offline calibration of the aequorin bioluminescence data into [Ca2+] values revealed that expression of CBV3, PV1, or HRV14 2B resulted in a significant decrease in intraluminal ER and Golgi complex [Ca2+] (Fig. 3). HAV 2B expression did not alter ER or Golgi complex [Ca2+], while EMCV 2B significantly decreased ER [Ca2+] but not Golgi complex [Ca2+] (Fig. 3). Unfortunately, we were unable to reliably measure the ER and Golgi complex [Ca2+] in FMDV 2B-expressing cells. Immunofluorescence analysis showed that the aequorin probes were aberrantly localized in cells expressing high levels of FMDV 2B (not shown). Since the aequorin probes measure the average ER or Golgi complex [Ca2+] of the total population of 2B-expressing cells, false results could be obtained due to mistargeting of the aequorin probes to low-[Ca2+] compartments in cells expressing high levels of FMDV 2B.
FIG. 3.
Analysis of ER and Golgi complex stored Ca2+ in cells expressing picornavirus 2B proteins. BGM cells were cotransfected with ER-targeted (A) or Golgi complex-targeted (B) aequorin and the indicated 2B-myc constructs. At 24 h posttransfection, ER and Golgi complex stores were depleted of Ca2+ and the aequorins were reconstituted with coelenterazine. Cells were perfused with HT medium without Ca2+ and subsequently with HT medium supplemented with 1 mM free Ca2+. The recorded aequorin luminescence data were calibrated offline into [Ca2+] values. Representative traces are shown for CBV3 2B. (A) ER Ca2+ levels were reduced in cells expressing CBV3, PV1, HRV14, or EMCV 2B but not in cells expressing HAV 2B. (B) Golgi complex Ca2+ levels were reduced in cells expressing CBV3, PV1, or HRV14 2B but not in cells expressing HAV or EMCV 2B.
Effects of picornavirus 2B proteins on protein trafficking through the secretory pathway.
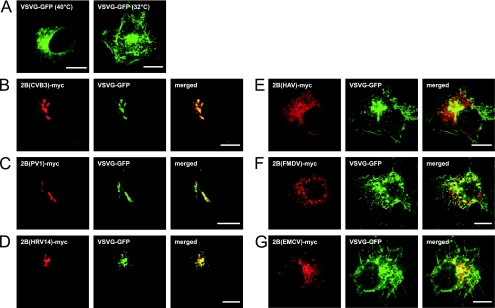
Next, we investigated whether the other picornavirus 2B proteins are able to interfere with protein trafficking through the secretory pathway. We used a temperature-sensitive mutant of the envelope glycoprotein of vesicular stomatitis virus (VSV-G-ts045; hereafter referred to as VSV-G) fused to fluorescent EGFP (VSV-G-GFP) to study protein trafficking. The temperature-sensitive VSV-G-GFP mutant is misfolded at 40°C, resulting in its accumulation in the ER. Upon shifting of the temperature to 32°C, the protein is refolded correctly and transported via the secretory pathway, resulting in its exposure on the plasma membrane (16).
BGM cells were cotransfected with expression constructs for VSV-G-GFP and the picornavirus 2B-myc proteins. Cells were incubated at 40°C for 18 h to accumulate VSV-G-GFP in the ER (Fig. 4A, left panel) and subsequently incubated at 32°C for 2 h to allow VSV-G-GFP transport out of the ER (Fig. 4A, right panel). Cells were fixed, stained with the anti-c-myc antiserum, and processed for CLSM analysis. Similar to CBV3 2B, the 2B proteins of PV1 and HRV14 efficiently inhibited protein trafficking through the Golgi complex (Fig. 4B to D). In cells expressing these 2B proteins, VSV-G-GFP accumulated in the juxtanuclear Golgi region, where it colocalized with the respective 2B proteins. This demonstrates that the ability of 2B to inhibit protein trafficking through the Golgi complex is conserved among these three closely related viruses. In contrast, expression of HAV, FMDV, or EMCV 2B did not result in the inhibition of protein trafficking. VSV-G-GFP was predominantly localized on the plasma membrane and showed no colocalization with these 2B proteins (Fig. 4E to G).
