Abstract
The hydrophobic effect, i.e., the poor solvation of nonpolar parts of molecules, plays a key role in protein folding and more generally for molecular self-assembly and aggregation in aqueous media. The perturbation of the water structure accounts for many aspects of protein hydrophobicity. However, to what extent the dispersion interaction between molecular entities themselves contributes has remained unclear. This is so because in peptide folding interactions and structural changes occur on all length scales and make disentangling various contributions impossible. We address this issue both experimentally and theoretically by looking at the force necessary to peel a mildly hydrophobic single peptide molecule from a flat hydrophobic diamond surface in the presence of water. This setup avoids problems caused by bubble adsorption, cavitation, and slow equilibration that complicate the much-studied geometry with two macroscopic surfaces. Using atomic-force spectroscopy, we determine the mean desorption force of a single spider-silk peptide chain as F = 58 ± 8 pN, which corresponds to a desorption free energy of ≈5 kBT per amino acid. Our all-atomistic molecular dynamics simulation including explicit water correspondingly yields the desorption force F = 54 ± 15 pN. This observation demonstrates that standard nonpolarizable force fields used in classical simulations are capable of resolving the fine details of the hydrophobic attraction of peptides. The analysis of the involved energetics shows that water-structure effects and dispersive interactions give contributions of comparable magnitude that largely cancel out. It follows that the correct modeling of peptide hydrophobicity must take the intimate coupling of solvation and dispersive effects into account.
Keywords: atomic-force microscopy, hydrophobic effect, molecular dynamics simulation, single molecules, protein adsorption
For scientists working with biological or soft matter systems, understanding what holds the world together largely means unraveling the mechanism behind the so-called hydrophobic effect. The term hydrophobic attraction (HA) was initially introduced to describe the attraction between small nonpolar molecules such as methane in water (1, 2). It is nowadays more broadly used to describe forces between all kinds of nonpolar objects in aqueous environments, implying a common mechanism for protein folding, micellization, self-assembly of lipids, oil–water demixing, and thus any supermolecular aggregation in water (3). For predicting protein structures and function the magnitude and nature of the HA acting between peptide segments is a central issue that has not been fully resolved. Much effort was put in force measurements between well defined model systems, for example mica surfaces made hydrophobic or micrometer-sized plastic beads. However, these systems are notoriously plagued by secondary effects, such as bubble adsorption and cavitation effects (4, 5) or compositional rearrangements (6). In simulations of interacting planar plates, similar effects give rise to severe equilibration problems (7, 8). Unfortunately, even if bubbles and other complications can be excluded, the very short-ranged portion of the HA between micrometer-sized surfaces can typically not be resolved experimentally because of mechanical instabilities of the measuring device (3). Likewise, the intricate scale dependence of the HA makes it nontrivial to relate the force between micrometer-sized particles to the one between molecules (9).
It transpires that for the understanding of hydrophobically driven protein and polymer collapse (10, 11) a model system on the molecular scale is needed. We introduce a hybrid system, where the force needed to pull a single hydrophobic peptide chain away from a synthetic flat hydrophobic surface is determined by single-molecule force spectroscopy. Because of the small scales involved, bubbles or cavitation constitute no barrier to equilibration, and the actual binding free energy can be determined in quasi-equilibrium. Apart from its value as a model system with well characterized and easily tunable parameters, this scenario is of direct relevance for a number of applications: (i) the initial stage of blood clotting in capillary vessels involves the specific adsorption of a large protein to the injured vessel wall (12), (ii) diamond surfaces can operate as ion-sensitive field-effect transistors and thus be used to monitor the adsorption of charged proteins in biosensor applications (13), and (iii) understanding how to prevent the adsorption of proteins to surfaces is the ultimate goal for engineering of biomedical implants and nonfouling surfaces. For the case of single-peptide desorption, faithful simulations can be performed with relative ease because the number of water molecules displaced is small and equilibration is fast. The excellent agreement between the desorption force found experimentally and in our simulations shows that classical simulations with explicit water do capture the essence of the HA. Experimentally only the sum of the various contributions to the total HA is measured. In the simulations, on the other hand, it is easy to disentangle interactions between water, peptide, and the surface from each other. The unexpected result is that almost complete cancellation of individual energy contributions takes place, meaning that the HA results from a subtle balancing act. Specifically, both the water structural forces and the direct dispersion interactions between peptide and surface contribute significantly to the total HA. Understanding the HA thus involves unraveling the interplay between those two factors.
