Abstract
The DNA-binding α/β-type small acid-soluble proteins (SASPs) are a major factor in the resistance and long-term survival of spores of Bacillus species by protecting spore DNA against damage due to desiccation, heat, toxic chemicals, enzymes, and UV radiation. We now report the crystal structure at 2.1 Å resolution of an α/β-type SASP bound to a 10-bp DNA duplex. In the complex, the α/β-type SASP adopt a helix–turn–helix motif, interact with DNA through minor groove contacts, bind to ≈6 bp of DNA as a dimer, and the DNA is in an A-B type conformation. The structure of the complex provides important insights into the molecular details of both DNA and α/β-type SASP protection in the complex and thus also in spores.
Keywords: α/β-type SASP, A-type DNA, DNA damage, molecular modeling, spore resistance
Spores of Bacillus and Clostridium species are extremely resistant to desiccation, heat, toxic chemicals, enzymes, and radiation. This high resistance is the major reason that spores of some species are agents of food spoilage and food poisoning and that B. anthracis spores are a biological weapon. Spore resistance is due to many factors, a major one being the protection of spore DNA against damage by the binding of small, acid-soluble proteins (SASPs) of the α/β-type (1, 2). These 59–75 residue proteins: (i) are encoded by multiple genes, (ii) comprise a protein family whose amino acid sequences are very highly conserved within and between species, (iii) are nonspecific DNA-binding proteins with apparent binding constants for random sequence DNA of 15–100 mM, (iv) are synthesized only within the developing forespore during sporulation; and (v) comprise 3–5% of total spore protein. The high levels of α/β-type SASPs in spores are sufficient to saturate the spore DNA, and the DNA within this nucleoprotein complex is protected from a variety of environmental insults. Indeed, the binding of α/β-type SASPs provides primary protection of spore DNA against damage by desiccation, heat, many genotoxic chemicals, enzymes, and UV radiation (1, 3).
In this study, we used x-ray crystallography to determine a 2.1 Å resolution structure of a complex between a 10-bp DNA duplex and an engineered α/β-type SASP originally obtained from B. subtilis. Analysis of this crystal structure provides important insight into the molecular mechanisms underlying the protection of both the protein and DNA components of the complex and the molecular details of their interactions. These results explain a great deal about the extreme resistance of the DNA in bacterial spores and thus explain much of bacterial spore resistance in atomic detail.
Results and Discussion
Overall Structure and Protection of the Protein in the Complex.
The α/β-type SASP chosen to form the complex with DNA for crystallization was B. subtilis SspCΔN11-D13K-C3, a 64-aa derivative engineered to bind tightly to DNA (4), which was an oligo(dG)·oligo(dC) 10-mer with single 3′-dA overhangs (5) The preparation and crystallization of the protein–DNA complex are described in ref. 6, and the structure was solved by multiwavelength anomalous dispersion (MAD) methods, using the selenomethionyl–protein–DNA complex [supporting information (SI) Fig. 5 and SI Table 1). In the asymmetric unit, the α/β-type SASP–DNA complex is composed of three protein molecules and one DNA, with the proteins binding to the DNA minor groove along the right-hand direction of the DNA helix (Fig. 1A). The size of the complex is 47 × 51 × 38 Å, and the protein is 65% α-helical. For every protein, seven residues at the C terminus were not visible in density maps and, therefore, were not included in the final structure. Two protein molecules bind to ≈6 bp of DNA as a dimer, and the third protein assumes a dimeric arrangement by interacting with another protein molecule from a crystallographic symmetry related complex, with this fourth protein bound to an adjacent symmetry related DNA molecule in the crystal. There are no significant structural differences between the three protomers in the complex; their pairwise structural comparisons give root mean square deviations (RMSDs) of 0.15 to 0.20 Å based on positions of all 56 Cα atoms included in the final model.
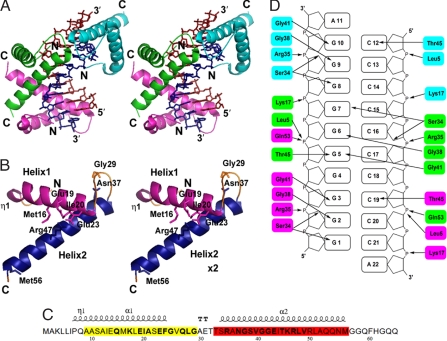
Fig. 1.
Crystal structure of the α/β-type SASP–DNA complex. (A) Stereo diagram of the complex. The positions of the N and C termini of each protomer are labeled. Green, SASP1; magenta, SASP2; cyan, SASP3; red, dG DNA strands; deep blue, dC DNA strands. (B) Stereo ribbon diagram of an α/β-type SASP protomer in the protein–DNA complex in a similar orientation as SASP1 in A. Magenta, helix1 (η1 denotes 310 helix); blue, helix2. The N- and C-terminal arms and the loop between helices1 and 2 are orange. The essential amino acids are shown as sticks and are labeled. (C) The amino acid sequence of the α/β-type SASP used for crystallization (6). Residues are numbered with the N-terminal Met (removed posttranslationally) as 1. Residues in regions highlighted in yellow and red are conserved in 45 α/β-type SASP from Bacillus species with emboldened residues exhibiting ≥95% identity. The helical structure is shown above the sequence, and the emboldened TT above the sequence denotes the region of a turn between the helices. (D) Summary of interactions between α/β-type SASPs and DNA (indicated by arrows). The color of amino acids from individual proteins is as in A.
