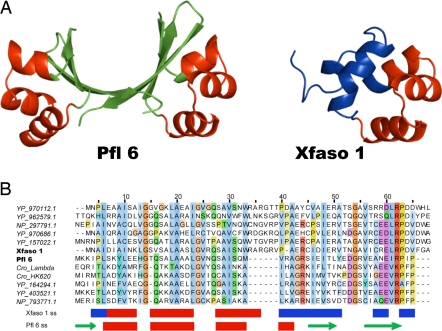
Most of us who teach protein structure have very likely stood in front of a class at some point and confidently stated that any two naturally occurring proteins displaying 40% sequence identity will be homologous and thus possess the same fold. A paper by Roessler et al. (1) in a recent issue of PNAS has definitively overturned this basic tenet by demonstrating that a pair of protein homologues displaying 40% identity exhibit markedly different folds. These proteins are both repressors of the Cro family and were identified in prophage sequences present in the genomes of the bacterial species, Pseudomonas fluorescens (Pfl 6) and Xylella fastidiosa (Xfaso 1). The atomic resolution structures of these proteins, solved by Roessler et al. using x-ray crystallography, reveal a similar N-terminal helix–turn–helix but widely diverging C-terminal regions; Xfaso 1 displays an all-helical monomeric fold, whereas the Pfl 6 C terminus forms an intertwined β-sheet dimer (Fig. 1A). The conclusion that these proteins are descended from a common ancestor is strongly supported. An alignment of homologues of each of these protein shows that many positions are conserved across both groups of proteins even in the C-terminal region where the structures diverge (Fig. 1B). This conservation pattern argues against a distinct C terminus being placed onto one of these proteins through a nonhomologous recombination event. The genomic context of the genes encoding these proteins with respect to other surrounding phage genes is also highly conserved, which implies a common ancestry and function.
Fig. 1.
Homologous proteins with 40% sequence identity and different folds. (A) The atomic resolution structures of Pfl 6 and Xfaso 1 as solved by x-ray crystallography are shown. The helix–turn–helix motif, which is seen in both proteins, is colored in red. The structurally divergent regions are shown in green (Pfl 6) and in blue (Xfaso 1). (B) Relatively close homologues (≈35–50% identity) of Xfaso 1 (above the Xfaso 1 sequence) and Pfl 6 (below the Pfl 6 sequence) are shown. The sequences are shaded by conservation by using ClustalX coloring as implemented in Jalview (18). Where noted, these sequences are from Cro repressor proteins of the named phages. All other sequences are from prophages found in a variety of bacterial species. These sequences are identified by their locus names. The secondary structure assignments for Xfaso 1 and Pfl 6 are shown below the alignment. Arrows denote a β-strand, and rectangles denote an α-helix, with coloring corresponding to the structures shown in A.
This work provides the potential for critical new insights into how protein folds may have evolved. Given that there are well over 1,000 folds in nature, most would agree that these folds could not have all arisen independently and that, at some point, many folds must have evolved from a small number of primordial folds. However, there have been few examples identified to demonstrate how proteins can dramatically change folds through a mutagenic process. Although previous studies have uncovered pairs of proteins that appear to be homologous yet possess different structures (2–4), these proteins display ≈30% sequence identity or less, and the case for homology is not clear-cut in most cases. From comparison of the Pfl 6 and Xfaso 1 sequences and those of their homologues (Fig. 1B), it is evident that the divergence of these structures has occurred primarily through point mutations, with the possible additional aid of an insertion/deletion at position 33 and the addition of a few extra residues at each end of the protein, as has been discussed in ref. 5. The identification of highly similar homologous proteins with different structures, such as Pfl 6 and Xfaso 1, opens up the feasibility of determining structure switching mechanisms through mutagenesis studies (6).