Results
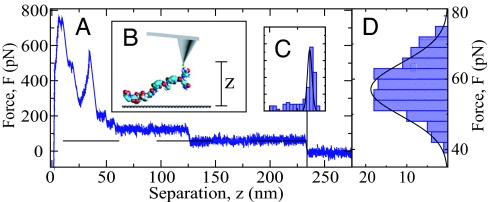
For our experiments, one or very few molecules of a mildly hydrophobic spider silk peptide consisting of 16 repeats of the 35-aa C-motif derived from Araneus diadematus fibroin [this C16 peptide shows a moderate solubility of maximally 8% (wt/wt) in water (14); for structure see supporting information (SI) Fig. 7], are covalently bound at their N terminus to the tip of an atomic-force microscope (AFM) by a short polymeric spacer. The AFM tip is immersed in a 20 mM NaCl solution and put in contact with a hydrophobized polished diamond substrate. After sufficient time for the peptide to adsorb onto the substrate (of the order of one second), the AFM tip is vertically moved away from the substrate at a speed of v0 = 0.5 μm/s. Fig. 1A shows a representative force–distance curve that exhibits a mean plateau force of 59 pN (indicated by a horizontal broken line) up to a tip–surface separation of zmax = 234 nm. For a separation smaller than 125 nm a plateau with almost doubled height is observed, which suggests that a second peptide chain is attached to the cantilever tip and also adsorbs on the substrate. Fig. 1B schematically shows the experimental setup. The distribution of plateau lengths zmax from 200 pulling experiments with the identical tip preparation and thus the same peptide chain, Fig. 1C, is quite sharply peaked around a polymer extension of 236 nm, which amounts to 91% of the estimated polymer contour length in the fully stretched conformation, in line with previous AFM measurements. The distribution of plateau forces (averaged over surface separations >150 nm) (Fig. 1D), gives an average desorption force of F = 58 ± 8 pN. Inserting the measured chain stretching, it amounts to a desorption free energy of 4.8 kBT per amino acid. Key to the experimental measurement is that the polymer is long enough so that direct interactions between substrate and AFM tip are negligible. In addition, the practically irreversible covalent chain attachment to the AFM tip ensures that only the attraction between chain and substrate contributes to the plateau force.
Fig. 1.
AFM experiment. (A) A typical experimental AFM force–extension trace for the desorption of a single C16 spider silk peptide chain from a hydrophobic diamond surface in 20 mM NaCl, exhibiting a force plateau in the extension range 125 nm < z < 234 nm. (B). Scheme of the setup. (C and D) Distribution for 200 traces with the identical peptide chain for the force plateau length (C) and the plateau height (D), giving a mean desorption force of F = 58 ± 8 pN.
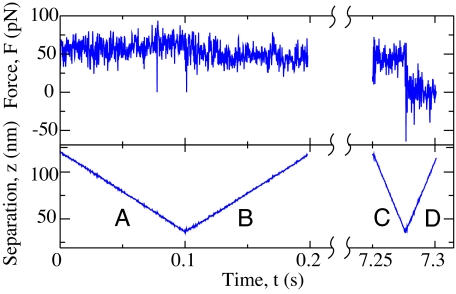
In Fig. 2 we demonstrate that the observed plateau force is independent of the pulling rate and the pulling direction. This observation points to a high mobility of the peptide on the diamond surface and implies that the adsorbed chain section reaches equilibrium on the experimental time scales (15–17). Our measurement thus corresponds to a high-precision determination of the equilibrium HA between a single peptide chain and a hydrophobic substrate. We deliberately call this interaction a free energy of desorption to distinguish it from the standard adsorption free energy. As will be discussed in more detail later on, the desorption force measured with the AFM includes the polymeric response in the stretched state, whereas the reference state of the ordinary polymer adsorption scenario is the stress-free polymeric configuration in solution.
Fig. 2.