Each α/β-type SASP protomer comprises two long helical segments connected by a turn region and extended N and C termini devoid of secondary structure (Fig. 1 B and C). The first helical region (helix1; Pro-7 to Phe-24) includes a 310 helix (Pro-7 to Ala-9) followed by an α-helix (Ala-10 to Phe-24), whereas the second helical region (helix2; Ser-34 to Met-56) is entirely α-helical. These two long helices encompass regions in α/β-type SASPs whose amino acid sequence is highly conserved not only in the proteins from Bacillus species (Fig. 1C) but also in the more distantly related anaerobic spore formers (1). The largest variation in sequence between the proteins from the aerobes and anaerobes is in the length of the extended amino acid chain region between helices 1 and 2; this length is constant in the proteins from the aerobes but varies considerably in the proteins from anaerobes. Helix1 lies on the edge of the DNA minor groove, and helix2 is located in the minor groove along the right-hand direction of the DNA helix. The major intramolecular interactions in protein molecules in the complex are hydrophobic interactions between residues of the two helices: Ile-13, Met-16, Ile-20, and Phe-24 of helix1 and Val-40, Ile-44, and Leu-48 of helix2 (Fig. 1 A and B). These contact residues are responsible for the packing of the protein molecules and are either conserved completely in α/β-type SASPs from Bacillus species or are replaced by homologous bulky hydrophobic residues. These helices form a helix–turn–helix (HTH) motif. The structure comparison, using the SSAP Server (7), revealed this structural resemblance to family of HTH motif DNA-binding proteins. The superposition of the α/β-type SASP with, for example, Escherichia coli diphtheria tox repressor (PDB ID code 1DDN) and trp repressor (PDB ID code 1TRR) as a prokaryotic HTH motif proteins and homeodomain protein (PDB ID code 2HDD) and transcription factor E2F-DP (PDB ID code 1CF7) as a eukaryotic HTH motif proteins, results in the RMSDs for the Cα-atoms of 1.95, 2.96, 4.74, and 3.37 Å, respectively. The classical HTH motif is composed of two α-helices and a short amino acid turn region (3–5 amino acid residues) ≈30 aa long. Despite the different lengths of helices and the turn region (8 amino acid residues), α/β-type SASP is more similar to diphtheria tox and trp repressors, but these are most often bound in the major groove of DNA not the minor one as is the case for α/β-type SASPs. However, several HTH-motif proteins bind to the minor groove of DNA by the so-called “hinge helix.” Among these are the lactose repressor LacI (PDB ID code 1LBG), purine repressor PurR (PDB ID code 2PUA), and catabolite control protein A, CcpA (PDB ID code 12VV). In these proteins, the DNA-binding domain contains a HTH motif that interacts with DNA major groove, and residues of hinge helices bind deeply in the minor groove, prying it open.
Also, several other DNA binding proteins bind to the minor groove of DNA, which often results in change in conformation of the minor groove (8), namely its opening and greater accessibility (9). Among these are the high mobility group (HMG) proteins, HMG-D (PDB ID code 1QRV), lymphoid enhancer factor (LEF-1) (PDB ID code 2LEF), and the male sex determining factor SRY (PDB ID code 1J46).
Other major intramolecular interactions within α/β-type SASPs protomers are between the OE2 oxygen of Glu-23 in helix1 and the NH2 group of Arg-47 in helix2 (hydrogen bond distances in SASP1, SASP2, and SASP3 are 2.9, 2.8, and 3.1 Å, respectively). The OD1 oxygen and ND2 nitrogen of Asn-37 in helix2 are hydrogen bonded to the backbone nitrogen and carbonyl oxygen, respectively, of the conserved Gly-29 of the extended amino acid chain region. (Both distances in SASP1 are 3.2 Å.) Corresponding bonds are present in SASP2 and 3 (length varies from 3.0 to 3.3 Å) (Fig. 1 A and B). These two examples of intramolecular interactions allow for the maintenance of the proper orientation of helix1 and 2 with respect to one another and of the turn between them.
The Asn-37-Gly-29 interactions (10) play an important role in preserving the HTH DNA-binding conformation in these proteins, because both the main chain conformation of the turn and specific side chains are required for the DNA-binding function of the HTH motif. Consequently, even small changes in the interactions in this region of the protein, for example, replacement of the two well defined hydrogen bonds in the Asn-37-Gly-29 interaction with just one as in the N37D variant (11), disrupts the structure of the turn and compromises DNA binding (Fig. 1B).
α/β-Type SASPs can undergo several chemical modifications that greatly decrease DNA binding, one being the rapid deamidation of Asn-37 in solution (11). However, this reaction is greatly slowed when the protein is bound to DNA in vitro or in spores, presumably in part due to the a-helical structure of this region of the DNA-bound protein. In addition, access of water to Asn-37 is restricted in the complex, as indicated by its high occluded surface packing (OSP) parameter (see DNA Protection by α/β-Type SASP) (12). Another amino acid occluded in the complex is Met-16, which is buried in the middle of helix1 (Fig. 1B). Surface-exposed Met residues are generally more sensitive to oxidation than buried ones (13), resulting in this Met being sensitive to oxidation in free but not DNA-bound α/β-type SASPs (14). Its high OSP parameter (see below) again indicates that Met-16 is occluded in the complex. The structure of the complex also explains why DNA-bound α/β-type SASP is protected against the spore germination protease (GPR) that cleaves only the conserved Glu-19-Ile-20 bond in this protein (Fig. 1C) but only in free α/β-type SASPs (15, 16). Glu-19 and Ile-20 are in helix1, and the Glu-19-Ile-20 bond in the protein–DNA complex is again inaccessible to the ≈160-kDa GPR, as indicated by OSP calculations (see below).
Protein–Protein Interactions in the Complex.
