Abstract
We describe the molecular structure of the collagen fibril and how it affects collagen proteolysis or “collagenolysis.” The fibril-forming collagens are major components of all mammalian connective tissues, providing the structural and organizational framework for skin, blood vessels, bone, tendon, and other tissues. The triple helix of the collagen molecule is resistant to most proteinases, and the matrix metalloproteinases that do proteolyze collagen are affected by the architecture of collagen fibrils, which are notably more resistant to collagenolysis than lone collagen monomers. Until now, there has been no molecular explanation for this. Full or limited proteolysis of the collagen fibril is known to be a key process in normal growth, development, repair, and cell differentiation, and in cancerous tumor progression and heart disease. Peptide fragments generated by collagenolysis, and the conformation of exposed sites on the fibril as a result of limited proteolysis, regulate these processes and that of cellular attachment, but it is not known how or why. Using computational and molecular visualization methods, we found that the arrangement of collagen monomers in the fibril (its architecture) protects areas vulnerable to collagenolysis and strictly governs the process. This in turn affects the accessibility of a cell interaction site located near the cleavage region. Our observations suggest that the C-terminal telopeptide must be proteolyzed before collagenase can gain access to the cleavage site. Collagenase then binds to the substrate's “interaction domain,” which facilitates the triple-helix unwinding/dissociation function of the enzyme before collagenolysis.
Keywords: cell attachment, collagenolysis, digestion, integrin, matrix metalloproteinase
Type I collagen is the most significant and abundant fibrillar collagen involved in promoting the activation of cell membrane proteases for collagenolysis (1), which in turn leads to cell migration and/or adhesion. Its structure is complex. Monomeric type I collagen consists of two α1 chains and one α2 chain intertwined into a triple helix (2–5). Each triple helix is staggered from its molecular neighbor by a multiple of 67 nm in the direction of the helix, and laterally, the helices are arranged quasi-hexagonally (6) with respect to each other within the fibril. Every five molecular packing neighbors are found in a right-handed microfibril, a higher-order supramolecular structure that resembles a helix and that interdigitates with neighboring microfibrils to form the basis of the fibril (7) (see Fig. 1). At neutral pH, the breakdown of type I collagen is initiated by interstitial collagenases belonging to the matrix metalloproteinase (MMP) family. They cleave the α chains of collagen at a specific Gly-Ile/Leu bond (8), yielding three-quarter and one-quarter length fragments. How the MMPs select this location in preference to other sites with the same, or a similar, amino acid sequence is not clear. Furthermore, although the collagen triple helix is thermally unstable at physiological temperatures (9), the relevance of a prior disassociation of the three peptide chains from the triple helix (see Fig. 2) to binding and catalysis (10) has not been assessed in the context of the fibrillar surface structure (see Fig. 3).
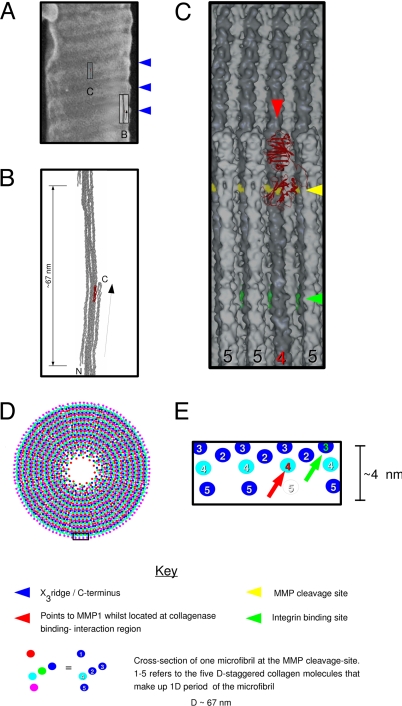
Fig. 1.
Molecular substructure of the D-periodic collagen fibril. (A) A field emission gun–scanning electron microscope image of nonstained, lyophilized fibrils, adapted from ref. 19. The blue arrows mark three successive “X3” ridges, which correspond to the C terminus of the collagen molecule (19). The tilted surface of the fibril surface and the location of the bulging X3 ridge are similar to that of the microfibril's tilt and bulging C terminus (B). To scale, Insets B and C correspond to elements shown in B and C (below). (B) Single D-period of a microfibril. The C terminus (folded structure marked with a “C”) points toward the outside of the fibril. The N terminus is marked with an “N.” (C) Expansion of box in A. Surface accessibility of two fibril–ligand interaction sites. The surface view shows a section of the fibril surface where one C-telopeptide, and its connecting triple helix, has been removed, allowing access for MMP1 to the B (α2) peptide chain (red arrow). The cleavage site is represented as a yellow band, whereas the rest of monomer 4 is dark gray. Although the cleavage site is partly solvent-accessible, MMP1 (red) is unable to approach it while the C-telopeptide remains intact because of steric hindrance. A central integrin binding site (S. Sweeney and J. San Antonio, personal communication, and ref. 29) is illustrated in green. The B (α2) chain is accessible within the 2- to 3-nm-deep solvent pit on the solvent surface (see C), but neither of the α1 chains are, unless further proteolysis were to fully expose monomer 3 (see D). This may explain the poor binding of integrin to intact fibrils (34). (D) Fibril model based on the parameters of refs. 7 and 17. See key for identification of each segment by color. (E) Schematic of a section of the fibril surface shown in C and outlined in the small box at the bottom of D. The red arrow indicates the location of the cleavage site shown in C. The green arrow points to the same monomer 3 as C. Monomer 3 carries the ligand attachment sites needed for cell surface interactions [an integrin binding site (S. Sweeney and J. San Antonio, personal communication, and ref. 29)]. Strong cell surface interaction may require a prior, limited proteolysis of the fibril (25, 26), because the site is not otherwise fully accessible without the removal of monomers 5 (clear) and 4 (red number).