Desorption force (Upper) and AFM tip position (Lower) versus time. The rate dependence is studied by the following modified experimental procedure: starting with a surface-adsorbed C16 chain, the AFM tip is moved toward the surface at a rate of 1 μm/s (A), pulled away from the surface at a rate of 1 μm/s (B), held at constant height for 7 s, then moved toward (C) and away from the surface at a rate of 4 μm/s (D). The plateau forces in A, B, and C are comparable. Between C and D the force on the cantilever drops suddenly to zero because the peptide detaches from the surface.
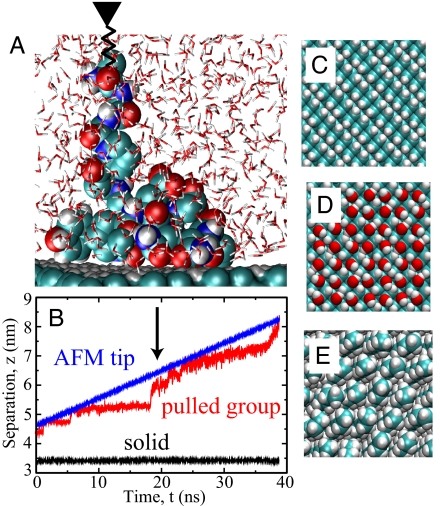
To gain insight into the microscopic mechanisms responsible for the HA, and to elucidate the interplay between dispersion interactions and water-structural effects, we perform all-atomistic molecular dynamics simulations of exactly the same system. We model the interactions among the peptide, the diamond surface, and water with the well established Gromos96 force field. A 6 nm × 3 nm diamond surface is put in contact with up to 3,000 simple point charge (SPC) water molecules, and simulations are performed at constant pressure and different fixed temperatures. Hydrophobic hydrogen-terminated (contact angle 106° as derived from simulations) and hydrophilic hydroxyl-terminated diamond surfaces as well as self-assembled monolayers (SAM, contact angle 92°) are studied, top views of which are presented in Fig. 3 C–E. To manipulate the peptide, we connect one end of a spring to the terminal peptide group and move the other end vertically away from the substrate at constant speed v0, similar to the AFM setup (see Fig. 3A for a simulation snapshot). The spring acts only in the direction normal to the interface and thus the lateral peptide motion is not restrained. Although there is no lateral peptide sliding on the surface required during desorption, we observe pronounced lateral peptide motion and diffusion. The positions of the two ends of the spring, plotted in Fig. 3B as a function of time, yield the force exerted on the peptide.
Fig. 3.
Simulation details. (A) Simulation snapshot of spider silk fragment 3 that is pulled by an attached spring away from a hydrophobic diamond surface. (B) Vertical position of the two ends of the spring, one attached to the terminal peptide group (red) and the other (blue) moved at constant velocity v0 = 0.1 m/s. The snapshot in A is indicated by an arrow. (C–E) The surfaces of hydrophobic H-terminated diamond (C), hydrophilic OH-terminated diamond (D), and the hydrophobic methyl-terminated self-assembled monolayer (E), as used in the simulations.
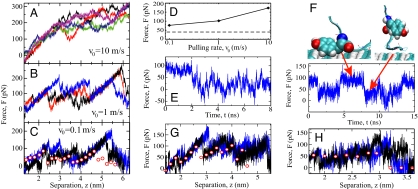
Because of the finite box size, the peptide C-motif is cut in three roughly equal-sized partially overlapping fragments. Force–extension traces for fragment 3 are shown for three different pulling speeds v0 in Fig. 4 A–C. As v0 decreases, friction effects diminish and the mean desorption force goes down. In Fig. 4C also static mean forces are shown (spheres), obtained from simulations where the peptide end is held fixed for 8 ns as shown for fragment 3 at 4-nm tip separation in Fig. 4E. In Fig. 4F we show the force at a tip separation of 4.6 nm, where pronounced force fluctuations due to switching of the tyrosine residue between adsorbed and desorbed states are present. It is the strong adsorption of tyrosine to the surface that causes the force spike in Fig. 4C in the tip separation range of z = 4–5 nm.
Fig. 4.