Although free α/β-type SASPs are monomers in solution, the complex's structure exhibits many intermolecular protein–protein interactions, because SASP1 and 2 form a dimer (Fig. 1A), whereas SASP3 forms a dimer with a symmetry related molecule (SASP3′, Fig. 2A). The accessible surface areas (ASA) of SASP1 and the SASP1–2 dimer are 4997 Å2 and 8149 Å2, respectively, and thus dimer formation alone decreases the ASA/monomer by 18.5%. The buried surface area between SASP1 and SASP2 is 1845 Å2 17, whereas that between SASP1 and 3 (between dimers) is 795 Å2, reflecting the more limited interactions between dimers. Of the buried surface residues, 61% are nonpolar, 28% are polar, and 11% are charged, indicating that the majority of the protein's dimer interface involves hydrophobic interactions. Indeed, helices2 from each subunit are located at the dimer interface and contribute hydrophobic interactions involving Leu-4, Leu-5, Ile-6, Pro-7, Val-49, Leu-48, Leu-51, and Ala-52 (Fig. 2C). Again, these amino acids, or homologous ones, are conserved in α/β-type SASPs. Dimerization of α/β-type SASPs appears important for DNA-binding, because most of the dimer interface is nonpolar, and therefore dimer formation is energetically favored. The only major interactions between SASP1 from one dimer and SASP3 from another are hydrogen bonds between SASP1's OE2 oxygen of Glu-31 and the NH1 group of Arg-35, with NH1 group of Arg-35 and the OE2 oxygen of Glu-31 from SASP3, respectively (distances of 2.8 and 2.5 Å). All residues involved in these interactions are within the corresponding HTH loop regions (Fig. 1A).
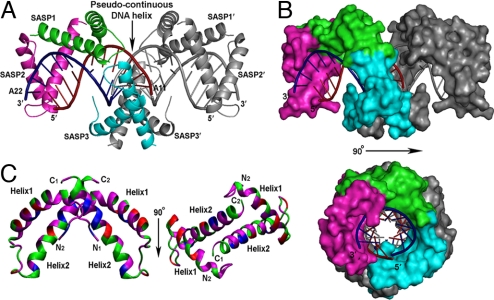
Fig. 2.
Molecular packing of the α/β-type SASP–DNA complex. (A) DNA helices consisting of 10 bp are stacked end-to-end in the crystal. The unpaired A11 on the dG strand is stabilized by stacking interactions with symmetry-related DNA to form a pseudocontinuous DNA helix. The unpaired A22 on the dC strand makes a Hoogsteen-like interaction with base pairs G1·C21. α/β-Type SASP protomers and the dG and dC DNA strands are colored as in Fig. 1A. The symmetry related SASP3′ and DNA are gray. (B) Space-filling surface diagram of the complex. Two perpendicular views are shown. (C) Ribbon diagram of the α/β-type SASP dimer structure. All helices from SASP1 and 2 are packed with nonpolar residues, forming a hydrophobic core between helices1 and 2. Helix2 from each subunit is located at the dimer interface contributing hydrophobic interactions. Green, polar residues; magenta, nonpolar residues; blue, basic residues; red, acidic residues. The N-and C-termini of each subunit are labeled N1/N2 and C1/C2, respectively. Two perpendicular views of the same dimer are shown.
DNA Conformation in the α/β-Type SASP–DNA Complex.
The analysis of DNA duplex in the complex exhibited the characteristic shape of A-type DNA with the base pair planes essentially parallel to each other and normal to the helix axis. Other parameters for the DNA duplex were also within the range typically observed for A-DNA (18), because the average value for twist was 31.5° (SI Table 2) and the sugar puckering were all in the C3′-endo conformation as in A-DNA (SI Table 3). One parameter of DNA in the complex deviated significantly from that found for A-DNA, in that the rise per base pair was essentially identical to that for B-DNA (SI Table 2). However, this latter observation is consistent with (i) the absence of any effect of α/β-type SASP binding on DNA length (19) and (ii) the rise per base pair seen for many short protein-bound A-like DNA segments, and some other A-like DNAs (20, 21). The width of the minor groove of the DNA in the complex is 16.7 ± 0.5 ŧ by direct P–P distances, similar to that in A-DNA fragments, whereas the major groove width (17.8 ± of 0.5 Å) is significantly greater than the average for A-type DNA (20). The majority of proteins that bind to DNA interact with the major groove because of the physical limitations of fitting secondary structure motifs such as an HTH into a narrow minor groove. However, in binding to DNA, the α/β-type SASPs in effect widen the minor groove to allow entry of this protein's helices, resulting in a transition to a DNA helix with features of both A- and B-DNA that we call “A-B-DNA” (SI Fig. 2). If binding had been in the major groove, the DNA might not have been distorted significantly and might have remained in a B-type conformation (21).
The DNA distortion in the α/β-type SASP–DNA complex is consistent with changes in the inclination angles of the base pairs along the global axis of the DNA compared with those in B-DNA. In particular, the inclination angles of base pairs G4:C18, G5:C17, G9:C13, and G10:C12 are significantly higher than in B-DNA (SI Fig. 6 and SI Table 2). In addition, the base pair twist is as small as 28°, and the roll is as large as 11° (SI Table 2), although the average value of base pair roll (5.7 ± 2.8°) is between mean values for B- and A-DNA (20). The large positive roll angle (11°) in the DNA in the complex makes the major groove narrower and deeper than in B-DNA, whereas the minor groove becomes shallower. This change in average roll associated with B- to A-DNA distortions of individual base pair steps generally shifts the positions of the DNA strands; in our case the agent causing the shifting is the binding of α/β-type SASPs.
The DNA structure in the complex is consistent with circular dichroism and Fourier transform infrared studies (22); cryoelectron microscopy also indicates a change in the pitch of α/β-type SASP-bound DNA to 3.2 nm (23). The structure of DNA in the complex is also consistent with the effects of α/β-type SASPs on DNA supercoiling (24), because the average number of basepairs per turn in the DNA in our structure is 11.5, similar to that in A-DNA, whereas this value is 10 in B-DNA 25 (SI Table 2).
