Abstract
A component of the cellular response to zinc deficiency operates via control of transcript abundance. Therefore, microarray analysis was employed to identify Schizosaccharomyces pombe genes whose mRNA levels are regulated by intracellular zinc status. A set of 57 genes whose mRNA levels were substantially reduced in response to zinc deficiency was identified, while the mRNA levels of 63 genes were increased by this condition. In order to investigate the mechanisms that control these responses, a genetic screen was employed to identify mutants with defective zinc-responsive gene expression. Two strains (II-1 and V7) that were identified by this screen harbor mutations that are linked to zrt1+, which encodes a putative Zrt/IRT-like protein (ZIP) zinc uptake transporter. Importantly, zrt1+ mRNA levels are increased in response to zinc deprivation, and cells lacking functional Zrt1 are highly impaired in their ability to proliferate at limiting zinc concentrations. Furthermore, zrt1 null cells were found to have severely reduced zinc contents, indicating that Zrt1 functions as a key regulator of intracellular zinc levels in fission yeast. The deletion of fet4+, another zinc-responsive gene encoding a putative metal ion transporter, exacerbated the phenotypes associated with the loss of Zrt1, suggesting that Fet4 also plays a role in zinc uptake under limiting conditions.
Zinc is a structural component of numerous transcription factors, enzymes, and cell signaling proteins. Indeed, more than 3% of identified human proteins contain zinc-binding motifs (26), and as a result, a wide variety of cellular processes are dependent upon zinc (1, 32). Therefore, it is not surprising that zinc deficiency in humans is associated with numerous conditions, including impaired immune function, gastrointestinal problems, behavioral abnormalities, growth retardation, delayed wound healing, and dermatitis (29). All organisms must maintain intracellular zinc at an acceptable level, and therefore, cells possess specific zinc uptake systems that mediate its acquisition even when it is scarce.
Studies of the budding yeast, Saccharomyces cerevisiae, have provided insight into the molecular basis of zinc uptake. In S. cerevisiae, this process is predominantly mediated by two Zrt/IRT-like protein (ZIP) transporters, Zrt1 and Zrt2, which comprise a high-affinity and a low-affinity transport system, respectively (44, 45). Over 90 ZIP or solute carrier 39 (SLC39) family members have now been identified and are present in a wide range of organisms (10, 21). Humans have at least 15 of these transporters, and although the precise biological role of many of them has yet to be determined, at least a subset (human Zip 1 [hZip1], hZip2, hZip3, hZip4, hZip5, and hZip7) has been implicated in zinc transport (8, 10, 18, 21, 37). hZip4 appears to play a major role in dietary zinc absorption, as it is predominantly expressed in the intestine, and mutations in ZIP4 are responsible for the zinc deficiency disorder acrodermatitis enteropathica (19, 38). ZIP transporters are also implicated in zinc transport in plants because the Arabidopsis thaliana ZIP1, ZIP2, and ZIP3 genes all confer zinc uptake when expressed in yeast (14).
The control of zinc uptake is exercised at multiple levels. In zinc-deficient S. cerevisiae cells, Zrt1 is stable and located at the plasma membrane, but exposure to elevated zinc concentrations results in rapid endocytosis and degradation in the vacuole (11-13). There is evidence that similar mechanisms operate in mammalian cells because the mouse Zip 1 (mZip1), mZip3, and mZip4 transporters are all subject to zinc-stimulated endocytosis (9, 36). Such mechanisms appear to function to protect cells from the overaccumulation of zinc.
The cellular response to zinc is also controlled at the level of transcript abundance. In response to zinc deficiency, S. cerevisiae ZRT1 and ZRT2 mRNA levels are induced by more than 10-fold (46). This is mediated by the Zap1 transcription factor, which binds zinc-responsive promoter elements and induces the coordinate expression of around 40 genes whose products confer an advantage under conditions of zinc limitation (2, 23). The importance of this transcriptional control is underscored by the finding that zap1 mutants have an impaired ability to grow under conditions of zinc limitation (46). There is evidence that zinc uptake is also regulated at the RNA level in both plant and mammalian cells. In the monocytic cell line THP-1, the level of hZIP2 mRNA can be markedly induced by zinc depletion and downregulated by excess (4). In addition, the mRNA level of mZip4 has been demonstrated to increase in adult mice fed a zinc-deficient diet and to decrease upon zinc supplementation (9). Furthermore, A. thaliana ZIP1, ZIP3, and ZIP4 mRNA levels are increased in zinc-limited plants (14). However, the mechanisms by which these responses are coordinated remain obscure, as homologues of S. cerevisiae Zap1 are not present in mammals or plants. Neither are Zap1 homologues present in the fission yeast Schizosaccharomyces pombe, which is evolutionarily divergent from S. cerevisiae (17). Thus, eukaryotic organisms from fission yeast to humans employ alternative mechanisms to regulate transcript abundance in response to zinc deficiency.
As S. pombe lacks a Zap1 homologue, we have used this system to investigate the control of mRNA levels in response to zinc limitation. Using RNA blot hybridization and transcript profiling, we have identified sets of genes whose mRNA levels are regulated in response to zinc deficiency. One highly induced gene was adh4+, which encodes a putative iron-dependent alcohol dehydrogenase. In order to understand how this response is regulated, we have performed a genetic screen for mutants with aberrant gene expression that is regulated by a low level of zinc. Through this screen, we isolated 19 mutants, 2 of which displayed hypersensitivity to zinc deprivation. This hypersensitivity was found to be linked to the zrt1 gene, which encodes a putative ZIP zinc uptake transporter. Cells lacking Zrt1 are highly impaired in their ability to proliferate under zinc-limiting conditions and furthermore have severely reduced zinc levels, indicating that Zrt1 mediates zinc uptake under limiting conditions.
MATERIALS AND METHODS
Strains and media.
The genotypes of strains used in this study were h− (972), h+ ade6-M210 leu1-32 ura4-D18 (NT4), h− ade6-M216 leu1-32 ura4-D18 (NT5), h− ade6-M210 leu1-32 ura4-D18 zip1::ura4+ (zip1Δ), h+ ade6-M210 leu1-32 ura4-D18 zrt1::ura4+ (SW227), h+ ade6-M210 leu1-32 ura4-D18 zrt1-II1 (SW538), h+ ade6-M210 leu1-32 ura4-D18 V7 (SW542), h− zrt1-II1 (SW511), h+ ade6− leu1-32 ura4-D18 fet4::kanMX4 (SW496), h+ ade6− leu1-32 ura4-D18 fet4::kanMX4 zrt1::ura4+ (SW500), and h− ade6-M216 leu1-32 ura4-D18 cta3-lacZ::ura4+ (HAI003). Cell culture was performed in YE5S medium and, where selection was required, EMM (27). EMM is a defined medium whose ZnSO4 concentration is 1.4 μM. Inductively coupled plasma mass spectrometry analysis of the YE5S medium indicated that its zinc concentration is approximately 11 μM. Chelex-treated synthetically defined (CSD) medium, which was used to limit zinc availability, was prepared as described previously (23), with some modifications. All glassware used for the preparation of the CSD medium was pretreated for approximately 12 h with 1% nitric acid and rinsed thoroughly in nanopure H2O. Twenty grams of glucose and 5.1 g of yeast nitrogen base, without divalent cations or potassium phosphate (Bio 101), were dissolved in 1 liter nanopure H2O. This was stirred overnight at 4°C with 25 g Chelex-100 ion-exchange resin (Sigma). After the removal of the resin, the pH of the solution was adjusted to 4.0 with HCl, and the following solutions were added: MnSO4 (0.4 mg/ml), FeCl3 (0.2 mg/ml), CuSO4 (0.04 mg/ml), CaCl2 (100 mg/liter), MgSO4 (500 mg/liter), and KH2PO4 monobasic (100 g/liter). The resulting solution was then filter sterilized into polycarbonate containers. Inductively coupled plasma mass spectrometry analysis of the filtered CSD medium estimated the final metal concentrations as follows: Zn67, 65 nM; Fe57, 18.5 μM; and Cu63, 174 μM. The culturing of the cells in CSD medium was carried out in polypropylene tubes (Falcon) and polycarbonate flasks (Nalgene). Cells were precultured overnight in CSD medium before being diluted into fresh CSD medium. To create a zrt1+ null strain, oligonucleotides Zrt1KOA (5′-GCGTACGTCGACAACCACTTTGGATTCCTAAGG-3′) and Zrt1KOB (5′-CCAGATGGAGATAGCATCC-3′) were used to amplify a 1.5-kb region of the zrt1+ open reading frame. The resulting DNA was digested with BamHI and SalI and ligated to the BamHI and SalI sites of pBluescript to yield pBSSK-zrt1. The 1.8-kb ura4+ cassette from pRep42 was then cloned into the HindIII site to give plasmid pGEM-zrt1::ura4+. Following digestion with BglII and BamHI, this plasmid was used to transform a strain to Ura+, and correct insertion was confirmed by PCR analysis. A strain from which the fet4+ gene was deleted was purchased from Bioneer.
Plasmids.
An adh4+ promoter-lacZ fusion plasmid was constructed by PCR amplifying a DNA fragment corresponding to positions −1380 to 115 (relative to the predicted ATG start codon) using primers Adh4BamHI (5′-TGGACTGGATCCCGGTTGATTGATGCTTTAAGCC-3′) and Adh4EcoRI (5′-GCAGCTGAATTCTTACTTTCGATATGATCGAGC-3′). The resulting product was digested with EcoRI and BamHI before being ligated into the BamHI and EcoRI sites of pSPE356 (20) to yield pSPE356-adh4.