FIG. 4.
Analysis of protein trafficking in cells expressing picornavirus 2B proteins. BGM cells were transfected with the VSV-G-GFP construct (A) or cotransfected with VSV-G-GFP and the picornavirus 2B-myc constructs (B to G). Cells were incubated at 40°C for 18 h to accumulate VSV-G-GFP in the ER and subsequently incubated at 32°C for 2 h (B to G). Cells were fixed, stained with the anti-c-myc antiserum, and processed for CLSM analysis. (A) Accumulation of VSV-G-GFP in the ER upon incubation at 40°C (left) and on the plasma membrane upon additional incubation at 32°C (right). VSV-G-GFP accumulated in the Golgi complex upon incubation with CBV3 2B (B), PV1 2B (C), and HRV14 2B (D). VSV-G-GFP was exposed on the plasma membrane in cells expressing HAV 2B (E), FMDV 2B (F), or EMCV 2B (G). Bars = 10 μm.
Is Golgi complex localization of 2B required for alterations in calcium homeostasis and protein trafficking?
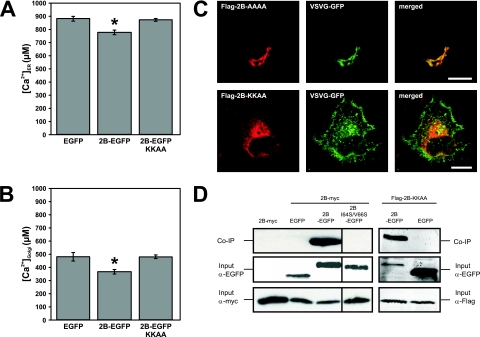
Thus far, we have demonstrated that the 2B proteins of PV1, CBV3, and HRV14 are closely related and that these proteins localize predominantly to the Golgi complex, reduce ER and Golgi complex Ca2+ levels, and inhibit protein trafficking. We asked ourselves whether the Golgi complex localization of enterovirus and rhinovirus 2B proteins is required for their ability to modify Ca2+ homeostasis and to inhibit protein trafficking or if 2B can also exert these functions from the ER. To this end, we made use of wild-type CBV3 2B-EGFP carrying a C-terminal KKAA dilysine motif (2B-EGFP-KKAA), which acts as an ER retention signal in mammalian cells (15). Cells were cotransfected with expression constructs for ER-targeted or Golgi complex-targeted aequorins and either 2B-EGFP-KKAA or the control plasmid EGFP or 2B-EGFP-AAAA, and the ER and Golgi complex Ca2+ concentrations were determined as described above. Figure 5A and B show that expression of 2B-EGFP-KKAA had no effect on the filling state of the ER and Golgi complex Ca2+ stores. The finding that 2B-EGFP-AAAA expression reduced both ER and Golgi complex Ca2+ levels demonstrates that the presence of a C-terminal tag does not inhibit 2B-EGFP function. Consistently, the ER-retained Flag-2B-KKAA did not alter VSV-G-GFP trafficking, while Flag-2B-AAAA expression resulted in the accumulation of VSV-G-GFP in the juxtanuclear Golgi area, where it colocalized with 2B-AAAA (Fig. 5C). The possibility that the functions of ER-retained 2B-EGFP-KKAA are impaired because the KKAA tag somehow interferes with 2B homomultimerization, which is a prerequisite for 2B to exert its functions, was considered (8, 12, 14, 15). To investigate multimerization, coimmunoprecipitation experiments were set up. First, multimerization of wild-type 2B was tested using this method. BGM cells coexpressing wild-type 2B-EGFP and 2B-myc were lysed at 24 h posttransfection, and the EGFP-tagged proteins were precipitated using an anti-EGFP antibody as described previously (15). Western blot analysis showed that 2B-myc was efficiently coimmunoprecipitated with 2B-EGFP but not with EGFP alone or the multimerization-deficient mutant 2B-I64S/V66S-EGFP (13) (Fig. 5D). To investigate homomultimerization of 2B-KKAA, BGM cells were cotransfected with Flag-2B-KKAA and 2B-EGFP-KKAA expression constructs. Figure 5D shows that Flag-2B-EGFP was coimmunoprecipitated with 2B-EGFP-KKAA but not with EGFP, indicating that the ability of 2B to homomultimerize was not affected by the presence of the KKAA tag. All proteins were efficiently expressed, excluding the possibility that differences in coprecipitation were due to different expression levels.