Interaction of α/β-Type SASPs with DNA.
The ASA of α/β-type SASPs and the DNA are 4,997 Å2 and 4,409 Å2, respectively, and 35–40% of the protein and DNA surfaces are buried in the complex. This large buried surface area indicates that α/β-type SASPs bind to DNA via many specific interactions (Fig. 1D). Binding to DNA facilitates protein dimerization in the complex, and both dimers' subunits point their HTH regions in opposing directions along DNA minor groove and directly interact through residues of helix2. This directional binding facilitates optimal packing of the protein on DNA. Although we focus below on interactions of the SASP1–2 dimer with DNA, similar interactions are present for the SASP3–3′ dimer (Fig. 2A).
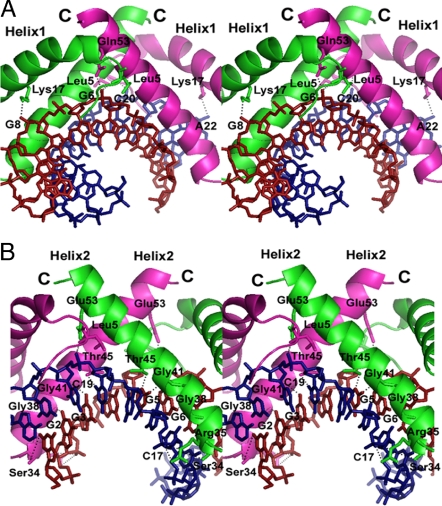
In the region of helix1 involved in DNA binding, each α/β-type SASP interacts with the DNA backbone on both edges of the DNA helix (Fig. 3A). In SASP1, the side chain NZ nitrogen of the conserved Lys-17 (Fig. 1C) forms a hydrogen bond with the phosphate O1P oxygen of base G8 (distance of 2.9 Å), and the backbone nitrogen of Leu-5 forms a hydrogen bond with the phosphate O1P oxygen of base G6 (distance of 3.1 Å). On the dC-DNA strand, the side chain NZ nitrogen of Lys-17 and the backbone nitrogen of Leu-5 from SASP2 form hydrogen bonds with the phosphate O1P oxygens of bases A22 and C20, respectively (distances of 3.7 and 3.3 Å). The interactions between the DNA backbone and Leu-5 and Lys-17 facilitate the arrangement of α/β-type SASPs on the DNA helix, and the majority of helix2 interacts closely with DNA minor groove (Fig. 3B). The side chain NH2 and NE nitrogen groups of Arg-35 from SASP1 form hydrogen bonds with the phosphate O1P oxygen of base C17 in a bifurcate interaction (distances of 3.0 and 3.7 Å, respectively). The corresponding distances of SASP2's Arg-35 to the phosphate O1P oxygen of base G3 are 4.8 and 4.3 Å, indicating that these interactions are weak or nonexistent. The NE2 group of Gln-53 from each protomer also forms hydrogen bonds with the phosphate O1P oxygens of bases G6 and C20 (distances of 3.8 Å and 3.9 Å), and the backbone nitrogen of Leu-5 from each protomer is hydrogen bonded with the same O1P oxygens (distances of 3.1 Å and 3.3 Å). The interactions between the DNA backbone with Arg-35 and Gln-53 of helix2 in SASP1 and 2 position helices2 on the opposite sides of the DNA minor groove (Fig. 1A). This arrangement fosters DNA interactions with helix1, and facilitates interactions between protein molecules in the dimer. Four conserved amino acids from each protomer (Ser-34, Gly-38, Gly-41, Thr-45) make hydrogen bonds to G:C base pairs, and are the only amino acids that directly interact with DNA bases. The OG oxygen of SASP1's Ser-34 is hydrogen bonded to the O4′ oxygen of the sugar on base C16 (distance of 3.4 Å), and the carbonyl oxygen of this amino acid is hydrogen bonded to the N2 nitrogen of base G7 (distance of 3.2 Å). The carbonyl oxygen of SASP2's Ser-34 is hydrogen bonded to the O4′ oxygen of the sugar on base G2 and the N2 nitrogen of base G1 (distances of 3.8 Å and 3.9 Å); the corresponding distances involving the OG oxygen of SASP2's Ser-34 are longer (4.2 and 4.8 Å). The OG1 oxygen of SASP1's Thr-45 is hydrogen bonded with the N2 nitrogen of base G5 (distance of 3.1 Å), and there is a similar interaction between the OG1 oxygen of SASP2's Thr-45 with the O2 oxygen of base C19 (distance of 2.6 Å). In addition, the carbonyl oxygens of SASP1's Gly-38 and Gly-41 form hydrogen bonds with the N2 nitrogens on bases G6 and G5 (distances of 3.0 and 3.4 Å). SASP2's same carbonyl oxygens also form hydrogen bonds with N2 nitrogens, but with those of bases G2 and G3 (distances of 3.3 and 3.4 Å) (Fig. 1D and 3B).
Fig. 3.
Stereo diagrams of the interaction between the α/β-type SASP dimer and DNA. (A) Interactions between helix1 and the DNA backbone on both edges of the DNA helix. (B) Interactions between helix2 and G:C base pairs and the DNA backbone. The color scheme is as in Fig. 1A. The DNA and amino acid side chains are shown as sticks. Hydrogen bonds are shown as dotted black lines.
It is notable that Gly-41 interacts directly with a DNA base, because changing this residue to Ala eliminates effects of α/β-type SASPs on DNA structure (26). There is not enough room for a methyl group on Gly-41 to be accommodated within the complex (Figs. 1A and 3B), and therefore the methyl group in Ala-41 would compromise the interaction of helix2 with DNA and this helix's ability to shift DNA structure from B to A-B. The severe effects of this small change in α/β-type SASP sequence highlight the importance of interactions between DNA and helix2 in changing DNA to an A-B structure.