UV mutagenesis and genetic screens.
NT4 cells transformed with pSPE356-adh4 were subjected to random mutagenesis. Exponentially growing cells were spread onto EMM agar supplemented with 100 μM ZnSO4 at a density of approximately 1 × 103 cells per plate. Cells were then subjected to UV irradiation using a Stratalinker UV cross-linker at a dosage that resulted in approximately 70% killing. Plates were incubated in the dark at 30°C for 4 to 5 days. The resulting colonies were transferred to filters and assayed for β-galactosidase activity as previously described (15). Quantitative β-galactosidase assays were also performed as previously described (34).
RNA analysis.
Cell pellets were washed in H2O and resuspended in 200 μl of RNA buffer (50 mM Tris HCl [pH 8.0], 100 mM NaCl, 50 mM EDTA [pH 8.0], 0.25% [wt/vol] sodium dodecyl sulfate) with 200 μl of phenol-chloroform in a 2-ml screw-cap Eppendorf tube. Cells were ruptured with 0.75 ml of 0.5-mm glass beads (Biospec) in a Ribolyser (Hybaid) using two 10-s bursts at full power. A further 0.75 ml of RNA buffer was added, followed by centrifugation in a microcentrifuge for 5 min. The aqueous layer was subjected to further phenol-chloroform extractions before the RNA was precipitated with 0.1 volume of sodium acetate (pH 5.2) and 0.6 volume of isopropanol. RNA pellets were washed in 70% (vol/vol) ethanol and resuspended in H2O. A 10- to 15-μg sample of total RNA was denatured with glyoxal, separated on either a 1.2% or 1.4% (wt/vol) agarose gel prepared in 15 mM sodium phosphate (pH 6.5), and transferred to a GeneScreen hybridization membrane (Dupont NEN Research Products). DNA probes were produced by PCR amplification from genomic DNA using the appropriate primers. All probes were labeled with [α-32P]dCTP by use of a Prime-a-Gene labeling kit (Promega).
Microarray analysis.
Wild-type (972) and zrt1-II1 (SW511) cells were grown to the mid-log phase in EMM at 30°C. RNA preparation, RNA labeling, and microarray analysis were performed as previously described (22). Microarray analysis was performed as two independent experiments. A gene was considered to be upregulated if its mRNA level for zrt1-II1 cells compared to that for wild-type cells was increased ≥1.5-fold in both experiments and had a mean increase of ≥2-fold. A gene was considered to be downregulated if its mean mRNA level (zrt1-II1/wild type) was ≤0.5. All normalized data sets are available from our website (http://www.sanger.ac.uk/PostGenomics/S_pombe/).
Metal content analysis.
Cell pellets from aliquots (10 ml) of cultures were washed in SSW (1 mM EDTA, 20 mM trisodium citrate [pH 4.2], 1 mM KH2PO4, 1 mM CaCl2, 5 mM MgSO4, 1 mM NaCl) and resuspended in 1 ml of 70% (vol/vol) HNO3. Zinc contents were determined by atomic absorption spectrophotometry.
Microarray data accession number.
Microarray data obtained in this study have also been submitted to ArrayExpress under accession number E-TABM-427.
RESULTS
As a first step in the identification of a zinc-sensing system in S. pombe, we sought to identify genes whose mRNA levels are increased by zinc deficiency. Transcript profiling of fission yeast stress responses has previously identified genes that are induced by the heavy metal cadmium but not by other environmental insults, such as oxidative and osmotic stress (5). Cadmium is known to displace zinc ions from proteins (33) and therefore may, to some degree, mimic zinc deprivation, suggesting that some of these cadmium-inducible genes may be regulated in response to zinc status. Consistent with this, a number of these genes are homologues of S. cerevisiae Zap1 targets. These include SPBC1348.06c, which is homologous to the budding yeast genes VEL1 and YOR387C, adh4+ (SPAC5H10.06c), which encodes a putative iron-dependent alcohol dehydrogenase homologous to S. cerevisiae ADH4 (31), and a gene (SPBC16D10.06) that we named zrt1+ because of its similarity to the budding yeast ZRT1 and ZRT2 genes (44, 45). We reasoned that the mRNA levels of these fission yeast genes may be regulated in response to cellular zinc status and so analyzed them in more detail. RNA blot hybridization confirmed that SPBC1348.06c, adh4+, and zrt1+ mRNA levels were induced in response to cadmium. In addition, adh4+ and zrt1+ transcripts were induced in response to high levels of copper (Fig. 1). Prolonged exposure (30 min) of cells to high levels of copper also resulted in the production of longer adh4+ transcripts. While the reason for this is not clear, it is interesting to note that we have previously observed a similar effect with the zym1+ gene (3). In contrast to what occurs with cadmium and copper, none of these genes was induced by high levels of zinc (Fig. 1). In order to test whether their mRNA levels are increased in response to zinc deficiency, intracellular zinc levels were limited by growing cells in rich medium (YE5S) supplemented with the metal chelator EDTA at 60 μM. Metal content analysis revealed that this treatment substantially reduced intracellular zinc levels (Fig. 2A). Furthermore, RNA blot hybridization demonstrated that the mRNA levels of both SPBC1348.06c and zrt1+ were significantly increased when cells were grown in the presence of EDTA. Cellular zinc levels were restored by further supplementing the EDTA-containing medium with ZnSO4 at 60 μM (Fig. 2A). Furthermore, this treatment also reduced transcripts to basal levels (Fig. 2B).
FIG. 1.
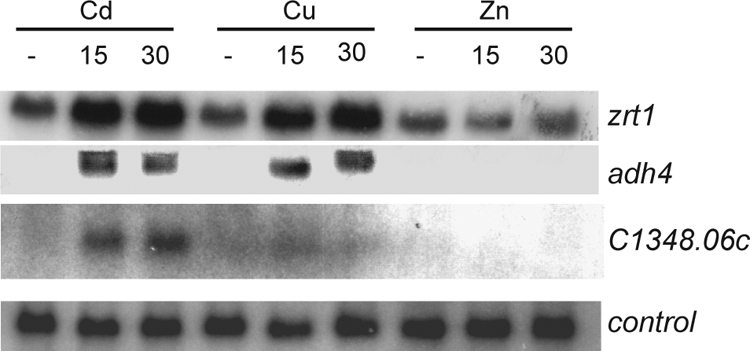
RNA was prepared from wild-type cells treated with 0.5 mM CdSO4, 2 mM CuSO4, or 2 mM ZnSO4 for 0, 15, or 30 min and was subjected to RNA blot hybridization using his3+ (control), zrt1+, SPBC1348.06c, and adh4+ probes.
FIG. 2.
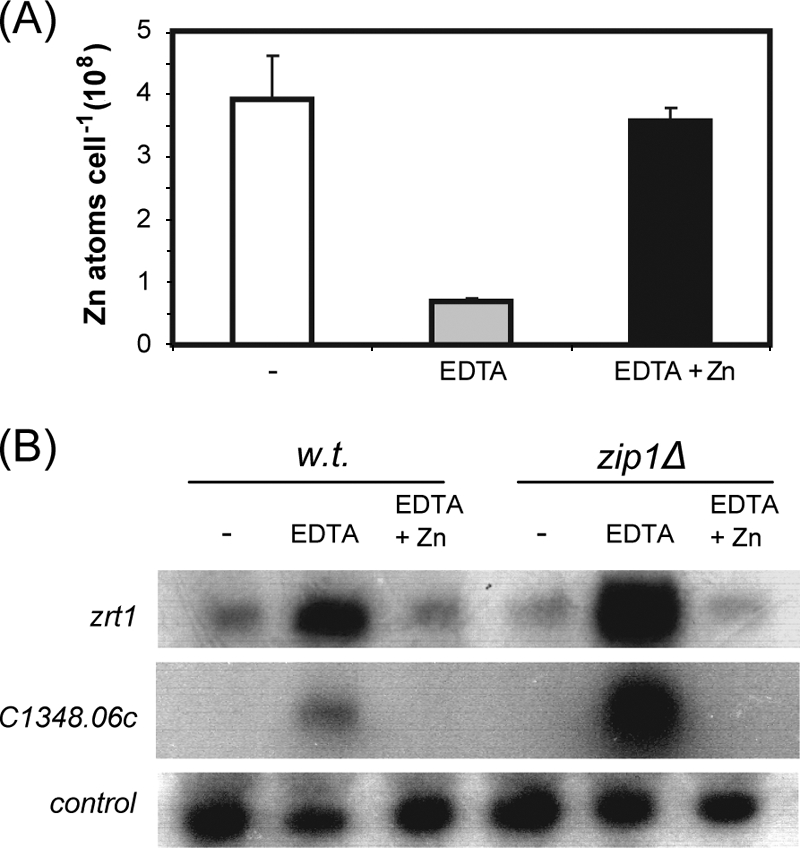
(A) Wild-type cells were grown to exponential phase in YE5S, YE5S supplemented with 60 μM EDTA, or YE5S supplemented with 60 μM EDTA and 60 μM ZnSO4. Cellular zinc contents were measured by atomic absorption spectrometry. Shown are the mean values from three experiments. Error bars indicate standard deviations. (B) Total RNA was prepared from cells grown as described for panel A and subjected to RNA blot hybridization using his3+ (control), zrt1+, and SPBC1348.06c probes. w.t., wild type.