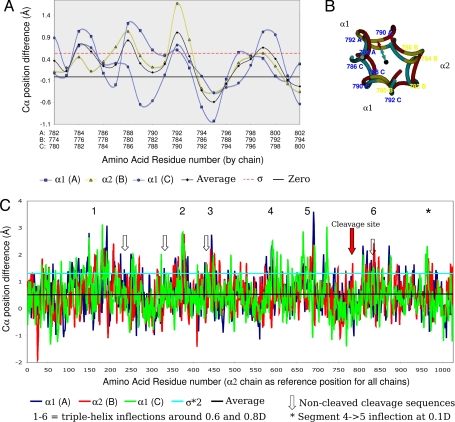
Fig. 2.
Disassociation of peptide chains: difference between the relaxed and stringent models of the collagen triple helix. Sequence numbering includes the N-telopeptide. A and C show the difference of the “from helix center” distances, a measure of triple-helix dissociation of the peptides. The magnitude of dissociation of the three peptide chains, A (α1), B (α2), and C (α1), are shown, along with the average, standard deviation (σ), two times standard deviation (2*σ), or zero, as indicated. (A) The cleavage site region (flanked by 10-aa residues N- and C-terminal). (B) End-on view of central section of cleavage site region. The black dot represents the triple-helix center, the cyan line is the radius of stringent model at 791A Cα, and the red line is the radius of the relaxed model 791A Cα. The difference between the two radiuses is used as the measure of native collagen's triple-helix disassociation (shown as graphs in A and C). (C) As A, except the entire triple helix.
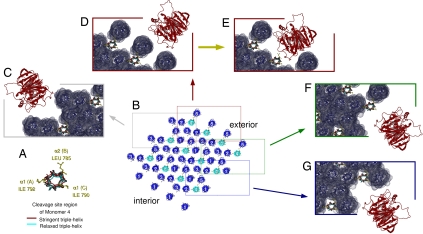
Fig. 3.
Molecular accessibility of the collagenase cleavage site at the fibril surface. (A) End-on view of the triple helix surrounding the cleavage site on monomer 4 (both stringent and relaxed models are shown). Monomer 4 is shown in the same orientation in all figure elements. (B) Lateral packing of collagen molecules in the region of the collagenase cleavage site (see Fig. 1E). The 3 × 4 microfibrils have been related by their crystal symmetry to give a representation of the fibril surface. It is apparent that there is no possibility of MMP1 interaction with monomer 4 from the N-terminal side of the fibril (monomer 1) (bottom left corner) because it is buried within the fibril interior (see also Fig. 1D). Four areas of possible collagenase–collagen fibril interaction have been marked with boxes, which are shown in greater detail in elements C–G. (C–G) Solvent-accessible surface views of the boxed regions of B and MMP1 attempting to dock with each area of the fibril. Monomers 1–3 and 5 are blue, and monomer 4 is as in A. MMP1 is red. MMP1 has been located as close to the monomer 4 cleavage site as possible, while avoiding steric clashes. For the areas represented in C, F, and G, MMP1 can come no closer than 15 Å from the cleavage site. (E) Prior removal of the C-terminal telopeptide allows access to the proteolytically vulnerable B (α2) chain of the cleavage site. The change in relative accessibility, by removal of the C terminus, is shown in the difference between E and F (marked by a yellow arrow). (F) Access to the less proteolytically vulnerable C (α1) chain would also become possible after the removal of one microfibril or at least the C-terminal fragment from one monomer (see also Fig. 1).