FIG. 5.
Importance of Golgi complex localization for the abilities of CBV3 2B to modify Ca2+ homeostasis and to inhibit protein trafficking. (A and B) ER and Golgi complex Ca2+ levels. BGM cells coexpressing ER-targeted (A) or Golgi complex-targeted (B) aequorin and the indicated 2B or control constructs were analyzed at 24 h posttransfection as described in the legend to Fig. 3. ER and Golgi complex Ca2+ levels were unchanged in cells expressing ER-localized 2B-EGFP-KKAA or the EGFP control, while 2B-EGFP expression decreased ER and Golgi complex Ca2+ levels. (C) Protein trafficking. BGM cells coexpressing VSV-G-GFP and either Flag-2B-KKAA or Flag-2B-AAAA were analyzed for VSV-G-GFP trafficking as described in the legend to Fig. 4. Flag-2B-KKAA did not alter VSV-G-GFP trafficking, while expression of control Flag-2B-AAAA resulted in the accumulation of VSV-G-GFP in the juxtanuclear Golgi area, where it colocalized with Flag-2B-AAA. Bars = 10 μm. (D) Homomultimerization. BGM cells (co)expressing the indicated proteins were lysed at 24 h posttransfection, and lysates were subjected to immunoprecipitation using an anti-EGFP antibody. Coimmunoprecipitation of 2B-myc with 2B-EGFP is demonstrated, while 2B-myc could not be precipitated from lysates of cells expressing 2B-myc alone or coexpressing 2B-myc and EGFP or 2B-I64S/V66S-EGFP. Flag-2B-KKAA was coimmunoprecipitated with 2B-EGFP-KKAA but not with EGFP. Analysis of the lysates that were used as input for the coimmunoprecipitation experiments demonstrated that all proteins were properly expressed.
Together, these results further emphasize the importance of the Golgi complex localization of enterovirus 2B, and most likely also of rhinovirus 2B, for its activities in the host cell.
DISCUSSION
This study was performed to gain more insight into the phylogenetic conservation of the structure and function of the picornavirus 2B proteins. Our results demonstrate that the structural and functional properties of enterovirus 2B (represented in this study by CBV3 and PV1 2B) and rhinovirus 2B (represented by HRV14 2B) are strongly conserved. Phylogenetic analysis revealed that CBV3, PV1, and HRV14 2B proteins are closely related and share approximately 50% identity and >70% similarity. Despite differences in amino acid sequence, CBV3, PV1, and HRV14 2B proteins share remarkable structural similarities. These proteins are similar in length (97 to 99 aa) and contain two hydrophobic regions, the first of which is predicted to form a cationic amphipathic α-helix typical for the group of lytic polypeptides. The amphipathic α-helices of these 2B proteins all contain three lysine residues in the (small) hydrophilic face of the helix. CBV3 2B is present at ER and Golgi complex membranes (15) and is responsible for the release of Ca2+ and H+ from these organelles, most likely by forming membrane-integral pores (8, 14). Moreover, CBV3 2B has been shown to inhibit protein trafficking through the Golgi complex and to increase hygromycin B entry (14, 40). Here we demonstrate that PV1 and HRV14 2B proteins also localize to the Golgi complex, reduce ER and Golgi complex Ca2+ levels, and inhibit VSV-G trafficking through the Golgi complex, whereas the more distantly related HAV, FMDV, and EMCV 2B proteins behave differently.