Another α/β-type SASP modification that highlights the role of helix2 in determining DNA structure is the K46Q change (26) that also hinders the ability of α/β-type SASPs to alter DNA structure from B to A-B. As was the case for Gly-41, Lys-46 is also in the middle of helix2, but Lys-46 does not interact with DNA. However, the side chain of Gln-46 (modeled compared with that of Lys) clashes significantly with DNA. As a consequence the K46Q mutation will shift helix2 out of its ideal position for binding to and causing a conformational change in DNA.
The α/β-type SASPs orientation within the DNA minor groove is determined primarily by the orientation of each subunit's helix2, and this orientation is determined by the location of basic amino acids within this helix and helix1 interactions with DNA along with the HTH conformation. The location of these basic amino acids within the protein dimer has a pseudosymmetric relationship within the areas of helix2 involved in interactions with the DNA minor groove (Fig. 2C). Contacts that position these basic amino acids on DNA include the conserved Lys-17 and Arg-35 donating hydrogen bonds (distances of 3.0 ± 0.1 Å) to oxygen atoms of the DNA backbone; contacts with the DNA backbone are essentially the same for all protein subunits. It is notable that the interface between α/β-type SASPs and DNA contains no water molecules. Because α/β-type SASPs play the major role in resistance of B. subtilis spore DNA to desiccation and dry heat and protect DNA against dry heat in vitro, these proteins must bind to DNA even without water present.
Another property of DNA bound to α/β-type SASPs is the huge increase in DNA persistence length (19), suggesting that DNA in the complex is rigid. Such a property is likely a consequence of the many interactions of these proteins with DNA, and this binding also minimizes sequence-dependent DNA bending. The relative rigidity of α/β-type SASP-bound DNA also helps explain the minimal generation of cyclobutane dimers (CPDs) and 6,4-photoproducts (64PP) between adjacent pyrimidines in DNA upon UV irradiation in vitro and in spores (ref. 3 and below).
DNA Protection by α/β-Type SASPs.
The binding of α/β-type SASPs results in the energetic stabilization and protection of DNA, presumably the major reason this complex is formed in spores and the increased rigidity of its components due to intermolecular interactions as discussed above. Because of changes in structure of both the DNA and protein molecules upon complexation, the precise energetics of this binding process is intricate. The reasons for the structural changes in DNA in the complex from B-type to A-B-type could well have been related to DNA protection against UV radiation, because A-type DNA is more UV resistant than its other forms. This could be due to past evolutionary pressure, presumably on early Earth, selecting for the structural changes in DNA in the complex. It is, however, unclear whether this selection would continue in spores in their current natural environments, because their UV exposure is limited, and protection against DNA depurination may be a reason for the maintenance of this selection at present. All reasons for the change in α/β-type SASP structure in the complex from unstructured to highly helical are still not clear, but this protein's sole role here may well be only to assume an appropriate structure when binding to DNA minor groove to cause the change in the structure of the DNA that results in its protection. In addition, the α/β-type SASPs create a protective coat around DNA, arguably the most important component of bacterial spores, creating a physical barrier. The α/β-type SASP–DNA complex also has a number of specific properties that result in DNA protection against damage, as discussed in detail below.
The major function of α/β-type SASPs is to protect spore DNA from a variety of types of damage, including desiccation, heat, toxic chemicals and enzymes, and UV radiation. As noted earlier, interactions of α/β-type SASPs with DNA do not involve water molecules, and thus, especially considering the energetic stability of the complex (see above), these proteins remain tightly bound to DNA even in dry spores to preserve DNA and its structure and, therefore, provide resistance against desiccation damage to DNA. DNA exposure to dry and/or wet heat normally results in a cleavage of the glycosylic bond. However, α/β-type SASPs when bound to DNA greatly slow cleavage of the glycosylic bond between a deoxyribose and a base in DNA, in particular a purine, under both wet and dry heat conditions (27, 28). Not surprisingly, the α/β-type SASP–DNA complex has a different environment around the glycosylic bond than does B-DNA, including tighter packing due to the presence of the protein; the sugar–phosphate backbone in the complex is also occluded. In addition, the bond angle from the deoxyribose to the base is −156 ± 10° in the complex but −110 ± 50° in B-DNA (21). It is reasonable to assume that this change greatly reduces the rate of glycosylic bond cleavage. This behavior is consistent with observation that DNA conformation can significantly affect the rate of this reaction (29), but, unfortunately, there have been no direct studies of the rate of cleavage of the glycosylic bond in A-DNA. The energetic stability of the α/β-type SASP–DNA complex noted above should also stabilize the complex against glycosylic bond cleavage.
A number of the effects, or lack of effects, of α/β-type SASPs on the reactivity of DNA with genotoxic chemicals in spores and in vitro are also readily explained by the structure of the α/β-type SASP–DNA complex. Helix2 of α/β-type SASP binds not in the major groove of DNA as do most HTH DNA binding proteins, but rather in the minor groove. The imino/amino groups of guanine in DNA are in the minor groove and are well shielded by interaction with protein amino acid residues (Ser-34, Gly-38, Gly-41, and Thr-45) from helix2, thus explaining the strong protection afforded by α/β-type SASPs binding against DNA damage by toxic chemicals, such as nitrous acid and formaldehyde in spores and in vitro. However, oxygen O6 and nitrogen N7 of guanine residues, prime targets for alkylating agents, such as ethylene oxide and methylmethanesulfonic acid, are in the major groove of the complex and thus are potentially accessible to such reagents as in B-DNA, consistent with the lack of effect of α/β-type SASPs on DNA modification by these and other alkylating agents both in spores and in vitro.