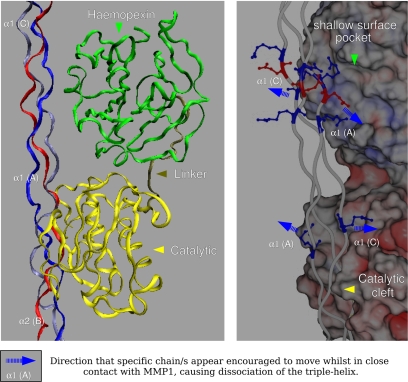
MMP1, the first member of the family to be characterized, consists of three domains required for cleavage site recognition and hydrolysis of the native triple helix: the catalytic domain, the substrate recognition (hemopexin-like) C-terminal domain, and a flexible, proline-rich hinge region that links the catalytic and hemopexin-like domains (see Fig. 4). Both the noncatalytic domains (hemopexin and linker) are involved in the binding and possibly the unwinding of triple-helical collagen (1, 11). The active site of the enzyme is a groove running across the surface of the catalytic domain. Structural studies of MMP1 indicate that the substrate-binding groove of the active site is only 5 Å wide and, therefore, unable to accommodate the intact collagen triple helix (≈3,000 Å in length and ≈15 Å in diameter) (12). Consequently, MMP1 is proposed to first bind to, and then locally unwind, the triple helix so that each peptide may fit into the active site binding groove, before hydrolyzing the peptide bonds of each chain in succession (12). This implicitly assumes that the binding/unwinding site on the substrate need not be limited to the cleavage sequence, but includes the surrounding residues of the triple helix, for one or more turns of the helix. This is implied in a recent study suggesting the direct, and perhaps simultaneous, involvement of MMP1's C-terminal hemopexin domain in unwinding the triple helix and in preferential binding to the α2 chain (12). Alternatively, MMP1 may simply take advantage of (thermal) fluctuations in helix structure to proteolyze it when it is rendered more vulnerable (10), i.e., when one or more of the peptide chains becomes more dissociated from the center of the triple helix, although this is still, in effect, triple-helix unwinding if collagenase binding assists in this process.
Fig. 4.
MMP1 at the collagenase-interaction domain (CID). The collagen B (α2) peptide chain is facing, and inside, the catalytic cleft while the two α1 chains interact with residues around, and in, the positively charged shallow surface pocket between modules 3 and 4 of the hemopexin domain (20). The relatively disassociated state of the triple helix represented here is the naturally occurring condition at approximately room temperature in situ (7). (Left) Domain organization of MMP1 while in contact with the fibrillar substrate. (Right) Expanded view rotated ≈30° from A, showing the solvent-accessible surface of MMP1, colored according to relative charge (spectrum toward red is acidic and toward blue is basic). The blue dashed arrows indicate the direction in which attraction/repulsion forces may operate to “pull” the two α1 chains in opposite directions, causing further dissociation on the triple helix at the cleavage site.
Previously, enzyme–collagen binding mechanisms have been proposed along the lines of the “sandwich model,” in which the C-terminal hemopexin domain folds over the catalytic domain thereby “trapping” the collagen molecule between the N- and C-terminal domains of the enzyme (13, 14). However, such models arbitrarily assume that the cleavage region of the collagen molecule is as readily accessible in the fibrillar form as it is in a single isolated collagen molecule. The sandwich model would require that the triple-helix region, carrying the cleavage site sequence, juts out of the fibril surface to allow it to be surrounded by the N- and C-terminal domains of MMP1. It is more logical to assume that only selected parts of the triple helix will be accessible from the surface of the fibril, be it for enzymatic degradation, or the location of sites suitable for cellular interaction (see Figs. 1, 3, and 4).
We have used MMP1 as a model that may be applicable for all MMPs with collagenolytic activity, membrane associated or not, to analyze collagenase activity within the fibrillar context. The basis of our fibril model is the structure of native in situ collagen in the rat tail tendon (7, 15, 16), its microfibrillar arrangement (7), observations of collagen packing in fibrils (7, 16–18), and electron microscopy observations of the fibril surface (18, 19). To our knowledge, no such study has been previously attempted and is only possible now because of the recent availability of in situ molecular resolution data on the conformations of the collagen molecule and microfibril.
Results and Discussion
Molecular Accessibility at the MMP1–Collagen Cleavage Site.
We present the structure of the collagen cleavage site within its native fibrillar environment (Figs. 1–4). The solvent-accessible surface of the fibril, which is a highly ordered liquid crystal (17), was calculated to assess the relative molecular accessibility of the collagenase cleavage site located on monomer 4 of the microfibril (Figs. 1 and 3). The extent of peptide chain disassociation from the center of the triple helix (which indicates vulnerability to proteolytic attack) was measured in the cleavage site region (see Materials and Methods and Figs. 2 and 4). This allows the viability and biological relevance of the “α2 chain first” hypothesis of collagen cleavage, to be assessed within the natural, fibrillar context. This context was established by attempting to fit a single MMP1 molecule at the cleavage site position in each of the representative regions of the intact, and/or partially proteolyzed fibril surface (Fig. 3). The domains of the MMP1 molecule were rearranged so that previous observations (12, 20, 21) were reflected in the relative positions of the catalytic and hemopexin domains (see Materials and Methods and Fig. 4). Even in this configuration, which slims down its profile, the MMP1 molecule can only fit within a few specific spaces on the fibril surface. However, these surfaces are obscured by the C-telopeptide, necessitating its removal before collagenolysis can occur.