Simulation results for spider silk at the hydrophobic diamond surface. (A–C) Individual force traces as fragment 3 is pulled away from the surface at different pulling rates v0. In C static simulation results are shown as spheres. (D) Average desorption force for fragment 3 as a function of v0; the broken line denotes the static result, F = 40 pN. (E) Force as a function of time for the static simulation of fragment 3 at z = 4-nm tip separation. (F) (Lower) Force for fragment 3 at a tip separation of z = 4.6 nm. (Upper) Snapshots of repeated switching between the adsorbed state (Left, high force) and the desorbed state (Right, low force) of a tyrosine residue. (G and H) Static and dynamic desorption forces at v0 = 0.1 m/s for fragment 2 (G) and fragment 1 (H).
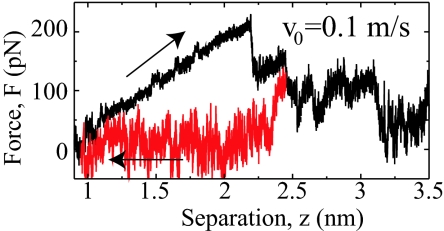
In Fig. 4D the averaged desorption force for fragment 3 is shown as a function of the pulling rate and compared with the static result (broken horizontal line). It is seen that the dissipative contribution due to friction is already quite small for the slowest dynamic pulling at a speed of v0 = 0.1 m/s, and the measured dynamic desorption forces extrapolate nicely to the static result in the limit of vanishing pulling speed. Proper equilibration is in principal a subtle issue, but in the present scenario peptide friction on a hydrophobic surface is small enough not to cause equilibration problems in the simulations, although the pulling rate is five orders of magnitude larger than in the experiment. The situation is clearly different for a peptide desorbing from hydrophilic diamond, where pronounced hysteresis is observed between desorption and adsorption (see Fig. 5). Here the presence of hydrogen bonds between surface and peptide dramatically increases the intrinsic time scales and equilibrium is impossible to reach in our simulations (SI Text and SI Fig. 8).
Fig. 5.
Force traces for fragment 3 on hydrophilic diamond showing large hysteresis between moving away (black) and toward (red) the surface, making equilibration impossible.
The static desorption forces of the three fragments averaged over the height of the peptide terminus are F = 54 pN (fragment 1, Fig. 4H), F = 64 pN (fragment 2, Fig. 4G), and F = 40 pN (fragment 3, Fig. 4C). The average over all three fragments, properly accounting for the region of overlap between fragments 2 and 3, yields on the hydrophobic diamond surface a static desorption force of F = 54 ± 15 pN for the complete C16-motif (see Table 1), which agrees well with the experimental result of F = 58 ± 8 pN. We take this agreement as an indication that the classical force fields and explicit water models used in the simulation quantitatively capture the hydrophobic effect. Desorption forces depend little on the specific type of substrate as long as it is hydrophobic; on a hydrophobic self-assembled monolayer, e.g., the static desorption force of fragment 3 is F = 35 pN, as compared with F = 40 pN on hydrophobic diamond.
Table 1.
Average desorption forces of peptide fragments as obtained in static long-time molecular dynamics simulations
| Surface | Peptide fragment | T, K | F, pN |
|---|---|---|---|
| H-terminated diamond | C16-1 | 300 | 54 |
| C16-2 | 300 | 64 | |
| C16-3 | 300 | 40 | |
| C16 total | 300 | 54 ± 15 | |
| C16-3 | 280 | 39 | |
| C16-3 | 320 | 44 | |
| SAM | C16-3 | 300 | 35 |
Discussion
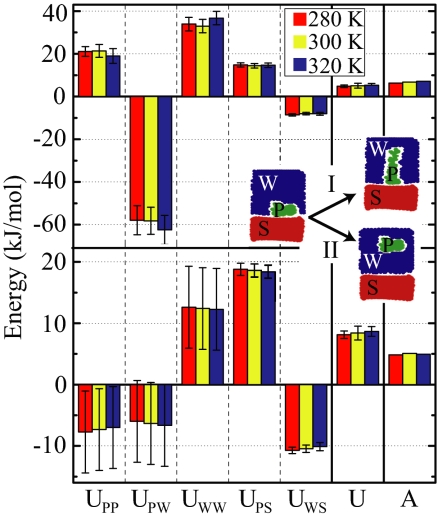
After having demonstrated that simulations are able to reproduce the experimental plateau forces and therefore the HA in a quantitative fashion, we analyze the microscopic mechanism behind HA. In the traditional Derjaguin–Landau–Verwey–Overbeek (DLVO) approach (3), the force between uncharged bodies immersed in water is explained solely in terms of dispersion or van der Waals interactions; nowadays, solvation effects are thought to play an equally or even more important role (3, 9). Insight into the interplay between both contributions can be gained from simulations: in Fig. 6 we show how the various energetic contributions stemming from the interactions between peptide (P), surface (S), and water (W) atoms sum up to the total desorption energy per peptide monomer, U, according to U = UPP + UPW + UWW + UPS + UWS. We distinguish process I, in which the peptide is brought from the adsorbed state to a state that is stretched between surface and terminal peptide group that closely mimics the AFM experiment.