The results suggest that the mRNA levels of these genes are regulated by zinc limitation. However, EDTA is not a zinc-specific metal chelator, and thus, we could not exclude the possibility that the observed effects were due to changes in the intracellular concentrations of other cations. Therefore, in order to conclusively demonstrate that the expression of SPBC1348.06c, adh4+, and zrt1+ is controlled by zinc deficiency, it was necessary to establish conditions for the culture of S. pombe cells in which zinc, but not other metal ions, was limited. Therefore, we employed CSD medium, which was originally developed for the culture of S. cerevisiae at limiting zinc concentrations (23). In this case, the metal availability of synthetic defined medium is first limited by treatment with Chelex-100 ion-exchange resin. Following this treatment, cations other than zinc (Cu2+, Fe3+, Mg2+, Mn2+, Ca2+, K+) are added back, resulting in a medium whose Zn2+ concentration is submicromolar (23). Indeed, analysis of a batch of CSD medium indicated a zinc concentration of 65 nM (data not shown). We found that CSD medium was able to support the growth of wild-type fission yeast cells without zinc supplementation. However, growth rates were clearly enhanced by the addition of 20 μM ZnSO4, indicating that CSD medium is indeed zinc limiting (Fig. 3A). This is reflected in the very low intracellular zinc content of cells grown in CSD medium compared to that of cells grown in CSD medium supplemented with 20 μM ZnSO4 (Fig. 3B). Growth rates were not further enhanced by the addition of zinc at concentrations up to 60 μM, and above this concentration, zinc impaired growth rates (data not shown). Importantly, the addition of iron or copper did not enhance the growth rate, indicating that this medium is not limited for these metal ions (data not shown). Having established the appropriate conditions, we compared the transcript levels from cells grown at “adequate” (CSD medium plus 20 μM ZnSO4) and “limiting” (CSD medium) zinc concentrations (Fig. 3C). This revealed that the mRNA levels of SPBC1348.06c, zrt1+, and adh4+ were highly increased by zinc limitation.
FIG. 3.
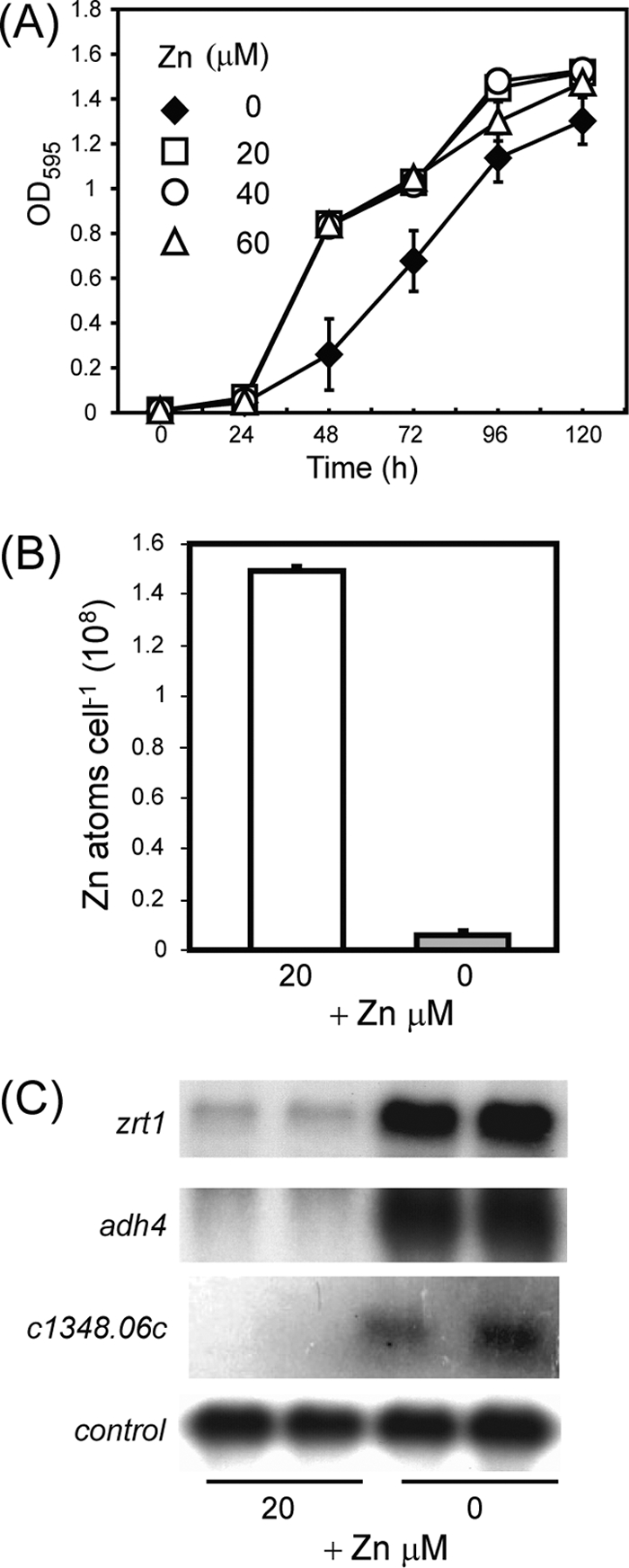
(A) Growth in CSD medium. Wild-type (wt) cells were precultured in CSD medium and then inoculated into CSD medium supplemented with 20 to 60 μM ZnSO4. Cultures were incubated at 30°C, and the optical densities at 595 nm (OD595) were measured every 24 h over a 120-h period. Shown are the mean values from three experiments. Error bars indicate standard deviations. (B) Zinc contents of wild-type cells cultured in CSD medium. Wild-type cells were cultured overnight in CSD medium and then inoculated into CSD medium with or without 20 μM ZnSO4 and incubated at 30°C until they proliferated exponentially. Total cellular zinc contents were measured by atomic absorption spectrometry. Shown are the mean values from three experiments. Error bars indicate standard deviations. (C) Gene expression in response to a low level of zinc. Wild-type cells were precultured in CSD medium and then inoculated into CSD medium with or without 20 μM ZnSO4. Total RNA was prepared after a 24-h incubation at 30°C and subjected to RNA blot hybridization with zrt1+, adh4+, SPBC1348.06c, and SPBPB2B2.08 (control) probes.
In fission yeast, the Sty1/Spc1 stress-activated protein kinase pathway controls the expression of numerous genes in response to a range of adverse conditions (35). However, microarray analysis has suggested that the induction of adh4+ and zrt1+ transcripts in response to cadmium is independent of this pathway (5), and this was confirmed by RNA blot hybridization (data not shown). This suggests that the increase in mRNA levels in response to zinc deficiency does not require the Sty1/Spc1 pathway. We therefore investigated the role of the Zip1 transcription factor, which mediates the activation of a set of genes in response to cadmium exposure (16). Indeed, the induction of SPBC1348.06c in response to cadmium has previously been shown to be Zip1 dependent (16). However, RNA blot hybridization demonstrated that the increase of SPBC1348.06c mRNA in response to the chelator EDTA occurred in a zip1Δ background (Fig. 2B). Furthermore, the deletion of zip1+ resulted in an increased level of SPBC1348.06c transcripts, suggesting that Zip1 also has a repressive effect on this gene (Fig. 2B). The induction of zrt1+ mRNA levels in response to EDTA was also found to be independent of Zip1.
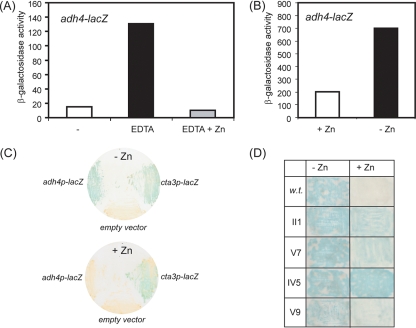
These findings suggest that Zip1 does not control the response to zinc limitation. Therefore, in order to investigate the mechanisms by which transcript levels are controlled in response to zinc deprivation, we constructed an adh4 promoter-lacZ fusion reporter and determined whether its expression was regulated in response to zinc availability (Fig. 4). The reporter was expressed at a low (basal) level when cells were grown in liquid medium (EMM). Supplementing the medium with excess zinc (ZnSO4, 200 μM) did not affect the expression of the reporter (see Fig. S1 in the supplemental material), suggesting that wild-type cells are “zinc replete” when grown in liquid EMM. In contrast, the expression of the adh4-lacZ reporter was markedly increased by growing cells in medium supplemented with the chelator EDTA, which reduces intracellular zinc levels (Fig. 4A). Consistent with this, when cells were grown in limiting zinc medium (CSD medium), high levels of reporter expression were observed, which were suppressed by the addition of ZnSO4 (20 μM) to the medium (Fig. 4B). Thus, the expression of the adh4-lacZ reporter is regulated in response to zinc deprivation. Furthermore, truncation analysis of the reporter indicated that adh4+ expression is controlled by a combination of activating and repressing sequence elements (see Fig. S2 in the supplemental material).
FIG. 4.