Prior Peptide Dissociation May Aid Proteolysis but Does Not Determine Cleavage Site Location.
The extent of peptide-chain disassociation was estimated by measuring the difference between the “stringent” and “relaxed” states of in situ collagen structure [Research Collaboratory for Structural Bioinformatics (RCSB) accession codes 1Y0F and 1YGV] (see Materials and Methods). The stringent model represents the tighter, ideal form of the triple helix, whereas the relaxed is the energy minimized, refined structure determined by x-ray diffraction (5, 7).
At room temperature (7), the triple helix is disassociated in the region of the α2 chain cleavage site centered at the Gly784-Leu785 bond [see supporting information (SI) Fig. 5 for clarification of sequence numbering]. The average extent of disassociation, above a standard deviation of 0.5 Å, for the cleavage site of the B (α2) chain, was calculated to be ≈1.1 Å. A significant but lesser extent of disassociation from the triple-helix center was observed around the Gly791-Ile792 bond region of the α1 chains, i.e., at the α1 cleavage site. Furthermore, the triple helix was found to be somewhat dissociated in the expanded region surrounding the cleavage site. Given the helix unwinding/peptide dissociation requirement of MMP1 catalysis, we suggest that the 774–801 (α2) and 782–809 (α1) sequences encompass a wider, binding-initiation region of the collagen molecule. This includes residues before and after the cleavage sites of the three α-peptides and covers approximately a single turn of the triple helix (Figs. 2 and 4).
Although there is no significant difference in the magnitude of triple-helix disassociation of the three peptide chains over the whole of the proposed enzyme interaction region, the α2 chain is more disassociated than the α1 chains at the actual cleavage position. The modest difference in relative melting temperatures of the chains in the region of the cleavage site may also indicate the greater proteolytic vulnerability of the B (α2) peptide compared with that of the two α1 peptide chains. These melting temperatures are 37.7 and 35.2°C for the α1 and α2 chains, respectively, when the relative stabilization of their sequence contexts is considered (5).
There are, however, regions other than the cleavage site where significantly greater disassociation occurs (Fig. 2C). These regions may even include the cleavage amino acid sequence in one or two of the peptides, but at which the MMPs do not cleave. The extent of peptide chain disassociation in these regions is up to 3.6 Å, whereas the average free energy difference between the relaxed and stringent triple-helical states (a measure of the relative stability of the triple helix) is −1.9 kJ/mol per amino acid residue. The free energy difference for the region represented by 774AGPPGPPGPQGLLGAPGFLGLPGSRGER801 (α2), i.e., the cleavage site region, is comparable at −1.8 kJ/mol per amino acid residue. The most disassociated regions are found at locations of relatively sharp inflection of the triple helix (marked 1–6 and * in Fig. 2C), six of which describe two cavities within the packing matrix of the collagen fibril at ≈0.6 and ≈0.8D. Although it should be expected that the peptide chains be more disassociated from the triple helix when the triple helix is bent, this is not the case at the cleavage site region where the triple helix is relatively straight, but still significantly disassociated.
Thus, collagenase substrate recognition and binding is not solely determined by the relative stability of the triple helix (transition between helical forms), because these calculations indicate that disassociated regions are relatively stable regardless of the actual extent of triple-helix disassociation. Similarly, binding is not solely dependent on the extent of helical disassociation because there are several regions of significant disassociation, equivalent to, or significantly greater than that found at the cleavage site. Also, the process is not initiated solely by the recognition of the tripeptide cleavage sequence at the three-quarter site, because there are other cleavage sequences that are not proteolyzed, even if they are more disassociated than at the cleavage site (see label 6 of Fig. 2C). It follows that it must be a combination of sequence and structural factors that determine the site of cleavage (22). In terms of sequence factors, the cleavage site is the only section of the triple helix where all three peptides contain the cleavage amino acid sequence. In addition, the cleavage site is unique compared with the other nonproteolysed cleavage sequences by the presence of the sequence “RGER” C-terminal to the site of proteolysis (the first R is residue 805 on α1 and 798 on α2). In our model of MMP1–fibril interaction, the RGER sequences of the cleavage site region are level with, and appear to interact with, the hemopexin domain (Fig. 4B). Finally, the most disassociated chain at the cleavage site, the α2 peptide, points toward the outside of the fibril. However, the entire cleavage site region is located within a narrow, solvent-accessible cleft, and the α2 chain is protected by the C-telopeptide, both features greatly restricting molecular access to MMP1 (Figs. 1 and 3).
At the Fibril Surface, the Narrow Cleavage Site Environment Affects Collagenolysis.