The 2B protein of HAV (a member of the hepatoviruses) shares little or no sequence and structural homology with the other picornavirus 2B proteins. HAV 2B had little, if any, effect on ER and Golgi complex Ca2+ homeostasis. Moreover, it did not inhibit protein trafficking through the secretory pathway. The latter finding is consistent with the hypothesis that HAV uses the secretory pathway to leave the cell, as virus release of the cell culture-adapted HAV strain from polarized epithelial cells was shown to be sensitive to trafficking-inhibiting drugs (6). Analysis of the subcellular localization of HAV 2B demonstrated that it localized predominantly in the ER and was absent from the Golgi complex. Immunoelectron microscopy analysis of the subcellular localization of HAV 2B in transfected cells (FRhK-4 and HeLa) demonstrated that the protein was present almost exclusively in a tubular-vesicular network, whose origin (i.e., the presence of organelle-specific markers) was not investigated (19). Our finding that HAV 2B largely colocalizes with a marker for the ER suggests that the tubular-vesicular structures may be derived, at least partly, from the ER.
The 2B proteins of FMDV and EMCV (members of the aphthoviruses and cardioviruses, respectively) are not related to any of the other picornavirus 2B proteins or to each other and showed a different localization from those of the other 2B proteins. In lowly to moderately expressing cells, both FMDV 2B and EMCV 2B appeared to be absent from the ER and the Golgi complex. Instead, they showed a punctate localization pattern in the cytosol, whose exact nature remains to be established. The localization of these proteins in highly expressing cells is less clear, and we cannot exclude the possibility that in these cells there is some degree of colocalization with ER markers, as seen for FMDV 2B (23, 24). Although EMCV 2B does not show a clear ER localization and lacks an amphipathic helix that is typical for the group of lytic polypeptides, expression of this protein was found to reduce the ER Ca2+ level (but not the Golgi complex Ca2+ level). The mechanism underlying this reduction remains to be established. Unfortunately, possible effects of FMDV 2B on ER and Golgi complex Ca2+ levels could not be investigated reliably because overexpression of this protein interfered with the correct localization of the calcium-sensitive aequorin probes. Neither FMDV 2B nor EMCV 2B interfered with protein trafficking through the secretory pathway. We are not aware of studies addressing protein trafficking in EMCV-infected cells, but early secretory pathway transport in FMDV-infected cells is inhibited (23). In agreement with our results, Moffat and coworkers recently showed that FMDV 2B alone does not inhibit transport but that coexpression of 2B and 2C or expression of the 2BC precursor protein is required for this effect (24).
Expression of the 2B proteins of both PV1 and CVB3 was previously shown to result in increased entry of hygromycin B, a drug that under normal conditions enters cells poorly (12, 15, 17, 40). The mechanism by which these enterovirus 2B proteins increase hygromycin B entry is still unknown. A possible explanation was recently provided by Cornell et al., who showed that endocytosis is upregulated upon individual expression of CVB3 2B and 2BC (10). There are strong indications that this effect is directly linked to the abilities of 2B to reduce ER and Golgi complex Ca2+ levels and to inhibit protein transport because mutations that disrupt these abilities also impair the ability of 2B to increase hygromycin B entry (8, 12, 14, 15, 40). Consistently, we found that expression of HRV 2B caused a similar strong increase in hygromycin B entry to that of CBV 2B, whereas expression of HAV, FMDV, and EMCV 2B proteins had little effect (data not shown).