Expression of an adh4+ promoter-lacZ reporter in response to limiting zinc. (A) Wild-type cells carrying the adh4+ promoter-lacZ reporter were cultured at 30°C in EMM (−), EMM supplemented with 10 μM EDTA, or EMM supplemented with 10 μM EDTA and 10 μM ZnSO4 until they proliferated exponentially. Cells were then harvested and processed for liquid β-galactosidase assays. Shown are the mean values (Miller units) from two experiments. (B) Cells were cultured at 30°C in CSD medium with or without 20 μM ZnSO4 and processed as described for panel A. Shown are the mean values (Miller units) from two experiments. (C) Cells containing the adh4+ promoter-lacZ fusion plasmid, an empty vector (pSPE356), or an integrated cta3+ promoter-lacZ reporter were cultured onto nitrocellulose membranes on EMM agar (−Zn) or EMM agar supplemented with 0.1 mM ZnSO4 (+Zn) for 2 days at 30°C. Filters were then subjected to β-galactosidase assays as described in Materials and Methods. (D) Examples of mutants isolated from the genetic screen. Cells were cultured onto EMM agar or EMM agar supplemented with 0.1 mM ZnSO4, transferred to filters, and processed for β-galactosidase assays. w.t., wild type.
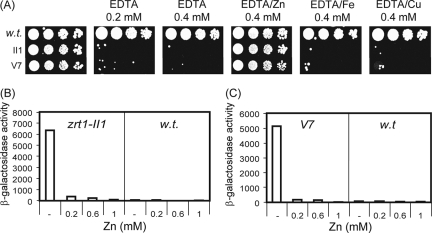
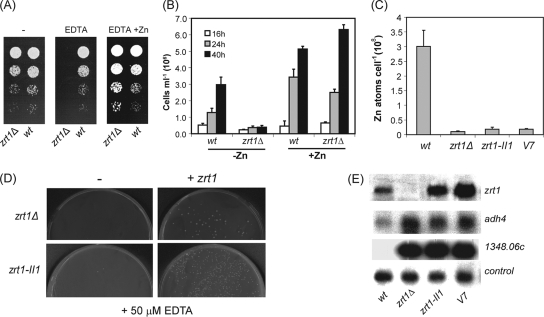
We next explored the possibility of using this reporter as the basis of a genetic screen. β-Galactosidase filter assays revealed that colonies of wild-type cells containing this reporter go blue rapidly when grown on minimal agar (EMM). This suggested that EMM agar is somewhat limiting for zinc, which is likely to be due to the ability of the agar to chelate metals and thus restrict zinc availability (Fig. 4C). Consistent with this, we found that supplementing the agar with ZnSO4 at 100 μM repressed the expression of the adh4+ reporter, because colonies grown under these conditions remained white for an extended period. It should be noted that wild-type fission yeast cells can tolerate relatively high levels of zinc (up to approximately 2 mM) (3), which suggested that the observed zinc-mediated repression of the adh4+-lacZ reporter is a specific effect. In support of this, the activity of an unrelated reporter (cta3-lacZ) was not detectably influenced by the addition of zinc to the medium (Fig. 4C). Therefore, we next sought to isolate mutants with an impaired ability to mediate the zinc-dependent repression of the adh4-lacZ reporter. Cells carrying the adh4-lacZ reporter (pSPE356-adh4) were plated onto zinc-supplemented EMM agar and subjected to random mutagenesis, and colonies with high levels of β-galactosidase activity were identified. Approximately 200,000 colonies were screened, from which 19 mutants with increased expression of the adh4-lacZ reporter were isolated. An example of some of these mutants is shown in Fig. 4D. Mutant strains were cured of their reporter plasmids, followed by the reintroduction of the reporter. This identified three strains with plasmid-associated (cis) mutations. The remaining strains with trans-acting mutations were analyzed in order to determine whether any of them possessed altered sensitivity to excess or limiting zinc. None of the strains was sensitive to excess zinc (data not shown). However, two mutant strains, II1 and V7, were hypersensitive to EDTA (Fig. 5A). Importantly, this sensitivity could be rescued by the addition of equimolar zinc but not iron or copper. Genetic analysis of these strains indicated that the EDTA hypersensitivity of these strains resulted from one recessive mutation (or several tightly linked recessive mutations). Quantitative β-galactosidase assays confirmed that the basal expression of the adh4-lacZ reporter was vastly increased (approximately 2 orders of magnitude) in these mutants compared to that in the wild-type background. However, the overexpression of the adh4-lacZ reporter in the mutant backgrounds was suppressed by supplementing the medium with increasing concentrations of ZnSO4 (Fig. 5B and C). This coupled with the finding that the V7 and II1 mutants are hypersensitive to EDTA suggested that these strains may have aberrant intracellular zinc contents resulting from impaired zinc uptake. As the zrt1+ gene encodes a putative zinc uptake transporter, we reasoned that these strains may harbor mutations in this gene. DNA sequencing revealed that the zrt1 gene from the II1 strain contained a G-to-A mutation which changes the Trp 141 codon to a stop codon and would therefore be predicted to result in a severely truncated protein. In order to confirm that zrt1-II1 encodes a nonfunctional protein, we constructed a strain in which zrt1+ was disrupted. As expected, zrt1Δ cells were also found to be hypersensitive to EDTA, a phenotype that could be rescued by adding back equimolar ZnSO4 (Fig. 6A). Furthermore, the zrt1Δ strain showed a very limited ability to proliferate in CSD medium unless it was supplemented with zinc (Fig. 6B), indicating that Zrt1 function is important for viability at limiting zinc concentrations. Indeed, these findings imply that Zrt1 plays a key role in maintaining intracellular zinc levels. Consistent with this, metal content analysis demonstrated that intracellular zinc levels were severely reduced in cells lacking functional Zrt1 (Fig. 6C). Next, we sought to reintegrate the wild-type zrt1+ allele into the zrt1Δ and zrt1-II1 mutants in order to complement their phenotypes. Indeed, when cells were transformed with a DNA fragment containing the wild-type zrt1+ gene, numerous colonies that had regained the ability to grow in the presence of EDTA were obtained (Fig. 6D). This demonstrates that the EDTA-sensitive phenotype is the result of mutation of the zrt1+ gene.
FIG. 5.
(A) Wild-type (w.t.), zrt1-II1, and V7 strains were grown to exponential phase, subjected to fivefold serial dilutions, and spotted onto YE5S supplemented with the indicated concentrations of EDTA, ZnSO4, FeCl3, and CuSO4. Plates were incubated at 30°C for 2 days. (B and C) The indicated strains were grown to exponential phase in YE5S or YE5S supplemented with the indicated concentrations of ZnSO4. Cells were then harvested and processed for liquid β-galactosidase assays. Shown are the mean values (Miller units) from duplicate experiments.
FIG. 6.
(A) Wild-type (wt) and zrt1Δ strains were grown to exponential phase, subjected to fivefold serial dilutions, and spotted onto YE5S agar supplemented with EDTA (200 μM) and ZnSO4 (200 μM) as indicated. Plates were incubated at 30°C for 2 days. (B) Wild-type and zrt1Δ strains were precultured in CSD medium and then inoculated into CSD medium (−Zn) or CSD medium supplemented with 20 μM ZnSO4 (+Zn). Cultures were incubated at 30°C, and cell titers were determined at the indicated time points. Shown are the mean values from three experiments. Error bars indicate standard deviations. (C) Total cellular zinc contents of the indicated strains were measured by atomic absorption spectrometry. Shown are the mean values from three experiments. Error bars indicate standard deviations. (D) Complementation of zrt1− mutants. Cultures of the zrt1Δ strain and zrt1-II1 were transformed with carrier DNA (−) or with carrier DNA and a DNA fragment containing the zrt1+ open reading frame (+ zrt1). Cells were plated onto EMM agar plates supplemented with EDTA (50 μM) and incubated at 30°C for 3 to 4 days. (E) Total RNA was prepared from the indicated strains and subjected to RNA blot hybridization using his3+ (control), zrt1+, adh4+, and SPBC1348.06c probes.
The V7 strain has phenotypes that are indistinguishable from those of the zrt1Δ and zrt1-II1 strains. Indeed, intracellular zinc content was also severely reduced in this mutant. Therefore, it was surprising that the sequencing of V7 did not reveal any mutations within the predicted zrt1+ open reading frame in this background. Nonetheless, genetic analysis suggested that the V7 mutation is linked to the zrt1 locus. Analysis of 13 tetrads resulting from a genetic cross between the zrt1Δ strain and V7 revealed that all of the progeny were hypersensitive to EDTA. Furthermore, a zrt1Δ/V7 diploid strain was also found to be hypersensitive to zinc limitation.
