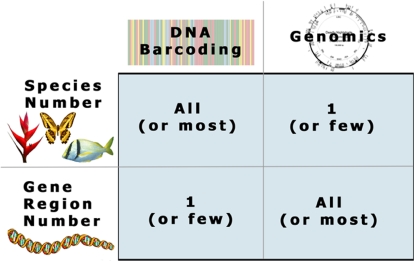
It is not a coincidence that DNA barcoding has developed in concert with genomics-based investigations. DNA barcoding (a tool for rapid species identification based on DNA sequences) and genomics (which compares entire genome structure and expression) share an emphasis on large-scale genetic data acquisition that offers new answers to questions previously beyond the reach of traditional disciplines. DNA barcodes consist of a standardized short sequence of DNA (400–800 bp) that in principle should be easily generated and characterized for all species on the planet (1). A massive on-line digital library of barcodes will serve as a standard to which the DNA barcode sequence of an unidentified sample from the forest, garden, or market can be matched. Similar to genomics, which has accelerated the process of recognizing novel genes and comparing gene function, DNA barcoding will allow users to efficiently recognize known species and speed the discovery of species yet to be found in nature. DNA barcoding aims to use the information of one or a few gene regions to identify all species of life, whereas genomics, the inverse of barcoding, describes in one (e.g., humans) or a few selected species the function and interactions across all genes (Fig. 1). The work of Lahaye et al. (2) reported in a recent issue of PNAS brings the application of DNA barcoding one step closer to implementation in plants.
Fig. 1.
The matrix of genetic information and taxonomic diversity, with DNA barcoding at one extreme (with high species diversity and limited genetic coverage) and genomics (with limited species diversity but complete gene description) at the other extreme.
The deceptively simple task of selecting an appropriate locus to serve as a plant barcode has been much more complex than expected and has engendered considerable debate. Despite the current lack of consensus on a universal plant barcode, taxonomists, ecologists, evolutionary biologists, and conservationists are already envisioning the application of a genetic identifier to a wide set of research and applied programs. Lahaye et al. (2) point out that plant DNA barcodes can be used to assess species identification in conservation biodiversity hotspots as well as hypothetically applied to monitoring the international trade in endangered species of orchids. Whole forest species inventories based on DNA barcodes are also now in progress in both the temperate zone (Plummers Island in Maryland and a park in New York) and the tropics (Forest Dynamics Plot in Panama and soon to be initiated at La Selva Biological Station in Costa Rica), which will allow the identification of plant tissue fragments in ecological investigations as well as quantitative comparisons of genetic diversity among forest sites. If the barcode marker is conservative enough (e.g., by including a well suited gene, such as rbcL, in a multilocus barcode), it will enable the construction of phylogenetic trees for all of the species in a forest, facilitating investigations of community structure (3) and functional trait evolution (4). The Forest Dynamics Plot is one of 20 sites located in tropical countries (Center for Tropical Forest Science; www.ctfs.si.edu/doc/index.php), which taken together encompass nearly 3.5 million trees representing 12% of all known tree species. A complete DNA barcode census is now planned for all of the woody plants at these sites. The resultant germplasm bank from this intercontinental application of DNA barcoding will open up new opportunities for DNA investigations ranging from community phylogenetics (5) to ecological genomics (6).
To be practical as a DNA barcode a gene region must satisfy three criteria: (i) contain significant species-level genetic variability and divergence, (ii) possess conserved flanking sites for developing universal PCR primers for wide taxonomic application, and (iii) have a short sequence length so as to facilitate current capabilities of DNA extraction and amplification. A short DNA sequence of 600 bp in the mitochondrial gene for cytochrome c oxidase subunit 1 (CO1) (7) has been accepted as a practical, standardized species-level barcode for animals (see www.barcoding.si.edu). The inability of CO1 to work as a barcode in plants (8) set off a race among botanists to find a more appropriate marker (9). A number of candidate gene regions have been suggested as possible barcodes for plants (10–14), but none have been widely accepted by the taxonomic community. This lack of consensus is in part due to the limitations inherent in a plastid marker relative to plant CO1, and also because a quantitative context for selecting a gene region as a barcode for plants has not been offered. Several factors must be considered and weighted in selecting a plant DNA barcode: (i) universal PCR amplification, (ii) range of taxonomic diversity, (iii) power of species differentiation, and (iv) bioinformatic analysis and application.