Another factor involved in α/β-type SASPs' protection of DNA against chemical and enzymatic attack is restriction of access of such agents to susceptible bonds or groups in DNA. The α/β-type SASP–DNA complex assumes a near pentagonal shape (Fig. 2B) with each DNA molecule tightly surrounded by α/β-type SASP dimers, and this tight packing should prevent (i) access of many chemicals to the DNA and (ii) recognition of the protein-bound DNA as a substrate by enzymes that act on free DNA. Even small reactive chemicals must cross this protein coat to reach DNA in the complex, and during this process would encounter many reactive amino acids. It seems reasonable that much of these reactive chemicals will thus be neutralized before reaching DNA.
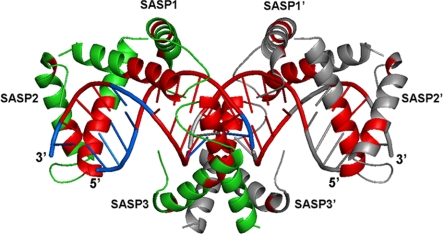
To quantify the accessibility of various regions of the α/β-type SASP–DNA complex, the atomic packing was analyzed. Studies of density within protein cores have implicated “packing efficiency” as an important determinant of stability (30). Consequently, the OSP for each amino acid and base in the complex was calculated (12) (Fig. 4 and SI Text) as a measure of the accessibility of solvent to moieties. Amino acids of α/β-type SASPs with high OSP values (≈0.5, compared with low values of ≈0.1) are arranged around the DNA minor groove, indicating that this region of the protein is tightly packed, in particular residues Ala-9, Glu-19–Ala-21 in helix1 and Ser-34–Thr-45 in helix2. The amino acids with low OSP values are at the N and C termini and in a few residues of the HTH turn (Figs. 1B and 4). The DNA bases showing high OSP values are G5–G10, C13–C16, and C19–C21. One example of shielding from enzymatic attack in high OSP regions is the Glu-19-Ile-20 bond that is cleaved by GPR only in free α/β-type SASPs (15, 31). The highly packed amino acids are localized at the interface between α/β-type SASPs and DNA and close to DNA minor groove that interacts with helix2 (Fig. 4). Individual OSP values clearly show a highly packed core of amino acids around the DNA. This complex can be stable in spores at temperatures >90°C (3, 27), and the complex's highly packed core is the logical cause of this extreme stability (30).
Fig. 4.
Ribbon representation of the α/β-type SASP–DNA complex indicating occluded regions with high OSP values. Amino acids with high OSP values (≈0.5; colored in red) are arranged around the minor groove of DNA, indicating that regions of SASP1, SASP2, and SASP3 that interact with DNA are tightly packed. The low OSP value (≈0.1) regions of α/β-type SASP and DNA are green and blue, respectively. The crystallographic symmetry related molecule of the complex, including SASP3′, SASP2′, and SASP1′, is gray.
Features of the complex's structure also suggest the mechanism of α/β-type SASPs' protection of DNA against UV radiation. These proteins contain few amino acids that absorb well at ≈260 nm and thus cannot shield DNA from UV. Consequently α/β-type SASPs must alter the effects of UV on DNA by modifying DNA structure. CPDs and 64PP formed between adjacent pyrimidines, the major UV photoproducts in B-DNA, are not generated in spores or an α/β-type SASP–DNA complex. Instead, the major UV photoproduct from these two targets is a thyminyl-thymine adduct termed the spore photoproduct (SP) that is repaired with high fidelity after spores germinate. A major factor determining DNA's UV photoreactivity and photochemistry is the ease with which adjacent base pairs adopt geometry necessary for a particular photoreaction, and B-DNA can readily adopt conformations favoring CPD and 64PP formation (32, 33). However, the UV photoreactivity of A-DNA is significantly lower because it is much more difficult for bases in A-DNA to adopt the geometry necessary for CPD and 64PP formation, mainly because of A-DNA's lower base pairs twist (32, 33).
The distances between atoms in adjacent C bases in our structure that could be involved in CPD or 64PP formation are 3.9–4.3 Å; the comparable distances between atoms in adjacent C bases in a GC containing B-DNA (PDB ID code 1GCC) (34) are 3.8–4.5 Å. However, B-DNA is significantly less rigid than our A-B-DNA, and DNA flexibility is essential to transiently adopt a conformation needed for CPD and 64PP formation. Presumably CPD and 64PP formation in rigid DNA would require shorter distances between relevant atoms than in B-DNA.
The DNA in our α/β-type SASP–DNA complex contained no T residues. To probe the formation of UV photoproducts between Ts in an α/β-type SASP–DNA complex, we modeled the complex with a 2-bp region in which G6:C16 and G7:C15 were changed to A6:T16 and A7:T15 (SI Text). The distance between carbons C7 of base T16 and C5 of base T15, the two atoms involved in covalent bond formation in SP, is 3.4 Å (SI Fig. 7), >1 Å less than in B-DNA (PDB ID code 2EFZ) (35). The distance between carbon C5 of base T15 to carbon C5 of base T16 and comparable distance between the C6 carbons in the model, all of which would be involved in CPD formation are 3.9 Å and 4.3 Å, respectively, and the distance between carbons C6 of base T16 to C4 of base T15 that would be involved in 64PP formation, is 4.9 Å. The significantly shorter distance between C7 of base T16 and C5 of base T15 in our modeled structure relative to that in B-DNA indicates that SP should be formed much more readily in our complex than in B-DNA and more readily than a CPD or 64PP. This difference in the relative ease of formation of these photoproducts is further increased by the rigidity of the A-B-DNA in the complex.