Fig. 6.
Decomposition of the total internal energy per monomer, U, into the contributions from interactions between peptide (P), surface (S), and water (W) for three different temperatures and compared with the free energy A. The upper histogram gives the energy price for desorbing peptide fragment 3 into a stretched conformation (process I), as applicable to AFM experiments, and the lower histogram has the totally relaxed and solvated peptide conformation as the reference state, as applicable to equilibrium adsorption from a bulk solution (process II). In both cases, the various internal energy contributions are larger in magnitude than the total internal energy U.
In process II the peptide is transferred from the adsorbed state to an unperturbed bulk state, which correctly describes the thermodynamic adsorption equilibrium of a peptide in solution. The resultant adsorption free energy of about A = 5 kJ/mol per monomer in process II is quite close to the value of A = 3.6 kJ/mol for phenylalanine as measured by adsorption isotherm analysis (18). For both processes I and II the water–water and peptide–surface contributions to the internal energy, UWW and UPS, are positive and larger than the resulting total energy U and free energy A. Both the van der Waals interaction between surface and peptide (included by means of the simulation force field in a heuristic fashion) and the solvation energy are thus important. However, they are considerably compensated by the water–surface and peptide–water contributions, UWS and UPW. Interestingly, the peptide–peptide interaction, UPP, often neglected in theoretical considerations, is quite large and differs in sign between the two desorption scenarios. From the free energy A it follows that the entropy S = (U − A)/T is negative for process I, i.e., the system loses entropy as the peptide is brought into the stretched conformation, but is positive for process II, i.e., the system gains entropy as the peptide leaves the surface-adsorbed state. This difference in entropy can be explained by the conformational entropy of the peptide, which is maximal in the bulk state, intermediate in the surface-adsorbed state, and minimal in the stretched state. The temperature dependence of the free energy A is small but consistent with the thermodynamic relation S = −dA/dT within the numerical uncertainty.
It is amusing to note that the sum of the peptide–surface interaction and the water–surface interaction, UPS + UWS, for both processes roughly equals the total internal energy change U. This finding should not be interpreted as suggesting that peptide solvation effects are irrelevant. Rather, it explains why the often used approximation of calculating effective solute–surface interactions from dispersion interactions among the surface, solute, and displaced solvent works well. The good agreement between UPS + UWS and U is most likely coincidental, and it is related to the fact that the peptide–peptide interaction UPP in the present case almost exactly cancels the water contributions UPW + UWW. Note that the separation into the various contributions is done for the internal energy only and not for the (more relevant) free energy, simply because it is difficult to analyze the entropy in such a discriminatory fashion. Presumably, similar features underlie the free energy as well.
The comparison of the experimental desorption forces with the simulations necessitates several simplifications and assumptions. (i) Because of simulation time constraints, pulling rates in the simulations are orders of magnitude higher than in experiments. First, we demonstrate that the experimental desorption force is rate-independent and free of hysteresis (see Fig. 2). By carefully considering different desorption rates and comparing with static simulations, we show that equilibrium is also reached in simulations on hydrophobic surfaces (whereas on hydrophilic substrates equilibration is impossible with present computer power). (ii) In the simulations the peptide strand is necessarily much shorter than in the experiment. This shortness means that strictly speaking the statistical ensembles are different. Whereas in the experiment the already desorbed peptide strand acts as a flexible tether that averages out the force acting on the AFM cantilever, leading to flat force plateaus, in the simulations the tether is much shorter and the sequence dependence of the adsorption strength is clearly resolved with a spiky force response that is related to the hydrophobicity variation. An extreme example is the strongly hydrophobic tyrosine residue, which shows pronounced and almost bistable switching (see Fig. 4F). However, the additional error introduced by the different averaging procedures is estimated to be less than the statistical errors already present in the data. (iii) Finally, we note that it is possible that a different force field would produce somewhat different desorption forces; it is plausible that the trends discussed in this paper are robust and do not depend on the specific force field used.