We compared the transcript levels in the zrt1Δ, zrt1-II1, and V7 backgrounds and found, as expected, that adh4+, zrt1+, and SPBC1348.06c mRNA levels were increased in the absence of functional Zrt1 protein (Fig. 6E). In order to identify other genes whose mRNA levels are regulated in response to zinc deficiency, we used microarray analysis to compare wild-type cells with zrt1-II1 cells, which have severely reduced intracellular zinc levels. This identified 57 genes whose mRNA levels were reduced by zinc deficiency (Table 1), and 63 genes were found to have increased mRNA levels under these conditions (Table 2). In order to validate the transcript profiling data, a number of genes were also analyzed by RNA blot hybridization (Fig. 7). Prominent among the genes with reduced transcript levels were those encoding proteins involved in protein synthesis, for instance, ribosomal subunits (rpl3002+, rpl31+, rpl34+, rps403+, rpl1101+, and rps801+) and proteins involved in amino acid uptake (SPBC359.03c, SPBC359.01) and biosynthesis (apt1+, eca39+, SPCC364.07, SPBC428.11, SPBPB2B2.05, SPAP8A3.07c). Furthermore, the mRNA levels of several genes whose products are involved in nucleotide metabolism (SPCC965.14c, SPCC1442.14c, ura1+) were also reduced under these conditions. These findings are consistent with the finding that zinc deficiency led to reduced growth rates. Zinc limitation also led to a decrease in the transcript abundance of genes involved in the acquisition of phosphate, such as pho1+ (acid phosphatase) and SPBC8E4.01c, which encodes an inorganic phosphate transporter. A further response to zinc deficiency was the downregulation of the mRNA levels of genes whose products are involved in iron and sulfur uptake (SPAC869.05c, str3+) and utilization (isu1+, sua1+, SPBPB10D8.02c). Thus, zinc limitation impacts a variety of cellular processes.
TABLE 1.
Genes downregulated by zinc limitation
| Gene | Functiona | Relative mRNA level (level in zrt1-II1 cells/level in wild-type cells)
|
||
|---|---|---|---|---|
| Expt 1 | Expt 2 | Mean | ||
| isp3+ | Upregulation of meiotic expression | 0.196 | 0.047 | 0.12 |
| pho1+ | Acid phosphatase | 0.113 | 0.176 | 0.14 |
| SPAC27D7.10c | Similar to SPAC27D7.09c, SPAC27D7.11c, and SPBC3D6.02 | 0.249 | 0.178 | 0.21 |
| str3+ | Siderophore-iron transporter | 0.218 | 0.210 | 0.21 |
| SPBPB10D8.02c | Acylsulfatase (predicted) | 0.310 | 0.144 | 0.23 |
| SPBC359.03c | Amino acid permease family (predicted) | 0.274 | 0.201 | 0.24 |
| SPCC965.14c | Cytosine deaminase (predicted) zinc metalloenzyme | 0.258 | 0.240 | 0.25 |
| SPAC27D7.09c | Similar to SPAC27D7.10c, SPAC27D7.11c, and SPBC3D6.02 | 0.332 | 0.169 | 0.25 |
| wis2+ | Cyclophilin | 0.311 | 0.269 | 0.29 |
| SPAC11D3.18c | Nicotinic acid plasma membrane transporter (predicted) | 0.262 | 0.334 | 0.30 |
| SPAC27D7.11c | Similar to SPAC27D7.09c, SPAC27D7.10c, and SPBC3D6.02 | 0.405 | 0.199 | 0.30 |
| SPAC869.05c | Sulfate transporter (predicted) | 0.367 | 0.248 | 0.31 |
| SPBC1347.11 | Sequence orphan | 0.442 | 0.207 | 0.32 |
| SPBC1539.07c | Glutathione-dependent formaldehyde dehydrogenase (predicted) | 0.371 | 0.302 | 0.34 |
| SPBC3E7.06c | Major facilitator superfamily membrane transporter | 0.335 | 0.355 | 0.35 |
| pan6+ | Pantoate-beta-alanine ligase | 0.360 | 0.336 | 0.35 |
| SPAC11D3.02c | ELLA family protein | 0.368 | 0.342 | 0.36 |
| SPBC21C3.08c | Ornithine aminotransferase | 0.432 | 0.294 | 0.36 |
| SPBPB10D8.01 | Membrane transporter | 0.405 | 0.371 | 0.39 |
| aes1+ | Enhancer of RNA-mediated gene silencing | 0.460 | 0.348 | 0.40 |
| SPCC364.07 | D-3 phosphoglycerate dehydrogenase (predicted) | 0.464 | 0.368 | 0.42 |
| SPBP8B7.05c | Carbonic anhydrase (predicted) | 0.501 | 0.343 | 0.42 |
| apt1+ | Adenine phosphoribosyltransferase (predicted) | 0.423 | 0.422 | 0.42 |
| SPAC23C4.06c | Methyltransferase (predicted) | 0.409 | 0.450 | 0.43 |
| SPBC8E4.01c | Inorganic phosphate transporter (predicted) | 0.513 | 0.366 | 0.44 |
| SPAC11D3.13 | ThiJ domain | 0.454 | 0.451 | 0.45 |
| rpl3002+ | 60S ribosomal protein L30 | 0.516 | 0.391 | 0.45 |
| rpl31+ | 60S ribosomal protein L31 | 0.508 | 0.399 | 0.45 |
| SPBC359.01 | Amino acid permease family | 0.538 | 0.375 | 0.46 |
| SPCC330.06c | Thioredoxin peroxidase | 0.566 | 0.349 | 0.46 |
| eca39+ | Branched-chain amino acid aminotransferase | 0.486 | 0.429 | 0.46 |
| SPCC622.12c | NADP-specific glutamate dehydrogenase | 0.462 | 0.454 | 0.46 |
| egd2 | Nascent polypeptide-associated complex (alpha subunit) | 0.549 | 0.374 | 0.46 |
| sks2+ | Heat shock protein 70 family | 0.539 | 0.387 | 0.46 |
| SPBPB2B2.05 | GMP synthase | 0.532 | 0.399 | 0.47 |
| rpb8+ | DNA-directed RNA polymerase I, II, and III subunit | 0.561 | 0.371 | 0.47 |
| SPCP20C8.03 | Pseudogene | 0.491 | 0.448 | 0.47 |
| SPAP8A3.07c | Phospho-2-dehydro-3-deoxyheptonate aldolase | 0.588 | 0.360 | 0.47 |
| SPBC428.11 | Homocysteine synthase | 0.562 | 0.389 | 0.48 |
| SPBC13A2.04c | PTR family peptide transporter | 0.518 | 0.434 | 0.48 |
| rpl34+ | 60S ribosomal protein L34 | 0.591 | 0.368 | 0.48 |
| SPBPB2B2.06c | Calcineurin-like phosphoesterase | 0.579 | 0.385 | 0.48 |
| SPCC330.07c | Membrane transporter | 0.437 | 0.530 | 0.48 |
| cdb4+ | Metallopeptidase | 0.537 | 0.431 | 0.48 |
| meu19+ | Noncoding RNA | 0.566 | 0.403 | 0.48 |
| hsp90+ | Hsp90 family | 0.555 | 0.420 | 0.49 |
| rps403+ | 40S ribosomal protein S4 | 0.598 | 0.383 | 0.49 |
| sod1+ | Superoxide dismutase | 0.525 | 0.462 | 0.49 |
| snz1+ | pyridoxine biosynthesis protein | 0.479 | 0.513 | 0.50 |
| rpl1101+ | 60S ribosomal protein L11 | 0.533 | 0.461 | 0.50 |
| ssa1+ | Heat shock protein 70 family | 0.563 | 0.433 | 0.50 |
| rps801+ | 40S ribosomal protein S8 | 0.544 | 0.452 | 0.50 |
| ubc15+ | Ubiquitin-conjugating enzyme | 0.502 | 0.497 | 0.50 |
| isu1+ | Iron-sulfur cluster assembly scaffold protein | 0.638 | 0.362 | 0.50 |
| SPCC1442.14c | Adenosine 5′-monophosphoramidase | 0.559 | 0.446 | 0.50 |
| sua1+ | Sulfate adenylyltransferase (ATP) | 0.610 | 0.396 | 0.50 |
| ura1+ | Carbamoyl-phosphate synthase | 0.525 | 0.482 | 0.50 |
The descriptions of the protein functions are based on information for S. pombe in GeneDB (http://www.genedb.org/genedb/pombe/index.jsp).
TABLE 2.