Lahaye et al. (2) report tests of the various loci and intergenic spacers that have already been proposed as plant barcodes against their favorite candidate: the plastid gene matK. Their article contains many of the right elements: a diverse sample of taxa in the flowering plants, a primer set for matK that increases universality, trials of their marker on species identification and discovery, and the application of barcodes to important environmental issues. This article is welcome, but as with many of the other publications that have proposed candidate plant barcodes, the authors omit quantitative criteria and standards that are necessary to compare the success and applicability of their favored locus against all others.
Successful universal PCR amplification across a wide range of plants must be the primary criterion for selecting a DNA barcode. A challenging tradeoff exists between universal PCR amplification and high rates of sequence divergence. This tradeoff, which is particularly problematic in coding loci, is less so in noncoding regions because universal primers are normally found in the highly conserved genes that flank the hypervariable intergenic spacers. The taxonomic community has wavered on setting a level of universal amplification (e.g., all land plants or just flowering plants?) and the simplicity of PCR conditions (one primer set for all taxa or multiple sets across taxonomic groups?) required for a barcode. DNA barcoding must be practical for a wide range of practitioners and, therefore, the methodology must be accessible and easily carried out by multiple users. The power of DNA barcoding is also directly proportional to the data available in the barcode library; building a very complete database will greatly increase the power of DNA barcoding (15). These considerations require a narrow, standard range of PCR conditions along with a limited (ideally one) set of PCR primers per locus that will provide a robust barcode marker for the widest range of taxa and users. Lahaye et al. (2) purport to have tested their barcode loci on the widest sample of taxa so far used in any published study. Although the number of species is the largest sample yet published on plants, 96% of those samples are in a single family, Orchidaceae. The other samples are spread across 23 families in 18 orders, which is less than half the families and orders sampled in earlier trials (12, 13). In addition, they report employing several primer pairs, rather than an optimal single pair, to successfully amplify matK across the samples. For broad universality and simplicity of use, matK has not yet been demonstrated to pass the test for a successful plant barcode.
A criterion related to PCR universality is the relative success of a barcode marker across the major lineages of land plants, including angiosperms, gymnosperms, ferns, and mosses. Lahaye et al. (2) tested matK only on angiosperms, explicitly stating that it is not important to select a barcode that works successfully across all land plants. In today's ecosystems in which the vast majority of plants are angiosperms, some might argue that markers should be chosen that work best for these dominant land plants. Yet given that the purpose of a DNA barcode is to facilitate identification of unknown samples, including small isolated fragments of tissue, then the selected loci should work easily on all groups of green land plants.
Conceptually, any consistent, nonzero sequence variation that distinguishes two species should work as a DNA barcode. Furthermore, DNA barcodes do not require any demonstration of the homology of mutations as would be needed in a phylogenetic marker. In other words, low levels of divergence may be sufficient to distinguish among species even if not adequate to estimate phylogenetic relationships. Relative to CO1 in animals, the mean divergence level between species in plants is usually quite low (13, 16). Curiously, Lahaye et al. (2) reject the gene region that showed the highest divergence value (trnH–psbA) in favor of matK, which showed nearly 50% less interspecific divergence. As of yet a quantitative metric that can be used to compare barcode candidates does not exist. The use of a simple statistic that could be calculated as the product of the levels of PCR universality and sequence divergence would allow for direct comparisons between putative DNA barcode markers. The proportion of taxa that are successfully amplified and sequenced across a targeted test set together with the percentage of species pairs that are differentiated by a particular locus are independent characters that can be combined as a product of the two values into a single metric for comparison.
The simple comparative statistic proposed is relevant only when other factors are considered, including the effort required to recover the PCR amplicon and the number of different primers and reaction conditions used for sequencing each putative barcode locus. The suitability of a locus for large-scale DNA barcoding could easily be evaluated by comparing loci across the same set of taxa under a designated set of reaction conditions. Although not an explicit measure of how well a DNA barcode will perform at identifying species within a bioinformatics context, this statistic takes into account the intrinsic tradeoff inherent in a DNA barcode marker between the ability to amplify a locus and the rate of divergence of that locus across a phylogenetic range of taxa.