Conclusions
Additional factors not accounted for in the structure, primarily because the environment of the complex in spores is different from in the crystal, also have a somewhat protective effect on DNA in spores but to a much lesser degree. Such factors include a low water content in the spore core, presumably resulting in severely restricted macromolecular movement (36), although the effect of the low water content on vibrational motion within core molecules is not clear. Perhaps even more importantly, in addition to α/β-type SASPs, the DNA in spores is associated with other primarily small chemical molecules such as Ca2+-dipicolinate (pyridine-2,6-dicarboxylate) in spores of all Bacillus species. However, how Ca2+-dipicolinate interacts with DNA or the α/β-type SASP–DNA complex is also not known.
Regardless of these shortcomings the structure of the α/β-type SASP–DNA complex that we have determined provides the major insights into the reasons for properties of DNA in bacterial spores that are important for spore resistance and longevity. In addition, this study also illustrates the associated molecular mechanisms responsible for such spore DNA protection and suggests evolutionary insights for their development dating as far in time as the early Earth. These insights should lead to advances in dealing with spores in general, and their extreme resistance, in applied situations.
Materials and Methods
Complex Production and Structure Determination.
Protein expression and purification, production of oligo(dG)·oligo(dC), complex formation and crystallization, and x-ray diffraction data collection have been described in ref. 6. For details and the diffraction data processing statistics, see SI Text.
The α/β-type SASP–DNA complex's crystal structure was determined by the MAD method, using the diffraction dataset from the SeMet–protein–DNA complex. The locations of Se atoms were obtained with the SnB program (37) at 3.5 Å resolution; density modification and initial model building were performed with RESOLVE (38). The initial model of the complex was refined and manually rebuilt with CNS (39) and COOT (40). The refined model of the α/β-type SASP–DNA complex gave Rfactor and Rfree values of 25.7 and 28.3%, respectively. The qualities of the final and intermediate models were analyzed with PROCHECK (41).
Modeling of TT Containing Complex and Packing Calculations.
To obtain information on the probability of formation of various UV photoproducts in DNA in the α/β-type SASP–DNA complex, a complex was modeled with two adjacent AT base pairs substituted for G:C base pairs, using COOT. The modeled structure was refined energetically with CNS. Calculations of the OSP values for each residue in the complex were performed with OSP software (12).
Supplementary Material
ACKNOWLEDGMENTS.
We thank B. de Groot, D. C. Reason, and D.J. Rigden for suggestions and the staff at Advanced Light Source beamlines 8.2.1 and 8.2.2. This work was supported in part by National Institutes of Health Grant GM19698 (to P.S. and M.J.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.G.B. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2Z3X).
This article contains supporting information online at www.pnas.org/cgi/content/full/0708244105/DC1.
Distances and other values that involve multiple measurements are reported as average values ± standard deviation.
References
- 1.Setlow P. I will survive: DNA protection in bacterial spores. Trends Microbiol. 2007;15:172–180. doi: 10.1016/j.tim.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Setlow P. Spores of Bacillus subtilis: Their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol. 2006;101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- 3.Nicholson WL, Schuerger AC, Setlow P. The solar UV environment and bacterial spore UV resistance: Considerations for Earth-to-Mars transport by natural processes and human spaceflight. Mutat Res. 2005;571:249–264. doi: 10.1016/j.mrfmmm.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 4.Kosman J, Setlow P. Effects of carboxy-terminal modifications and pH on binding of a Bacillus subtilis small, acid-soluble spore protein to DNA. J Bacteriol. 2003;185:6095–6103. doi: 10.1128/JB.185.20.6095-6103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayes CS, Alarcon-Hernandez E, Setlow P. N-terminal amino acid residues mediate protein–protein interactions between DNA-bound alpha/beta-type small, acid-soluble spore proteins from Bacillus species. J Biol Chem. 2001;276:2267–2275. doi: 10.1074/jbc.M007858200. [DOI] [PubMed] [Google Scholar]
- 6.Bumbaca D, Kosman J, Setlow P, Henderson RK, Jedrzejas MJ. Crystallization and preliminary X-ray analysis of the complex between a Bacillus subtilis alpha/beta-type small acid-soluble spore protein and DNA. Acta Crystallogr F Struct Biol Cryst Commun. 2007;63:503–506. doi: 10.1107/S1744309107022750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orengo CA, et al. CATH–a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 8.Bewley CA, Gronenborn AM, Clore GM. Minor groove-binding architectural proteins: Structure, function, and DNA recognition. Annu Rev Biophys Biomol Struct. 1998;27:105–131. doi: 10.1146/annurev.biophys.27.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zacharias M. Minor groove deformability of DNA: A molecular dynamics free energy simulation study. Biophys J. 2006;91:882–891. doi: 10.1529/biophysj.106.083816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Efimov AV. A novel super-secondary structure of proteins and the relation between the structure and the amino acid sequence. FEBS Lett. 1984;166:33–38. doi: 10.1016/0014-5793(84)80039-3. [DOI] [PubMed] [Google Scholar]
- 11.Hayes CS, Setlow P. Analysis of deamidation of small, acid-soluble spore proteins from Bacillus subtilis in vitro and in vivo. J Bacteriol. 1997;179:6020–6027. doi: 10.1128/jb.179.19.6020-6027.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pattabiraman N, Ward KB, Fleming PJ. Occluded molecular surface: analysis of protein packing. J Mol Recognit. 1995;8:334–344. doi: 10.1002/jmr.300080603. [DOI] [PubMed] [Google Scholar]