Genes upregulated by zinc limitation
| Gene | Functiona | Relative mRNA level (level in zrt1-II1 cells/level in wild-type cells)
|
||
|---|---|---|---|---|
| Expt 1 | Expt 2 | Mean | ||
| SPBC1348.06c | Similar to SPBPB2B2.15 and SPAC977.05c | 165.91 | 194.24 | 180.1 |
| SPAC977.05c | Similar to SPBC1348.06C and SPBPB2B2.15 | 117.46 | 179.50 | 148.5 |
| adh4+ | Alcohol dehydrogenase | 42.52 | 56.47 | 49.5 |
| SPBPB2B2.15 | Similar to SPBC1348.06C and SPAC977.05c | 35.28 | 22.88 | 29.1 |
| zrt1+ (SPBC16D10.06) | ZIP zinc transporter | 9.72 | 14.04 | 11.9 |
| dak2+ | Dihydroxyacetone kinase | 5.14 | 10.26 | 7.7 |
| SPBP26C9.03c | Metal ion transporter similar to S. cerevisiae FET4 | 4.86 | 7.52 | 6.2 |
| SPCC18B5.02c | Cinnamoyl-coenzyme A reductase pseudogene | 3.27 | 4.65 | 4.0 |
| SPBC12C2.03c | Flavin adenine dinucleotide binding protein (predicted) | 3.30 | 4.40 | 3.9 |
| tlh1+ | RecQ type DNA helicase | 4.51 | 2.92 | 3.7 |
| SPAPB1A10.14 | F-box protein | 2.73 | 4.57 | 3.7 |
| SPAC23H3.15c | Sequence orphan | 5.64 | 1.53 | 3.6 |
| SPBC1652.01 | Predicted Sin3-interacting protein | 3.58 | 3.40 | 3.5 |
| SPAC977.04 | Pseudogene | 2.70 | 3.80 | 3.3 |
| SPAC212.09c | Pseudogene | 2.92 | 3.45 | 3.2 |
| SPAC750.08c | NAD-dependent malic enzyme | 2.88 | 3.28 | 3.1 |
| SPAPB18E9.04c | Glycoprotein (predicted) | 2.86 | 3.02 | 2.9 |
| mae2+ | Malate dehydrogenase | 2.71 | 3.15 | 2.9 |
| SPCC663.08c | Short-chain dehydrogenase (predicted) | 2.51 | 3.21 | 2.9 |
| SPAC57A10.06 | Sequence orphan | 2.54 | 3.16 | 2.9 |
| SPBPB7E8.01 | Glycoprotein (predicted) neutral zinc metallopeptidases | 3.12 | 2.54 | 2.8 |
| SPBC1348.05 | MFS family membrane transporter | 2.25 | 3.21 | 2.7 |
| SPAC22A12.06c | Serine hydrolase | 1.94 | 3.43 | 2.7 |
| obr1+ | Flavodoxin-like protein | 2.51 | 2.83 | 2.7 |
| SPCC663.06c | Short-chain dehydrogenase (predicted) | 2.48 | 2.81 | 2.6 |
| SPAC2H10.01 | Zn2-Cys6 transcription factor | 3.31 | 1.90 | 2.6 |
| SPAC977.07c | Glycoprotein | 1.50 | 3.64 | 2.6 |
| tpx1+ | Thioredoxin peroxidase | 2.48 | 2.60 | 2.6 |
| php2+ | CCAAT-binding factor complex subunit | 2.83 | 2.24 | 2.5 |
| SPBCPT2R1.07c | Malic pseudogene | 2.43 | 2.59 | 2.5 |
| SPAC13F5.03c | Glycerol dehydrogenase | 2.85 | 2.17 | 2.5 |
| SPCC794.01c | Glucose-6-phosphate 1-dehydrogenase | 2.27 | 2.74 | 2.5 |
| map2+ | Pheromone | 2.24 | 2.75 | 2.5 |
| rds1+ | Stress-induced protein | 3.42 | 1.56 | 2.5 |
| SPAC23C11.06c | Membrane protein | 3.13 | 1.77 | 2.5 |
| pex7+ | Peroxisomal signal receptor | 2.43 | 2.42 | 2.4 |
| gst2+ | Glutathione S-transferase | 2.36 | 2.45 | 2.4 |
| SPAC4F10.06 | Conserved fungal protein | 2.14 | 2.63 | 2.4 |
| srx1+ | Sulfiredoxin | 3.05 | 1.72 | 2.4 |
| SPAC4G8.03c | Pumilio family RNA binding protein | 2.31 | 2.39 | 2.4 |
| SPBC8E4.02c | Sequence orphan | 2.20 | 2.49 | 2.3 |
| SPAC22A12.17c | Short-chain dehydrogenase (predicted) | 2.36 | 2.31 | 2.3 |
| SPAC16.05c | Transcription factor similar to those of S. cerevisiae SFP1 | 2.29 | 2.32 | 2.3 |
| SPAC1786.02 | Phospholipase (putative) | 2.12 | 2.42 | 2.3 |
| SPBC1773.13 | Aromatic aminotransferase (predicted) | 2.34 | 2.13 | 2.2 |
| SPCC1235.01 | Glycoprotein (predicted) | 2.05 | 2.41 | 2.2 |
| SPCC569.05c | Spermidine family transporter (predicted) | 2.11 | 2.28 | 2.2 |
| SPAC5H10.04 | NADPH dehydrogenase | 2.03 | 2.31 | 2.2 |
| SPAP27G11.08c | Meiotic expression upregulated | 1.72 | 2.62 | 2.2 |
| SPBC16E9.16c | Sequence orphan | 2.44 | 1.88 | 2.2 |
| SPCC1223.13 | Transcription factor (predicted) | 2.27 | 2.05 | 2.2 |
| meu26+ | Conserved fungal protein | 2.00 | 2.29 | 2.1 |
| SPBPB2B2.19c | Membrane protein | 2.01 | 2.28 | 2.1 |
| SPAC19B12.10 | AMSH protein homolog | 2.11 | 2.16 | 2.1 |
| pof1+ | F-box protein | 2.10 | 2.16 | 2.1 |
| caf5+ | Spermine transporter family | 1.89 | 2.34 | 2.1 |
| SPBC409.08 | Spermine transporter family | 1.89 | 2.22 | 2.1 |
| tht1+ | Nuclear membrane protein involved in karyogamy | 2.05 | 2.05 | 2.1 |
| gst1+ | Glutathione S-transferase | 2.15 | 1.93 | 2.0 |
| atf31+ | bZIP transcription factor | 2.01 | 2.02 | 2.0 |
| spd1+ | Ribonucleotide reductase inhibitor | 1.96 | 2.06 | 2.0 |
| SPCC1020.03 | Mitochondrial ion transporter | 1.59 | 2.37 | 2.0 |
| SPBC1348.02 | S. pombe-specific protein | 1.88 | 2.08 | 2.0 |
The descriptions of the protein functions are based on information for S. pombe in GeneDB (http://www.genedb.org/genedb/pombe/index.jsp).
FIG. 7.
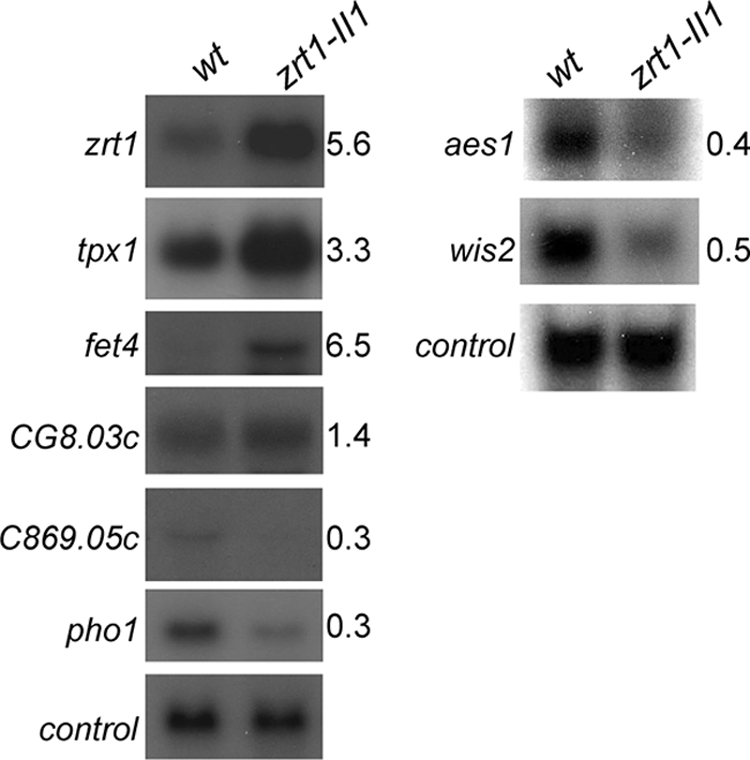
Genes regulated by zinc deficiency. Wild-type (wt) and zrt1-II1 cells were grown to exponential phase in EMM medium at 30°C. RNA was prepared and subjected to RNA blot hybridization with the indicated probes, with his3+ serving as a loading control. Blots were quantified using a PhosphorImager. The relative mRNA levels (zrt1-II1 cells compared to wild-type cells) are indicated.
Interestingly, the S. pombe genes whose mRNA levels were most highly increased by zinc deficiency have homologues in S. cerevisiae that are similarly regulated. These include SPBC1348.06c and the closely related genes SPBPB2B2.15 and SPAC977.05c, which are homologous to the budding yeast Zap1-regulated genes VEL1 and YOR387C. At present, the functions of the proteins encoded by these genes remain obscure, but they are predicted to be located at the cell surface. Many of the upregulated genes identified by this analysis have previously been shown to be induced in response to environmental stresses (5; see also http://www.sanger.ac.uk/PostGenomics/S_pombe/projects/). In particular, the mRNA levels of genes encoding known antioxidants, such as tpx1+, srx1+, gst1+, and gst2+, were increased in response to zinc limitation, and consistent with this, a number of limiting zinc-responsive genes are also induced in response to H2O2 (e.g., obr1+, rds1+, and SPCC663.08c) (5). There is evidence suggesting that zinc deficiency results in oxidative stress in mammalian cells (29), and our microarray data suggest that this may also be the case in fission yeast cells.