The protected location of the cleavage site within its narrow cleft at the fibril surface may be the most significant constraint on MMP1 catalyzed collagenolysis. It may explain the resistance of fibrillar collagen to collagenolysis relative to that of the monomer (22), providing protection from premature digestion. Although the cleavage site (located on monomer 4 of each D-period) is partly solvent accessible, it is not molecularly accessible to collagenase (Figs. 1 and 3). The proximity of neighboring microfibrils and, in particular the C-terminal telopeptide, creates a narrow, solvent-accessible cleft at the fibril surface, too small to accommodate either the catalytic or hemopexin domains of MMP1. However, studies on stromelysin-1 (MMP3) show that telopeptide cleavage appears to be a critical early event in fibril depolymerization and may be the initiating event (23, 24). Our observations indicate that C-telopeptide removal via a telopeptidase or by MMP1 itself (23, 25, 26) would significantly widen the entrance to the surface-accessible cleft (Fig. 3E) and directly expose the B (α2) peptide at the cleavage site. Subsequent removal of what remains of the microfibril would begin the process of digesting the outer fibril monomer layer (SI Fig. 6 E and F), because enzyme accessibility would be greatly improved.
This may represent a design feature of the fibril. For instance, when the fibril suffers damage in exercise-related or traumatic injury, changes in fibril structure, such as breakage of cross-linkages at the C terminus, may expose the B (α2) chain, leading to an initial cleavage event (Fig. 3 and SI Fig. 6). After the first peptide chain is cleaved, the remaining two peptides would have a greater freedom of movement, perhaps allowing them to be twisted to face the catalytic domain so that all three are processed. The cleaved collagen monomer would then be further digested by the gelatinases (26), assisted by collagen's tendency to disassociate at physiological temperatures (9) after the first cleavage by collagenase (27). Collagen molecules distal to the original proteolysis would become accessible and allow the larger damaged area to be removed (SI Fig. 6).
MMP1-Catalyzed Collagenolysis May Resemble a Molecular Ratchet Motor.
Removal of the C-terminal end of the initially cleaved molecule would expose the most proteolytically vulnerable peptide chain (α2) of its packing neighbor in the next D-period (SI Fig. 6). This initial cleavage event would also expose the C (α2) peptide of the neighboring collagen molecule in the same D-period, and the process of fibril depolymerization would gradually allow access to sites N-terminal to the initial cleavage. However, because of the greater vulnerability and accessibility of the (α2) peptide chain, C-terminal to the initial cleavage, it seems reasonable to suggest that the process of collagenolysis would proceed more rapidly in this (preferred) direction (N- to C-terminal), leaving behind a groove as a single microfibrillar track is proteolyzed. The process may be viewed as analogous to a snowplow traveling along a groove that it makes as it moves forward. This description is consistent with the Brownian Ratchet (burnt bridge) model (28). This model is based on the observation that there appears to be a preferred direction of MMP1 diffusion on fibrillar collagen, and the hypothesis that, in “changing tracks” (i.e., lateral diffusion), would proceed significantly more slowly. In the reaction mechanism proposed here, when collagenolysis moves laterally to neighboring microfibrils (the track change), it must do so by the “slow” reaction route via the C (α1) chain, and thus only moves in the C- to N-terminal direction very slowly (SI Figs. 6 and 7). One should note, however, that we describe a process of collagenolysis that resembles a molecular motor (diffusion), when in fact it may simply be that fibril architecture dictates the order in which random fibril MMP1 interactions lead to collagen digestion within the fibril.
Significance of Fibril Structure in Extracellular Matrix (ECM) and Tissue Cell Interactions.
The process described is likely to be overly simplified (see SI Fig. 7 for schematic). Interactions between the fibril and cell membrane-associated collagenases may be significantly more complicated: they may be accelerated, may pause, or stop altogether because of conformational changes induced by cell attachment to interaction sites exposed during the initial digestion of the fibril (Fig. 1 C and E). For instance, strain on the tight packing arrangement of collagen monomers within the fibril, induced through fibril bending, may expose the vulnerable sections of the MMP interaction domain to proteolysis at sites distal to any actual cell attachment. However, cellular attachment to the cell interaction domain (S. Sweeney and J. San Antonio, personal communication, and ref. 29) may simultaneously inhibit further proteolysis within that section of the fibril because of the cleavage sites of the monomers being obscured by ligand–protein interactions. That being said, it is clear that the conformation of the intact fibril surface, as presented here, covers or protects the major cell interaction sites of the collagen monomers. This suggests that partial proteolysis of the fibril surface is a requirement for ECM–cellular adhesion and strongly supports the hypothesis that the collagen fibril and the ECM appear to have crypto-biological functions (discussed below). It may also indicate why the collagen fibril exhibits feedback control on its proteolysis as it would appear to be (in part) through the exposure of cell interaction sites that were masked before collagenolysis (Fig. 1 C and E).
Mechanism of MMP1 Collagenolysis.