To investigate the importance of the Golgi complex localization of the enterovirus 2B protein for its function(s), we tested a (wild-type) CBV3 2B protein that is retained in the ER by virtue of an ER retention signal. Through this approach, evidence was obtained that the ability of 2B to reduce ER and Golgi complex Ca2+ levels as well as to inhibit protein trafficking critically depends on its Golgi complex localization. The observation that this ER-retained protein was unable to reduce the ER Ca2+ level was surprising. Coimmunoprecipitation experiments showed that this was not due to impaired formation of multimers in the ER. The finding that formation of 2B multimers in the ER is not sufficient to trigger the efflux of Ca2+ from this store indicates that 2B multimerization is not sufficient for pore formation. A possible explanation is that the mode of membrane interaction of 2B in the ER differs from that in the Golgi complex, possibly due to differences in lipid composition of these organelles that may affect the architecture of 2B multimers. Previously, we showed that the ER-retained CVB 2B also failed to increase the entry of hygromycin B (15). Although its exact function in the viral life cycle remains to be elucidated, the observations reported here point to the Golgi complex as the major target organelle of the enterovirus/rhinovirus 2B proteins, from where they exert (part of) their function(s).
Taken together, our results have demonstrated that the abilities of the enterovirus 2B protein to reduce ER and Golgi complex stored Ca2+ and to inhibit protein trafficking through the Golgi complex are conserved in the closely related rhinovirus 2B protein but not in the more distantly related 2B proteins of HAV, FMDV, and EMCV. We conclude that the 2B proteins of enterovirus and rhinovirus are closely related and most likely exhibit the same function in the viral life cycle.
Acknowledgments
We thank P. Keller and K. Simons for the kind gift of the pVSV-G(ts045)-GFP construct, R. Andino for the kind gift of the pXpA plasmid, R. Rueckert for the pWR3.26 plasmid, S. Emerson for the pHAV/7 plasmid, M. Ryan for the pMR15 plasmid, and A. Palmenberg for the pM16.1 plasmid.
This work was partly supported by grants from The Netherlands Organization for Scientific Research (NWO-VIDI-917.46.305), the M. W. Beijerink Virology Fund from the Royal Netherlands Academy of Sciences, and the European Communities (INTAS 2012).
Footnotes
Published ahead of print on 23 January 2008.
REFERENCES
- 1.Agirre, A., A. Barco, L. Carrasco, and J. L. Nieva. 2002. Viroporin-mediated membrane permeabilization. Pore formation by nonstructural poliovirus 2B protein. J. Biol. Chem. 27740434-40441. [DOI] [PubMed] [Google Scholar]
- 2.Barrero, M. J., M. Montero, and J. Alvarez. 1997. Dynamics of [Ca2+] in the endoplasmic reticulum and cytoplasm of intact HeLa cells. A comparative study. J. Biol. Chem. 27227694-27699. [DOI] [PubMed] [Google Scholar]
- 3.Reference deleted.
- 4.Bienz, K., D. Egger, and L. Pasamontes. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160220-226. [DOI] [PubMed] [Google Scholar]
- 5.Bienz, K., D. Egger, Y. Rasser, and H. Loeffler. 1978. Differential inhibition of host cell RNA synthesis in several picornavirus-infected cell lines. Intervirology 10209-220. [DOI] [PubMed] [Google Scholar]
- 6.Blank, C. A., D. A. Anderson, M. Beard, and S. M. Lemon. 2000. Infection of polarized cultures of human intestinal epithelial cells with hepatitis A virus: vectorial release of progeny virions through apical cellular membranes. J. Virol. 746476-6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burrage, T., E. Kramer, and F. Brown. 2000. Structural differences between foot-and-mouth disease and poliomyelitis viruses influence their inactivation by aziridines. Vaccine 182454-2461. [DOI] [PubMed] [Google Scholar]