In conclusion, two final factors that may strongly affect how well barcode markers work at species identification and discovery are database design and sequence search strategies. To date the exact method or algorithm to be used in searching the barcode database has not been thoroughly investigated nor debated, particularly as regards a multilocus DNA barcode (14, 17). The algorithms used in the most commonly used databases, GenBank and the Barcode of Life Database (BOLD), are quite different. However, a plethora of additional sequence alignment methodologies are available, which can be evaluated for use in DNA barcodes with regards to the following: (i) the application of confidence limits to species assignment, (ii) the use of partial sequences in database searches, and (iii) the impact on search algorithms of sequence length variation due to insertion/deletion events and the informative nature of these mutations. Clearly, DNA barcoding has great potential for enhancing ecological and evolutionary investigations if the right genetic markers are selected. The issues raised here, if carefully considered and implemented, will allow a rational selection of a plant DNA barcode based on a comparative and quantitative analysis.
Footnotes
The authors declare no conflict of interest.
See companion article on page 2923.
References
- 1.Savolainen V, Cowan RS, Vogler AP, Roderick GK, Lane R. Phil Trans R Soc London Ser B. 2005;360:1805–1811. doi: 10.1098/rstb.2005.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lahaye R, van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, Maurin O, Duthoit S, Barraclough TG, Savolainen V. Proc Natl Acad Sci USA. 2008;105:2923–2928. doi: 10.1073/pnas.0709936105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kembel SW, Hubbell SP. Ecology. 2006;87:S86–S99. doi: 10.1890/0012-9658(2006)87[86:tpsoan]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 4.Westoby M, Wright IJ. Trends Ecol Evol. 2006;21:261–268. doi: 10.1016/j.tree.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. Annu Rev Ecol Syst. 2002;33:475–505. [Google Scholar]
- 6.van Straalen NM, Roelofs D. An Introduction to Ecological Genomics. London: Oxford Univ Press; 2006. [Google Scholar]
- 7.Hebert PDN, Ratnasingham S, deWaard JR. Proc Roy Soc B. 2003;270(suppl):S96–SS99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho Y, Mower JP, Qiu YL, Palmer JD. Proc Natl Acad Sci USA. 2004;101:17741–17746. doi: 10.1073/pnas.0408302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pennisi E. Science. 2007;318:190–191. doi: 10.1126/science.318.5848.190. [DOI] [PubMed] [Google Scholar]
- 10.Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Proc Natl Acad Sci USA. 2005;102:8369–8374. doi: 10.1073/pnas.0503123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taberlet P, Coissac E, Pompanon F, Gielly L, Miquel C, Valentini A, Vermat T, Corthier G, Brochmann C, Willerslev E. Nucleic Acids Res. 2007;35(3):e14. doi: 10.1093/nar/gkl938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chase MW, Cowan RS, Hollingsworth PM, van den Berg C, Madriñán S, Petersen G, Seberg O, Jorgsensen T, Cameron KN, Carine M, et al. Taxon. 2007;56:295–299. [Google Scholar]
- 13.Kress WJ, Erickson DL. PLoS ONE. 2007;2(6):e508. doi: 10.1371/journal.pone.0000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sass C, Little DP, Stevenson DW, Specht CD. PLoS ONE. 2007;2(11):e1154. doi: 10.1371/journal.pone.0001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekrem T, Willassen E, Stur E. Mol Phylogenet Evol. 2007;43:530–542. doi: 10.1016/j.ympev.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 16.Shaw J, Lickey EB, Schilling EE, Small RL. Am J Bot. 2007;94:275–288. doi: 10.3732/ajb.94.3.275. [DOI] [PubMed] [Google Scholar]
- 17.Little D, Stevenson DW. Cladistics. 2006;22:1–21. doi: 10.1111/j.1096-0031.2006.00126.x. [DOI] [PubMed] [Google Scholar]