The α2 peptide is the most vulnerable of the three to proteolysis. This is because of its greater dissociation from the triple helix at the cleavage site and its location facing the outside of the fibril after the removal of the C-telopeptide. These observations support the “binding–unwinding” hypothesis, which proposes that the α2 peptide is cleaved first (12). It may, however, also support the model suggesting that collagen susceptibility to MMP proteolysis is due to thermal fluctuations from normal to “vulnerable” states (10). It is possible that both models are at least partially correct. Normal thermal fluctuations may leave the triple helix partially unwound and it is this vulnerable conformation that the MMP hemopexin domain recognizes, and then “locks in” or further unwinds (Fig. 4). This allows the catalytic domain to dock and hydrolyze the exposed backbone of the α2 peptide. However, the presence of significantly more disassociated regions, including ones containing the specific cleavage sequence, suggests that it may be necessary for all three chains to possess the cleavage sequence and the sequence RGER, located C-terminal to the cleavage sequence (Fig. 4). It would seem significant that the cleavage site is unique with respect to the presence of the RGER sequences compared with the other nonfunctional cleavage sequences and it is worth noting that “GER” is also the core of the integrin binding sequence (30). We note the apparent interactions between RGER and the amino acid residues within and surrounding the positively charged pocket between modules 3 and 4 of the hemopexin domain. It is possible that this further encourages the disassociation of the peptides from the triple helix allowing the peptide backbone of the α2 chain to be more fully inserted into the catalytic cleft (Fig. 4).
In conclusion, we have identified a binding-initiation region surrounding the cleavage site within which the three peptide chains are partially unwound from the triple helix. This supports the hypothesis that collagenase disassociates (12) or targets sections of the peptides that are normally disassociated (10) and therefore more vulnerable to attack by a proteolytic enzyme (Figs. 1–4). Furthermore, our visualization of the fibril surface confirms and illuminates the observation that the collagen fibril possesses crypto-biological features (26), i.e., strategically hidden facets that, when revealed, provide instructions or advantageous features to the cells of the tissue. In this case, “peeling” away parts of the outer layer of the fibril reveals cell adhesion sites in addition to the collagenase cleavage site (Fig. 1). In the intact fibril, molecular accessibility of collagenase to the cleavage site is restricted by the C-telopeptide and the arrangement of neighboring molecules to monomer 4 of the D-periodic microfibril, where collagenolysis occurs (Figs. 1 and 3). We propose that removing the C-telopeptide initiates the proteolysis of collagen within the fibril. We observe that the molecular packing of collagen molecules adjacent to the cleavage site narrows the possible modes of enzyme–substrate interaction and, in addition, may give the appearance of molecular-motor behavior.
Regardless of the specific mechanism of collagenolysis, the limited accessibility of the cleavage site at the fibril surface presents the intriguing possibility that collagenolysis in vivo is regulated or facilitated by the collagen monomers that line the enzyme accessible regions of the fibril surface. These regions and/or partially digested sites (Figs. 1 and 3) may present attractive targets for the design of inhibitory agents to combat collagenase-associated cancer and arthritis.
Materials and Methods
Models of collagen structure (RCSB deposited structures 1Y0F and 1YGV), determined by x-ray diffraction, were used to study “peptide chain from triple helix” disassociation and generate the fibril surface models. The MMP1 molecular coordinates were obtained from RCSB file 1FBL.
Disassociation of Peptides from the Triple Helix and Free Energy Calculation.
The statistically determined secondary structure of short collagen-like peptide models (5) was used to represent the “stringent,” or idealized model of the triple helix. In comparison, the natural (and energy minimized) native structure, experimentally determined by x-ray diffraction (7), was used to represent the “relaxed” form of the triple helix. The determination of both secondary structures has been described (5, 7). The centers of the stringent and relaxed triple helices were superimposed to closely evaluate the magnitude of chain disassociation from the center of each triple helix (stringent and relaxed). The extent of disassociation was thus quantified as the difference between the model peptide-derived (stringent) triple-helix Cα atom positions, and the experimentally constrained energy-minimized (relaxed) Cα atom positions (XYZCα relaxed − XYZCα stringent). Free energy differences were calculated by using the web-based software program FOLD-X (31).
Fibril Structure.
A model of the natural crystallite that forms the structural basis of the native collagen fibril (17) was constructed from a convolution of the microfibrillar structure (7) with the fibrillar crystal lattice (7, 16, 17). Hulmes et al. (18) showed that the 3.8-nm lattice is orientated toward the surface of the fibril, as indicated in SI Fig. 8 showing our representation of the same. This structure provides the fibril surface conformation, which was used to calculate the solvent-accessible face of the fibril in the axial region of the collagenase cleavage site. The quasi-hexagonal packing relationship between neighboring collagen molecules was maintained across the whole of the fibril thus constructed.
Solvent and Molecular Accessibility of the Fibril Surface.
The surface conformation was determined from the x-ray diffraction structure within the context of the molecular packing scheme of the fibril (see above and Figs. 1 and 3). The solvent-accessible surface was calculated and the graphical scene rendered with the program Spock (32) using the default parameters for each of the regions shown in Fig. 3.