- 8.Campanella, M., A. S. De Jong, W. J. G. Melchers, P. H. G. M. Willems, P. Pinton, R. Rizzuto, and F. J. M. Van Kuppeveld. 2004. The coxsackievirus 2B protein suppresses apoptotic host cell responses by manipulating intracellular Ca2+ homeostasis. J. Biol. Chem. 27918440-18450. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, J. I., B. Rosenblum, J. R. Ticehurst, R. J. Daemer, S. M. Feinstone, and R. H. Purcell. 1987. Complete nucleotide sequence of an attenuated hepatitis A virus: comparison with wild-type virus. Proc. Natl. Acad. Sci. USA 842497-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornell, C. T., W. B. Kiosses, S. Harkins, and J. L. Whitton. 2007. Coxsackievirus B3 proteins directionally complement each other to downregulate surface major histocompatibility complex class I. J. Virol. 816785-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuconati, A., W. Xiang, F. Lahser, T. Pfister, and E. Wimmer. 1998. A protein linkage map of the P2 nonstructural proteins of poliovirus. J. Virol. 721297-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Jong, A. S., W. J. G. Melchers, D. H. R. F. Glaudemans, P. H. G. M. Willems, and F. J. M. Van Kuppeveld. 2004. Mutational analysis of different regions in the coxsackievirus 2B protein: requirements for homo-multimerization, membrane permeabilization, subcellular localization, and virus replication. J. Biol. Chem. 27919924-19935. [DOI] [PubMed] [Google Scholar]
- 13.De Jong, A. S., I. W. J. Schrama, P. H. G. M. Willems, J. M. D. Galama, W. J. G. Melchers, and F. J. M. Van Kuppeveld. 2002. Multimerization reactions of coxsackievirus proteins 2B, 2C and 2BC: a mammalian two-hybrid analysis. J. Gen. Virol. 83783-793. [DOI] [PubMed] [Google Scholar]
- 14.De Jong, A. S., H. J. Visch, F. de Mattia, M. M. van Dommelen, H. G. Swarts, T. Luyten, G. Callewaert, W. J. G. Melchers, P. H. G. M. Willems, and F. J. M. van Kuppeveld. 2006. The coxsackievirus 2B protein increases efflux of ions from the endoplasmic reticulum and Golgi, thereby inhibiting protein trafficking through the Golgi. J. Biol. Chem. 28114144-14150. [DOI] [PubMed] [Google Scholar]
- 15.De Jong, A. S., E. Wessels, H. B. Dijkman, J. M. Galama, W. J. Melchers, P. H. Willems, and F. J. Van Kuppeveld. 2003. Determinants for membrane association and permeabilization of the coxsackievirus 2B protein and the identification of the Golgi complex as the target organelle. J. Biol. Chem. 2781012-1021. [DOI] [PubMed] [Google Scholar]
- 16.De Silva, A. M., W. E. Balch, and A. Helenius. 1990. Quality control in the endoplasmic reticulum: folding and misfolding of vesicular stomatitis virus G protein in cells and in vitro. J. Cell Biol. 111857-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doedens, J. R., and K. Kirkegaard. 1995. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14894-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duke, G. M., and A. C. Palmenberg. 1989. Cloning and synthesis of infectious cardiovirus RNAs containing short, discrete poly(C) tracts. J. Virol. 631822-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gosert, R., D. Egger, and K. Bienz. 2000. A cytopathic and a cell culture adapted hepatitis A virus strain differ in cell killing but not in intracellular membrane rearrangements. Virology 266157-169. [DOI] [PubMed] [Google Scholar]
- 20.Henikoff, S., and J. G. Henikoff. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 8910915-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, W. M., S. S. Monroe, and R. R. Rueckert. 1993. Role of maturation cleavage in infectivity of picornaviruses: activation of an infectosome. J. Virol. 672110-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leong, L. E. C., C. T. Cornell, and B. L. Semler. 2002. Processing determinants and functions of cleavage products of picornavirus polyproteins, p. 187-197. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, DC.