Just as protein folds have evolved, so to have our perceptions of fold evolution in recent years. Although deep down we all knew that protein structures had to evolve somehow, demonstration of fold evolution by using contemporary proteins seemed impossible because the tertiary structure of each protein appeared to be extremely “overdetermined” by its sequence. In other words, one could mutate a protein and still not change the tertiary structure at all [e.g., 22 of 60 residues in a homeodomain were replaced with Ala, yet the mutant still possessed native tertiary structure and could still function (7)]. The apparent fixity of protein folds in sequence space provoked Rose and Creamer in 1994 to issue the “Paracelsus Challenge,” whereby the protein folding community was charged with the task of designing two proteins that were at least 50% identical but possessed different folds (8). Amazingly, this goal was fully achieved in only 3 years, when Dalal et al. (9) designed a sequence with 50% identity to a mostly β-sheet protein that folded into a four-helix bundle. Since then, several others have achieved similarly impressive feats of design (10). Most spectacularly, Alexander et al. (11) recently designed two proteins with 88% identity that fold into completely different tertiary structures (one is all-α-helix and the other is an α/β-fold) and maintain their original functions. Although these studies have convinced us that proteins with very similar sequences can indeed possess different folds, which certainly opens up the possibility of fold evolution, these proteins have been generated by design protocols involving the introduction of many amino acid substitutions simultaneously. Obviously, evolution could not work in this manner.
At this juncture, the location of the evolutionary routes connecting the various regions of fold space are mostly a matter for speculation. It does seem clear, however, that most of the well established mechanisms of gene evolution, such as deletion/insertion, nonhomologous recombination, and gene duplication, do come into play (2, 4). One particularly intriguing question is how single amino acid changes can change a protein fold without producing completely unstable intermediate structures. In Darwinian terms, what are the missing links between protein folds? The possibility that limited changes in a protein fold can be induced by one or only a few substitutions has been demonstrated in several studies (12–14). For example, two substitutions in the N-terminal β-strand of the Arc repressor convert this region into a right-handed helix (12). Intriguingly, a single residue substitution in this same region results in a protein that can display either a strand or a helix, depending on conditions. This mutant could be seen as a true “missing link,” although not a naturally occurring one (15). The existence of “chameleon” sequences, identical sequences that can be found in completely different secondary and tertiary structures in different proteins, also hints at mechanisms for fold evolution. Such sequences of up to eight residues can be identified by searching the protein structure database (16). Strikingly, an 11-residue sequence that could fold into either an α-helix or a β-strand, depending on its location within the same protein, has been designed (17). In a similar vein, a comparative study of the Cro repressors from phages λ and P22, which adopt the same two divergent structures that Pfl 6 and Xfaso 1 do, demonstrated that a single, designed 18-residue sequence could fold as a β-hairpin when incorporated into the λ Cro and as a pair of α-helices when incorporated into the P22 Cro (6). The viability of chameleon sequences in various protein structures demonstrates that the low stability of proteins and the degeneracy of residue-encoded folding information may be sufficient to allow “missing link” proteins (i.e., ones able to simultaneously adopt two different folds) to actually function in nature.
Further investigation of the key sequence changes that allow Pfl 6 and Xfaso 1 to adopt different folds and the identification of other examples of highly similar homologous proteins adopting different folds will certainly be crucial to our understanding of fold evolution. This point brings us to a final important question: how difficult will it be to find these other examples? Or to put it another way, why have more cases like Pfl 6 and Xfaso 1 not been identified? A simple answer to the latter question is that, with the exception of the Cro repressor studies discussed here, no other systematic effort has been made to find and experimentally investigate examples of evolutionary fold switching. The Cordes group (5, 6) arrived at the observations described in a recent issue of PNAS through an impressive series of investigations over the past 5 years that were specifically aimed at identifying a mechanism for fold switching in the Cro repressor family. Evolutionary fold switching had long been suspected in this case because of functional and structural similarities between the β-sheet-containing λ Cro repressor and the all-helical cI repressor N-terminal domains. A tremendous advantage of the Cro system is the large number of highly diverse examples that could be definitively identified as true members of the family by both distant sequence similarity and by conserved position within phage or prophage genomes. The Cordes group (5) was able to use this collection of Cro sequences to trace a sequence-similarity-based path from the β-strand containing the Cro repressor of phage λ to the all-helical Cro repressor of phage P22, the structure of which they solved. This path was discovered through “transitive sequence comparison,” in which dissimilar sequences are linked through intermediate sequences that are closer in sequence to one or the other of the targets of interest. By using this method, a route through Cro sequence space was found from λ to P22 that involved three intermediaries, each of which was ≈40% identical to its closest neighbor. If a transitive path can be found between two putative homologues with different structures, then there is a good chance to discover two proteins with similar sequence but different structure, like Pfl 6 and Xfaso 1. Because there are many diverse sequence families in bacterial and phage genomes for which common evolutionary origin can be deduced both from sequence similarity and genome position, it seems probable that many more examples of fold evolution will be found as long as bold researchers are willing to embark on the appropriate journeys through sequence and folding space.