Solvent-Accessible Surface and Electrostatic Calculations of MMP1.
The coordinates of the full-length porcine synovial collagenase (MMP1) (RCSB code 1FBL) were modified by rearranging the hemopexin-like domain to be top-most [and so that the positively charged residues of modules 3 and 4 contact the triple helix (20)] and the catalytic domain bottom-most. This was so that both domains were able to make contact with the same collagen molecule while still part of the fibril and still reasonably model that approximated in refs. 1 and 8. In Fig. 4, the MMP surface has been colored in the standard way (red is increasing acidity, blue increasing basicity) by using the default electrostatic calculations for Spock (32). The triple helix amino acid residues are colored white, or as follows: acidic, red; basic, blue; polar, green. The conformation of the two MMP domains was energy minimized >1,000 steps after being manually docked with the collagen triple helix (which remained static) by using the default options for the NAMD (33) extension of VMD.
Supplementary Material
ACKNOWLEDGMENTS.
We thank James San Antonio and Hideaki Nigase for their valuable feedback and encouragement during the drafting of this manuscript. This work was supported by American Heart Association Greater Midwest Affiliate Grant 0435339Z, National Science Foundation Grant MCB-0644015 CAREER, and National Institutes of Health Grant RR08630.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710588105/DC1.
References
- 1.Tam EM, Wu YI, Butler GS, Stack MS, Overall CM. Collagen binding properties of the membrane type 1 matrix metalloproteinase (MT1-MMP) hemopexin C domain. The ectodomain of the 44-kDa autocatalytic product of MT1-MMP inhibits cell invasion by disrupting native type I collagen cleavage. J Biol Chem. 2002;277:39005–39014. doi: 10.1074/jbc.M206874200. [DOI] [PubMed] [Google Scholar]
- 2.Rich A, Crick FH. The molecular structure of collagen. J Mol Biol. 1961;3:483–506. doi: 10.1016/s0022-2836(61)80016-8. [DOI] [PubMed] [Google Scholar]
- 3.Okuyama K, Takayanagi M, Ashida T, Kakudo M. A new structural model for collagen. Polymer J. 1971;9:341–343. [Google Scholar]
- 4.Kramer RZ, Bella J, Brodsky B, Berman HM. The crystal and molecular structure of a collagen-like peptide with a biologically relevant sequence. J Mol Biol. 2001;311:131–147. doi: 10.1006/jmbi.2001.4849. [DOI] [PubMed] [Google Scholar]
- 5.Rainey JK, Goh MC. A statistically derived parameterization for the collagen triple-helix. Protein Sci. 2002;11:2748–2754. doi: 10.1110/ps.0218502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hulmes DJ, Miller A. Quasi-hexagonal molecular packing in collagen fibrils. Nature. 1979;282:878–880. doi: 10.1038/282878a0. [DOI] [PubMed] [Google Scholar]
- 7.Orgel JP, Irving TC, Miller A, Wess TJ. Microfibrillar structure of type I collagen in situ. Proc Natl Acad Sci USA. 2006;103:9001–9005. doi: 10.1073/pnas.0502718103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Visse R, Nagase H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: Structure, function, and biochemistry. Circ Res. 2003;92:827–839. doi: 10.1161/01.RES.0000070112.80711.3D. [DOI] [PubMed] [Google Scholar]
- 9.Leikina E, Mertts MV, Kuznetsova N, Leikin S. Type I collagen is thermally unstable at body temperature. Proc Natl Acad Sci USA. 2002;99:1314–1318. doi: 10.1073/pnas.032307099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stultz CM. Localized unfolding of collagen explains collagenase cleavage near imino-poor sites. J Mol Biol. 2002;319:997–1003. doi: 10.1016/S0022-2836(02)00421-7. [DOI] [PubMed] [Google Scholar]
- 11.De Souza SJ, Pereira HM, Jacchieri S, Brentani RR. Collagen/collagenase interaction: Does the enzyme mimic the conformation of its own substrate? FASEB J. 1996;10:927–930. doi: 10.1096/fasebj.10.8.8666171. [DOI] [PubMed] [Google Scholar]
- 12.Chung L, et al. Collagenase unwinds triple-helical collagen prior to peptide bond hydrolysis. EMBO J. 2004;23:3020–3030. doi: 10.1038/sj.emboj.7600318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bode W. A helping hand for collagenases: the haemopexin-like domain. Structure (London) 1995;3:527–530. doi: 10.1016/s0969-2126(01)00185-x. [DOI] [PubMed] [Google Scholar]
- 14.Gomis-Ruth FX, et al. The helping hand of collagenase-3 (MMP-13): 2.7 A crystal structure of its C-terminal haemopexin-like domain. J Mol Biol. 1996;264:556–566. doi: 10.1006/jmbi.1996.0661. [DOI] [PubMed] [Google Scholar]