- 23.Moffat, K., G. Howell, C. Knox, G. J. Belsham, P. Monaghan, M. D. Ryan, and T. Wileman. 2005. Effects of foot-and-mouth disease virus nonstructural proteins on the structure and function of the early secretory pathway: 2BC but not 3A blocks endoplasmic reticulum-to-Golgi transport. J. Virol. 794382-4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moffat, K., C. Knox, G. Howell, S. J. Clark, H. Yang, G. J. Belsham, M. Ryan, and T. Wileman. 2007. Inhibition of the secretory pathway by foot-and-mouth disease virus 2BC protein is reproduced by coexpression of 2B with 2C, and the site of inhibition is determined by the subcellular location of 2C. J. Virol. 811129-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman, J. F., and F. Brown. 1997. Foot-and-mouth disease virus and poliovirus particles contain proteins of the replication complex. J. Virol. 717657-7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nuesch, J., S. Krech, and G. Siegl. 1988. Detection and characterization of subgenomic RNAs in hepatitis A virus particles. Virology 165419-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinton, P., T. Pozzan, and R. Rizzuto. 1998. The Golgi apparatus is an inositol 1,4,5-trisphosphate-sensitive Ca2+ store, with functional properties distinct from those of the endoplasmic reticulum. EMBO J. 175298-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Porter, A. G. 1993. Picornavirus nonstructural proteins: emerging roles in virus replication and inhibition of host cell functions. J. Virol. 676917-6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Racaniello, V. R., and D. Baltimore. 1981. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science 214916-919. [DOI] [PubMed] [Google Scholar]
- 30.Rizzuto, R., M. Brini, C. Bastianutto, R. Marsault, and T. Pozzan. 1995. Photoprotein-mediated measurement of calcium ion concentration in mitochondria of living cells. Methods Enzymol. 260417-428. [DOI] [PubMed] [Google Scholar]
- 31.Rust, R. C., L. Landmann, R. Gosert, B. L. Tang, W. Hong, H. P. Hauri, D. Egger, and K. Bienz. 2001. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 759808-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryan, M. D., G. J. Belsham, and A. M. King. 1989. Specificity of enzyme-substrate interactions in foot-and-mouth disease virus polyprotein processing. Virology 17335-45. [DOI] [PubMed] [Google Scholar]
- 33.Sandoval, I. V., and L. Carrasco. 1997. Poliovirus infection and expression of the poliovirus protein 2B provoke the disassembly of the Golgi complex, the organelle target for the antipoliovirus drug Ro-090179. J. Virol. 714679-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Savolainen, C., S. Blomqvist, M. N. Mulders, and T. Hovi. 2002. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J. Gen. Virol. 83333-340. [DOI] [PubMed] [Google Scholar]
- 35.Schlegel, A., T. H. Giddings, M. S. Ladinsky, and K. Kirkegaard. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 706576-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toomre, D., P. Keller, J. White, J. C. Olivo, and K. Simons. 1999. Dual-color visualization of trans-Golgi network to plasma membrane traffic along microtubules in living cells. J. Cell Sci. 11221-33. [DOI] [PubMed] [Google Scholar]
- 38.Van Kuppeveld, F. J. M., J. M. D. Galama, J. Zoll, P. J. J. C. van den Hurk, and W. J. G. Melchers. 1996. Coxsackie B3 virus protein 2B contains cationic amphipathic helix that is required for viral RNA replication. J. Virol. 703876-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Kuppeveld, F. J. M., J. G. J. Hoenderop, R. L. L. Smeets, P. H. G. M. Willems, H. B. P. M. Dijkman, J. M. D. Galama, and W. J. G. Melchers. 1997. Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J. 163519-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Kuppeveld, F. J. M., W. J. G. Melchers, K. Kirkegaard, and J. R. Doedens. 1997. Structure-function analysis of coxsackie B3 virus protein 2B. Virology 227111-118. [DOI] [PubMed] [Google Scholar]
- 41.Van Kuppeveld, F. J. M., W. J. G. Melchers, P. H. G. M. Willems, and T. W. J. Gadella. 2002. Homomultimerization of the coxsackievirus 2B protein in living cells visualized by fluorescence resonance energy transfer microscopy. J. Virol. 769446-9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wimmer, E., C. U. Hellen, and X. Cao. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27353-436. [DOI] [PubMed] [Google Scholar]