- 13.Keck RG. The use of t-butyl hydroperoxide as a probe for methionine oxidation in proteins. Anal Biochem. 1996;236:56–62. doi: 10.1006/abio.1996.0131. [DOI] [PubMed] [Google Scholar]
- 14.Hayes CS, Illades-Aguiar B, Casillas-Martinez L, Setlow P. In vitro and in vivo oxidation of methionine residues in small, acid-soluble spore proteins from Bacillus species. J Bacteriol. 1998;180:2694–2700. doi: 10.1128/jb.180.10.2694-2700.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ponnuraj K, Rowland S, Nessi C, Setlow P, Jedrzejas MJ. Crystal structure of a novel germination protease from spores of Bacillus megaterium: Structural arrangement and zymogen activation. J Mol Biol. 2000;300:1–10. doi: 10.1006/jmbi.2000.3849. [DOI] [PubMed] [Google Scholar]
- 16.Jedrzejas MJ. Three-dimensional structure and molecular mechanism of novel enzymes of spore-forming bacteria. Med Sci Monit. 2002;8:183–190. [PubMed] [Google Scholar]
- 17.Liddington RC. Structural basis of protein–protein interactions. Methods Mol Biol. 2004;261:3–14. doi: 10.1385/1-59259-762-9:003. [DOI] [PubMed] [Google Scholar]
- 18.Kennard O, Hunter WN. Oligonucleotide structure: a decade of results from single crystal X-ray diffraction studies. Q Rev Biophys. 1989;22:327–379. doi: 10.1017/s0033583500002997. [DOI] [PubMed] [Google Scholar]
- 19.Griffith J, Makhov A, Santiago-Lara L, Setlow P. Electron microscopic studies of the interaction between a Bacillus subtilis alpha/beta-type small, acid-soluble spore protein with DNA: Protein binding is cooperative, stiffens the DNA, induces negative supercoiling. Proc Natl Acad Sci USA. 1994;91:8224–8228. doi: 10.1073/pnas.91.17.8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu XJ, Shakked Z, Olson WK. A-form conformational motifs in ligand-bound DNA structures. J Mol Biol. 2000;300:819–840. doi: 10.1006/jmbi.2000.3690. [DOI] [PubMed] [Google Scholar]
- 21.Jones S, van Heyningen P, Berman HM, Thornton JM. Protein–DNA interactions: A structural analysis. J Mol Biol. 1999;287:877–896. doi: 10.1006/jmbi.1999.2659. [DOI] [PubMed] [Google Scholar]
- 22.Mohr SC, Sokolov NV, He CM, Setlow P. Binding of small acid-soluble spore proteins from Bacillus subtilis changes the conformation of DNA from B to A. Proc Natl Acad Sci USA. 1991;88:77–81. doi: 10.1073/pnas.88.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frenkiel-Krispin D, et al. Structure of the DNA-SspC complex: implications for DNA packaging, protection, and repair in bacterial spores. J Bacteriol. 2004;186:3525–3530. doi: 10.1128/JB.186.11.3525-3530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholson WL, Setlow B, Setlow P. Binding of DNA in vitro by a small, acid-soluble spore protein from Bacillus subtilis and the effect of this binding on DNA topology. J Bacteriol. 1990;172:6900–6906. doi: 10.1128/jb.172.12.6900-6906.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saenger W. Principles of Nucleic Acid Structure 220–41. New York: Springer; 1984. [Google Scholar]
- 26.Tovar-Rojo F, Setlow P. Effects of mutant small, acid-soluble spore proteins from Bacillus subtilis on DNA in vivo and in vitro. J Bacteriol. 1991;173:4827–4835. doi: 10.1128/jb.173.15.4827-4835.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fairhead H, Setlow B, Setlow P. Prevention of DNA damage in spores and in vitro by small, acid-soluble proteins from Bacillus species. J Bacteriol. 1993;175:1367–1374. doi: 10.1128/jb.175.5.1367-1374.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Setlow B, Setlow P. Small, acid-soluble proteins bound to DNA protect Bacillus subtilis spores from killing by dry heat. Appl Environ Microbiol. 1995;61:2787–2790. doi: 10.1128/aem.61.7.2787-2790.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nejedly K, Chladkova J, Vorlickov M, Hrabcova I, Kypr J. Mapping the B-A conformational transition along plasmid DNA. Nucleic Acids Res. 2005;33:e5. doi: 10.1093/nar/gni008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards FM, Lim WA. An analysis of packing in the protein folding problem. Q Rev Biophys. 1993;26:423–498. doi: 10.1017/s0033583500002845. [DOI] [PubMed] [Google Scholar]
- 31.Setlow P. Spore germination. Curr Opin Microbiol. 2003;6:550–556. doi: 10.1016/j.mib.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Becker MM, Wang Z. B-A transitions within a 5 S ribosomal RNA gene are highly sequence-specific. J Biol Chem. 1989;264:4163–4167. [PubMed] [Google Scholar]
- 33.Kundu LM, Linne U, Marahiel M, Carell T. RNA is more UV resistant than DNA: The formation of UV-induced DNA lesions is strongly sequence and conformation dependent. Chemistry. 2004;10:5697–5705. doi: 10.1002/chem.200305731. [DOI] [PubMed] [Google Scholar]
- 34.Heinemann U, Alings C, Bansal M. Double helix conformation, groove dimensions and ligand binding potential of a G/C stretch in B-DNA. EMBO J. 1992;11:1931–1939. doi: 10.1002/j.1460-2075.1992.tb05246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huth JR, et al. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat Struct Biol. 1997;4:657–665. doi: 10.1038/nsb0897-657. [DOI] [PubMed] [Google Scholar]
- 36.Cowan AE, Koppel DE, Setlow B, Setlow P. A soluble protein is immobile in dormant spores of Bacillus subtilis but is mobile in germinated spores: Implications for spore dormancy. Proc Natl Acad Sci USA. 2003;100:4209–4214. doi: 10.1073/pnas.0636762100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller R, Gallo SM. SnB: Crystal structure determination via shake-and-bake. J Appl Cryst. 1994;27:613–621. [Google Scholar]
- 38.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Laskowski RA, Luscombe NM, Swindells MB, Thornton JM. Protein clefts in molecular recognition and function. Protein Sci. 1996;5:2438–2452. doi: 10.1002/pro.5560051206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.