Many folds must have evolved from a small number of primordial folds.
ACKNOWLEDGMENTS.
The research of A.R.D. is funded by the Canadian Institutes of Health Research.
Footnotes
The author declares no conflict of interest.
See companion article on page 2343 in issue 7 of volume 105.
References
- 1.Roessler CG, Hall BM, Anderson WJ, Ingram WM, Roberts SA, Montfort WR, Cordes MHJ. Proc Natl Acad Sci USA. 2008;105:2343–2348. doi: 10.1073/pnas.0711589105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreeva A, Murzin AG. Curr Opin Struct Biol. 2006;16:399–408. doi: 10.1016/j.sbi.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, Grishin NV, Vassylyev DG, Artsimovitch I. Mol Cell. 2007;26:117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grishin NV. J Struct Biol. 2001;134:167–185. doi: 10.1006/jsbi.2001.4335. [DOI] [PubMed] [Google Scholar]
- 5.Newlove T, Konieczka JH, Cordes MH. Structure. 2004;12:569–581. doi: 10.1016/j.str.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 6.Van Dorn LO, Newlove T, Chang S, Ingram WM, Cordes MH. Biochemistry. 2006;45:10542–10553. doi: 10.1021/bi060853p. [DOI] [PubMed] [Google Scholar]
- 7.Shang Z, Isaac VE, Li H, Patel L, Catron KM, Curran T, Montelione GT, Abate C. Proc Natl Acad Sci USA. 1994;91:8373–8377. doi: 10.1073/pnas.91.18.8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rose GD, Creamer TP. Proteins. 1994;19:1–3. doi: 10.1002/prot.340190102. [DOI] [PubMed] [Google Scholar]
- 9.Dalal S, Balasubramanian S, Regan L. Nat Struct Biol. 1997;4:548–552. doi: 10.1038/nsb0797-548. [DOI] [PubMed] [Google Scholar]
- 10.Ambroggio XI, Kuhlman B. Curr Opin Struct Biol. 2006;16:525–530. doi: 10.1016/j.sbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 11.Alexander PA, He Y, Chen Y, Orban J, Bryan PN. Proc Natl Acad Sci USA. 2007;104:11963–11968. doi: 10.1073/pnas.0700922104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cordes MH, Walsh NP, McKnight CJ, Sauer RT. Science. 1999;284:325–328. doi: 10.1126/science.284.5412.325. [DOI] [PubMed] [Google Scholar]
- 13.Tidow H, Lauber T, Vitzithum K, Sommerhoff CP, Rosch P, Marx UC. Biochemistry. 2004;43:11238–11247. doi: 10.1021/bi0492399. [DOI] [PubMed] [Google Scholar]
- 14.Yang WZ, Ko TP, Corselli L, Johnson RC, Yuan HS. Protein Sci. 1998;7:1875–1883. doi: 10.1002/pro.5560070904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cordes MH, Burton RE, Walsh NP, McKnight CJ, Sauer RT. Nat Struct Biol. 2000;7:1129–1132. doi: 10.1038/81985. [DOI] [PubMed] [Google Scholar]
- 16.Sudarsanam S. Proteins. 1998;30:228–231. doi: 10.1002/(sici)1097-0134(19980215)30:3<228::aid-prot2>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 17.Minor D, Jr, Kim PS. Nature. 1996;380:730–734. doi: 10.1038/380730a0. [DOI] [PubMed] [Google Scholar]
- 18.Clamp M, Cuff J, Searle SM, Barton GJ. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]