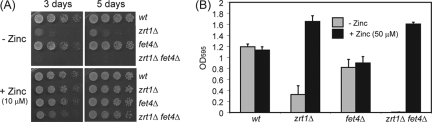
As expected, zrt1+ mRNA was significantly increased by zinc deficiency, as was the mRNA of another gene, SPBP26C9.03c, which we named fet4+ based on its homology to S. cerevisiae FET4. S. cerevisiae Fet4 was originally identified as a low-affinity iron uptake transporter, although more-recent evidence suggests that it is also capable of transporting zinc (39). Therefore, we examined the role of its fission yeast counterpart. Deletion of the fet4+ gene alone did not result in any increased sensitivity to limiting zinc concentrations (Fig. 8), nor did it influence intracellular zinc concentrations (data not shown). However, we found that the deletion of fet4+ exacerbated the defects associated with the loss of Zrt1. As described above, minimal agar (EMM) is somewhat limiting for zinc. Accordingly, zrt1Δ cells grew slowly on minimal agar (EMM), and furthermore a zrt1Δ fet4Δ double mutant strain was unable to grow on this medium even after an extended period (5 days) (Fig. 8A). Moreover, the deletion of fet4+ also exacerbated the slow-growth phenotype of zrt1Δ cells in liquid culture (Fig. 8B). In both cases, the phenotypes were suppressed by supplementing the media with zinc. Thus, these findings indicate that Fet4 contributes to viability when zinc is limiting.
FIG. 8.
(A) The indicated strains were grown to exponential phase, subjected to fivefold serial dilutions, and spotted onto EMM agar or EMM agar supplemented with ZnSO4 (10 μM) and incubated at 30°C for the indicated times. (B) The indicated strains were grown for 25 h at 30°C in EMM or EMM supplemented with ZnSO4 (50 μM). Shown are the mean values from three experiments. Error bars indicate standard deviations. Note that the wild-type (NT4) and fet4Δ (SW496) strains are ura4−, whereas the zrt1Δ (SW227) and fet4Δ zrt1Δ (SW500) strains are ura4+. wt, wild type.
DISCUSSION
Here we have employed both transcript profiling and a genetic screen to characterize the response of fission yeast cells to zinc deficiency. We have identified sets of genes whose mRNA levels are regulated by zinc availability, and we have also identified Zrt1, a ZIP transporter that is a central component of zinc homeostasis. The importance of Zrt1 is underscored by the inability of cells lacking this transporter to proliferate when zinc is scarce. Furthermore, cells lacking Zrt1 function have severely reduced intracellular zinc contents compared to wild-type cells. These findings suggest that S. pombe Zrt1 is a high-affinity zinc uptake transporter that is analogous to S. cerevisiae Zrt1. Sequence analysis of the S. pombe genome indicates that it lacks an obvious counterpart to S. cerevisiae Zrt2, the low-affinity ZIP zinc uptake transporter. However, S. pombe does encode a homologue of the S. cerevisiae Fet4 transporter, and our data are consistent with a role for this transporter in zinc uptake. Furthermore, there are a number of uncharacterized proteins (such as SPCC126.09, SPAP8A3.03, and SPAC17D4.03c) that have the potential to contribute to zinc uptake.
S. cerevisiae cells also respond to zinc deficiency by increasing the expression of Zrt3, another ZIP transporter that is responsible for the mobilization of zinc from the vacuole (24). Paradoxically, budding yeast cells also upregulate the expression of Zrc1, a vacuolar zinc influx cation diffusion facilitator transporter, and it has been suggested that zinc flux through the vacuole is important under conditions of deficiency (23). It is therefore intriguing that while S. pombe encodes homologues of Zrt3 and Zrc1 (SPCC126.09 and Zhf1, respectively), our microarray data indicate that the mRNA level of neither of these fission yeast genes is substantially influenced by intracellular zinc status. However, it is possible that these transporters are subject to zinc-dependent, posttranslational regulation.
Differential gene expression is predicted to aid the adaptation to the stress imposed by zinc deprivation. A potential example of this is the mitochondrial alcohol dehydrogenase Adh4 (6), which is similar to an alcohol dehydrogenase from Zymonas mobilis (and also S. cerevisiae Adh4) and is predicted to employ iron rather than zinc as a cofactor. As such, it has been suggested that the upregulation of this iron-dependent isoform may compensate for the loss of zinc-dependent alcohol dehydrogenase activity (23). However, the nature of the cofactor employed by these enzymes remains controversial. Despite being closely related to bacterial iron-dependent alcohol dehydrogenases, the activity of S. cerevisiae Adh4 was found to be stimulated by zinc rather than iron (7). Other genes that are upregulated by zinc-limiting conditions are also known to be induced in response to other adverse environmental conditions. These include a number of antioxidants and also other genes that are known to be induced upon exposure to hydrogen peroxide. Consistent with this, a number of studies have linked zinc deficiency to increased levels of reactive oxygen species in mammalian systems (28, 29, 42, 43). Furthermore, recent evidence suggests that zinc deficiency also results in oxidative stress in budding yeast (41). Although zinc is not redox active, a number of possible roles for zinc in antioxidant defense have been postulated. These include being a constituent of antioxidant enzymes, replacing redox-active metals from membrane binding sites, and protecting sulfhydryls (29). It is also noteworthy that work with human neuroblastoma cells has demonstrated that zinc status influences their sensitivity to iron-induced oxidative stress (25). It is therefore interesting that our data indicate that fission yeast cells downregulate some genes involved in iron uptake and utilization in response to zinc deficiency.
The function of other genes whose mRNA levels are increased under conditions of zinc deficiency is less obvious. SPBC138.06c, SPAC977.05c, and SPBPB2B2.15 are closely related genes that have arisen through the duplication of subtelomeric regions. These genes are homologues of the S. cerevisiae genes VEL1 and YOR387C, which are regulated by Zap1. Although the functions of the proteins encoded by these genes are not understood, they are predicted to be cell surface glycoproteins. Whether or not they confer any selective advantage under zinc-limiting conditions remains to be determined.
The requirement of zinc for numerous cellular processes dictates that zinc deficiency will have an adverse impact upon growth rate. This clearly has been observed, as the doubling time of zinc-limited (zrt1-II1) cells in minimal medium (EMM) is increased relative to that of wild-type cells. This effect of zinc deficiency is reflected at the level of mRNA because a large number of downregulated genes have products that are involved in cell growth, including ribosomal proteins and amino acid transporters, and in nucleotide metabolism. A potential candidate for mediating this process is SPAC4G8.03c, a Pumilio family RNA binding protein, which is induced by low intracellular zinc concentrations (Table 2). Pumilio family members are known to negatively regulate gene expression either through inhibition of translation or by enhancing mRNA turnover (40). It will be interesting to identify the mRNAs that are regulated by this RNA binding protein.
Our studies indicate that 2.5% of S. pombe genes have mRNA levels that are regulated in response to zinc deficiency. Comparison studies of other organisms have revealed much more profound changes in global transcript profiles. For instance, the study of Lyons et al. revealed that zinc limitation led to changes in more than 15% of S. cerevisiae genes (23). It is possible that some of this difference may reflect different approaches used to induce zinc deficiency. Whereas Lyons et al. employed Chelex-treated (CSD) zinc-limiting medium, we have exploited the zrt1-II1 mutation. However, a recently published study also finds substantial differences in the responses to copper and iron between budding and fission yeast (30). Nonetheless, there is a significant overlap between the S. pombe genes that are highly induced (greater than sixfold) by zinc deficiency and the S. cerevisiae Zap1 regulon. All but one (dak2+) have S. cerevisiae homologues that are upregulated by zinc limitation. For instance, both organisms strongly increase the mRNA levels of genes encoding zinc uptake transporters, mitochondrial alcohol dehydrogenases, and cell surface proteins. However, S. pombe, like mammals and plants, does not encode a homologue of the S. cerevisiae zinc-sensing transcriptional activator Zap1. Therefore, different regulatory mechanisms must be used by S. pombe to regulate mRNA levels in response to zinc availability. Indeed, preliminary evidence suggests that the expression of adh4+ is regulated by a combination of activating and repressing promoter elements. Furthermore, it is possible that posttranscriptional controls may also play a role in the response to zinc deprivation.
Supplementary Material
Acknowledgments
We thank Nigel Robinson for assistance with the metal content analysis, Nic Jones and Caroline Wilkinson for strains, and Elizabeth Veal for comments on the manuscript.
This work was funded by a BBSRC project grant (BB/C004752/1) to S.K.W. and by a Cancer Research UK program grant (C9546/A6517) to J.B.