- 15.Orgel JP, Wess TJ, Miller A. The in situ conformation and axial location of the intermolecular cross-linked non-helical telopeptides of type I collagen. Structure Fold Des. 2000;8:137–142. doi: 10.1016/s0969-2126(00)00089-7. [DOI] [PubMed] [Google Scholar]
- 16.Orgel JP, et al. The in situ supermolecular structure of type I collagen. Structure (London) 2001;9:1061–1069. doi: 10.1016/s0969-2126(01)00669-4. [DOI] [PubMed] [Google Scholar]
- 17.Hulmes DJ, Wess TJ, Prockop DJ, Fratzl P. Radial packing, order, and disorder in collagen fibrils. Biophys J. 1995;68:1661–1670. doi: 10.1016/S0006-3495(95)80391-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hulmes DJ, Jesior JC, Miller A, Berthet-Colominas C, Wolff C. Electron microscopy shows periodic structure in collagen fibril cross sections. Proc Natl Acad Sci USA. 1981;78:3567–3571. doi: 10.1073/pnas.78.6.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raspanti M, Alessandrini A, Gobbi P, Ruggeri A. Collagen fibril surface: TMAFM, FEG-SEM and freeze-etching observations. Microsc Res Tech. 1996;35:87–93. doi: 10.1002/(SICI)1097-0029(19960901)35:1<87::AID-JEMT8>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 20.Bigg HF, Shi YE, Liu YE, Steffensen B, Overall CM. Specific, high affinity binding of tissue inhibitor of metalloproteinases-4 (TIMP-4) to the COOH-terminal hemopexin-like domain of human gelatinase A TIMP-4 binds progelatinase A, the COOH-terminal domain in a similar manner to TIMP-2. J Biol Chem. 1997;272:15496–15500. doi: 10.1074/jbc.272.24.15496. [DOI] [PubMed] [Google Scholar]
- 21.Li J, et al. Structure of full-length porcine synovial collagenase reveals a C-terminal domain containing a calcium-linked, four-bladed beta-propeller. Structure (London) 1995;3:541–549. doi: 10.1016/s0969-2126(01)00188-5. [DOI] [PubMed] [Google Scholar]
- 22.Netzel-Arnett S, Fields GB, Birkedal-Hansen H, Van Wart HE. Sequence specificities of human fibroblast and neutrophil collagenases. J Biol Chem. 1991;266:6747–6755. [PubMed] [Google Scholar]
- 23.Wu JJ, Lark MW, Chun LE, Eyre DR. Sites of stromelysin cleavage in collagen types II, IX, X, and XI of cartilage. J Biol Chem. 1991;266:5625–5628. [PubMed] [Google Scholar]
- 24.Eyre D. Collagen of articular cartilage. Arthritis Res. 2002;4:30–35. doi: 10.1186/ar380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basbaum CB, Werb Z. Focalized proteolysis: Spatial and temporal regulation of extracellular matrix degradation at the cell surface. Curr Opin Cell Biol. 1996;8:731–738. doi: 10.1016/s0955-0674(96)80116-5. [DOI] [PubMed] [Google Scholar]
- 26.Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bank RA, et al. A simplified measurement of degraded collagen in tissues: Application in healthy, fibrillated and osteoarthritic cartilage. Matrix Biol. 1997;16:233–243. doi: 10.1016/s0945-053x(97)90012-3. [DOI] [PubMed] [Google Scholar]
- 28.Saffarian S, Collier IE, Marmer BL, Elson EL, Goldberg G. Interstitial collagenase is a Brownian ratchet driven by proteolysis of collagen. Science. 2004;306:108–111. doi: 10.1126/science.1099179. [DOI] [PubMed] [Google Scholar]
- 29.Di Lullo GA, Sweeney SM, Korkko J, Ala-Kokko L, San Antonio JD. Mapping the ligand-binding sites and disease-associated mutations on the most abundant protein in the human, type I collagen. J Biol Chem. 2002;277:4223–4231. doi: 10.1074/jbc.M110709200. [DOI] [PubMed] [Google Scholar]
- 30.Xu Y, et al. Multiple binding sites in collagen type I for the integrins alpha1beta1 and alpha2beta1. J Biol Chem. 2000;275:38981–38989. doi: 10.1074/jbc.M007668200. [DOI] [PubMed] [Google Scholar]
- 31.Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: A study of more than 1,000 mutations. J Mol Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 32.Christopher JA, Swanson R, Baldwin TO. Algorithms for finding the axis of a helix: fast rotational and parametric least-squares methods. Comput Chem. 1996;20:339–345. doi: 10.1016/0097-8485(95)00075-5. [DOI] [PubMed] [Google Scholar]
- 33.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jokinen J, et al. Integrin-mediated cell adhesion to type I collagen fibrils. J Biol Chem. 2004;279:31956–31963. doi: 10.1074/jbc.M401409200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.