Footnotes
Published ahead of print on 18 January 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Berg, J. M., and Y. Shi. 1996. The galvanization of biology: a growing appreciation for the roles of zinc. Science 2711081-1085. [DOI] [PubMed] [Google Scholar]
- 2.Bird, A., M. V. Evans-Galea, E. Blankman, H. Zhao, H. Luo, D. R. Winge, and D. J. Eide. 2000. Mapping the DNA binding domain of the Zap1 zinc-responsive transcriptional activator. J. Biol. Chem. 27516160-16166. [DOI] [PubMed] [Google Scholar]
- 3.Borrelly, G. P., M. D. Harrison, A. K. Robinson, S. G. Cox, N. J. Robinson, and S. K. Whitehall. 2002. Surplus zinc is handled by Zym1 metallothionein and Zhf endoplasmic reticulum transporter in Schizosaccharomyces pombe. J. Biol. Chem. 27730394-30400. [DOI] [PubMed] [Google Scholar]
- 4.Cao, J., J. A. Bobo, J. P. Liuzzi, and R. J. Cousins. 2001. Effects of intracellular zinc depletion on metallothionein and ZIP2 transporter expression and apoptosis. J. Leukoc. Biol. 70559-566. [PubMed] [Google Scholar]
- 5.Chen, D., W. M. Toone, J. Mata, R. Lyne, G. Burns, K. Kivinen, A. Brazma, N. Jones, and J. Bähler. 2003. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14214-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crichton, P. G., C. Affourtit, and A. L. Moore. 2007. Identification of a mitochondrial alcohol dehydrogenase in Schizosaccharomyces pombe: new insights into energy metabolism. Biochem. J. 401459-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drewke, C., and M. Ciriacy. 1988. Overexpression, purification and properties of alcohol dehydrogenase IV from Saccharomyces cerevisiae. Biochim. Biophys. Acta 95054-60. [DOI] [PubMed] [Google Scholar]
- 8.Dufner-Beattie, J., Y. M. Kuo, J. Gitschier, and G. K. Andrews. 2004. The adaptive response to dietary zinc in mice involves the differential cellular localization and zinc regulation of the zinc transporters ZIP4 and ZIP5. J. Biol. Chem. 27949082-49090. [DOI] [PubMed] [Google Scholar]
- 9.Dufner-Beattie, J., F. Wang, Y. M. Kuo, J. Gitschier, D. Eide, and G. K. Andrews. 2003. The acrodermatitis enteropathica gene ZIP4 encodes a tissue-specific, zinc-regulated zinc transporter in mice. J. Biol. Chem. 27833474-33481. [DOI] [PubMed] [Google Scholar]
- 10.Eide, D. J. 2004. The SLC39 family of metal ion transporters. Pflugers Arch. 447796-800. [DOI] [PubMed] [Google Scholar]
- 11.Gitan, R. S., and D. J. Eide. 2000. Zinc-regulated ubiquitin conjugation signals endocytosis of the yeast ZRT1 zinc transporter. Biochem. J. 346329-336. [PMC free article] [PubMed] [Google Scholar]
- 12.Gitan, R. S., H. Luo, J. Rodgers, M. Broderius, and D. Eide. 1998. Zinc-induced inactivation of the yeast ZRT1 zinc transporter occurs through endocytosis and vacuolar degradation. J. Biol. Chem. 27328617-28624. [DOI] [PubMed] [Google Scholar]
- 13.Gitan, R. S., M. Shababi, M. Kramer, and D. J. Eide. 2003. A cytosolic domain of the yeast Zrt1 zinc transporter is required for its post-translational inactivation in response to zinc and cadmium. J. Biol. Chem. 27839558-39564. [DOI] [PubMed] [Google Scholar]
- 14.Grotz, N., T. Fox, E. Connolly, W. Park, M. L. Guerinot, and D. Eide. 1998. Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc. Natl. Acad. Sci. USA 957220-7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guarente, L. 1983. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 101181-191. [DOI] [PubMed] [Google Scholar]
- 16.Harrison, C., S. Katayama, S. Dhut, D. Chen, N. Jones, J. Bähler, and T. Toda. 2005. SCF(Pof1)-ubiquitin and its target Zip1 transcription factor mediate cadmium response in fission yeast. EMBO J. 24599-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hedges, S. B. 2002. The origin and evolution of model organisms. Nat. Rev. Genet. 3838-849. [DOI] [PubMed] [Google Scholar]
- 18.Huang, L., C. P. Kirschke, Y. Zhang, and Y. Y. Yu. 2005. The ZIP7 gene (Slc39a7) encodes a zinc transporter involved in zinc homeostasis of the Golgi apparatus. J. Biol. Chem. 28015456-15463. [DOI] [PubMed] [Google Scholar]
- 19.Kury, S., B. Dreno, S. Bezieau, S. Giraudet, M. Kharfi, R. Kamoun, and J. P. Moisan. 2002. Identification of SLC39A4, a gene involved in acrodermatitis enteropathica. Nat. Genet. 31239-240. [DOI] [PubMed] [Google Scholar]
- 20.Lafuente, M. J., T. Petit, and C. Gancedo. 1997. A series of vectors to construct lacZ fusions for the study of gene expression in Schizosaccharomyces pombe. FEBS Lett. 42039-42. [DOI] [PubMed] [Google Scholar]
- 21.Liuzzi, J. P., and R. J. Cousins. 2004. Mammalian zinc transporters. Annu. Rev. Nutr. 24151-172. [DOI] [PubMed] [Google Scholar]
- 22.Lyne, R., G. Burns, J. Mata, C. J. Penkett, G. Rustici, D. Chen, C. Langford, D. Vetrie, and J. Bahler. 2003. Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyons, T. J., A. P. Gasch, L. A. Gaither, D. Botstein, P. O. Brown, and D. J. Eide. 2000. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc. Natl. Acad. Sci. USA 977957-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacDiarmid, C. W., L. A. Gaither, and D. Eide. 2000. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 192845-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackenzie, G. G., C. L. Keen, and P. I. Oteiza. 2002. Zinc status of human IMR-32 neuroblastoma cells influences their susceptibility to iron-induced oxidative stress. Dev. Neurosci. 24125-133. [DOI] [PubMed] [Google Scholar]
- 26.Maret, W. 2001. Zinc biochemistry, physiology, and homeostasis—recent insights and current trends. Biometals 14187-190. [Google Scholar]
- 27.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194795-823. [DOI] [PubMed] [Google Scholar]
- 28.Oteiza, P. I., M. S. Clegg, M. P. Zago, and C. L. Keen. 2000. Zinc deficiency induces oxidative stress and AP-1 activation in 3T3 cells. Free Radic. Biol. Med. 281091-1099. [DOI] [PubMed] [Google Scholar]
- 29.Oteiza, P. I., and G. G. Mackenzie. 2005. Zinc, oxidant-triggered cell signaling, and human health. Mol. Aspects Med. 26245-255. [DOI] [PubMed] [Google Scholar]
- 30.Rustici, G., H. van Bakel, D. H. Lackner, F. Holstege, C. Wijmenga, J. Bähler, and A. Brazma. 2007. Global transcriptional responses of fission and budding yeast to changes in copper and iron levels: a comparative study. Genome Biol. 8R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakurai, M., H. Tohda, H. Kumagai, and Y. Giga-Hama. 2004. A distinct type of alcohol dehydrogenase, adh4+, complements ethanol fermentation in an adh1-deficient strain of Schizosaccharomyces pombe. FEMS Yeast Res. 4649-654. [DOI] [PubMed] [Google Scholar]
- 32.Stefanidou, M., C. Maravelias, A. Dona, and C. Spiliopoulou. 2006. Zinc: a multipurpose trace element. Arch. Toxicol. 801-9. [DOI] [PubMed] [Google Scholar]
- 33.Stohs, S. J., and D. Bagchi. 1995. Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 18321-336. [DOI] [PubMed] [Google Scholar]
- 34.Takeda, T., T. Toda, K. Kominami, A. Kohnosu, M. Yanagida, and N. Jones. 1995. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 146193-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toone, W. M., and N. Jones. 1998. Stress-activated signalling pathways in yeast. Genes Cells 3485-498. [DOI] [PubMed] [Google Scholar]
- 36.Wang, F., J. Dufner-Beattie, B. E. Kim, M. J. Petris, G. Andrews, and D. J. Eide. 2004. Zinc-stimulated endocytosis controls activity of the mouse ZIP1 and ZIP3 zinc uptake transporters. J. Biol. Chem. 27924631-24639. [DOI] [PubMed] [Google Scholar]
- 37.Wang, F., B. E. Kim, M. J. Petris, and D. J. Eide. 2004. The mammalian Zip5 protein is a zinc transporter that localizes to the basolateral surface of polarized cells. J. Biol. Chem. 27951433-51441. [DOI] [PubMed] [Google Scholar]
- 38.Wang, K., B. Zhou, Y. M. Kuo, J. Zemansky, and J. Gitschier. 2002. A novel member of a zinc transporter family is defective in acrodermatitis enteropathica. Am. J. Hum. Genet. 7166-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waters, B. M., and D. J. Eide. 2002. Combinatorial control of yeast FET4 gene expression by iron, zinc, and oxygen. J. Biol. Chem. 27733749-33757. [DOI] [PubMed] [Google Scholar]
- 40.Wickens, M., D. S. Bernstein, J. Kimble, and R. Parker. 2002. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 18150-157. [DOI] [PubMed] [Google Scholar]
- 41.Wu, C. Y., A. J. Bird, D. R. Winge, and D. J. Eide. 2007. Regulation of the yeast TSA1 peroxiredoxin by ZAP1 is an adaptive response to the oxidative stress of zinc deficiency. J. Biol. Chem. 2822184-2195. [DOI] [PubMed] [Google Scholar]
- 42.Zago, M. P., G. G. Mackenzie, A. M. Adamo, C. L. Keen, and P. I. Oteiza. 2005. Differential modulation of MAP kinases by zinc deficiency in IMR-32 cells: role of H(2)O(2). Antioxid. Redox Signal. 71773-1782. [DOI] [PubMed] [Google Scholar]
- 43.Zago, M. P., and P. I. Oteiza. 2001. The antioxidant properties of zinc: interactions with iron and antioxidants. Free Radic. Biol. Med. 31266-274. [DOI] [PubMed] [Google Scholar]
- 44.Zhao, H., and D. Eide. 1996. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl. Acad. Sci. USA 932454-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao, H., and D. Eide. 1996. The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J. Biol. Chem. 27123203-23210. [DOI] [PubMed] [Google Scholar]
- 46.Zhao, H., and D. J. Eide. 1997. Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 175044-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.