In this paper we demonstrate that the forced desorption of a single peptide chain from a well defined flat surface constitutes an ideal system to disentangle and understand the various factors that combine to yield the hydrophobic attraction. This tractability is connected to the absence of bubbles and cavitation effects, which in more macroscopic systems prevent equilibration. The excellent agreement between the experimental AFM plateau force (F = 58 ± 8 pN) and the simulation (F = 54 ± 15 pN) shows that classical force fields are capable of quantitatively reproducing peptide solvation and interaction. The total desorption energy U results from an almost complete cancellation of the individual interaction contributions among water, surface, and peptide, each of which is larger in magnitude than U itself. It follows that there is not a single mechanism for the HA, but that water structural effects are as important as the dispersion interaction among substrate, water, and peptide segments.
Materials and Methods
Peptide Synthesis and Characterization.
Naturally occurring spider silk is mainly composed of large proteins with apparent molecular masses between 180 and 720 kDa. Spider silk proteins reveal a highly repetitive primary structure with alanine- and glycine-rich motifs. The repeating units exhibit high amphiphilicity and give rise to an intrinsically unstructured conformation in solution (19). It is striking that usually a pair of silk proteins is required to form a silk fiber, one being hydrophilic and one being slightly hydrophobic (14). Here, we study a genetically engineered and recombinantly produced spider silk protein based on a hydrophobic silk protein, ADF-4, derived from the garden spider Araneus diadematus. It consists of 16 repeats of the so-called C-unit with a sequence of GSSAAAAAAAASGPGGYGPENQGPSGPGGYGPGGP. A model of the repeat unit is shown in SI Fig. 7. The protein is recombinantly produced in Escherichia coli and purified as described in detail elsewhere (14).
Hydrophobic Diamond Surface Fabrication.
The diamond substrates used in this work are polished (root-mean-square roughness <0.3 nm, measured in an area of 5 μm × 5 μm) polycrystalline diamond films (Element Six) of size 5 mm × 5 mm × 0.5 mm. Hydrophobic diamond surfaces are obtained after hydrogen-termination of the surface (13). The hydrogenation is performed in a vacuum chamber by using a hydrogen gas flow of 300 standard cm3/min over a hot (2,000°C) tungsten filament, placed at a distance of 4 cm from the substrate. During the process, the sample temperature (approximately 700°C) is determined with a thermocouple. Static contact angle experiments, using a 1.5-μl deionized water droplet placed on top of the diamond substrate, are carried out for the characterization of the surface hydrophobicity. The measured water contact angle for a hydrogen-terminated diamond surface is 90° ± 1° as obtained from six measurements.
AFM Methods.
Amino-functionalized tip surfaces for the covalent attachment of C16 spider silk peptide molecules are obtained by applying Vectabond Reagent (Axxora) to activated silicon nitride tips (Veeco Probes). C16 spider silk was attached by keeping the tip for 30 min in an aqueous polyethylene glycol solution (mixture of 6-kDa PEG-α,ω-di-NHS ester and 5-kDa CH3O-PEG-NHS ester, in which NHS is N-hydroxysuccinimidyl; Rapp-Polymere) and afterward in a C16 solution (≈6 mg/ml) for 2 h. The PEG-α,ω-di-NHS polymer provides for the covalent attachment of the spider silk peptide chain; the CH3O-PEG-NHS polymer passivates the tip and thus prevents unspecific adhesion of the peptide chain to the tip (20). A stable amide bond is formed by the reaction of the succinimidyl active ester with the amine-containing N terminus of the spider silk. This way the contour length of the attached peptide chain is uniquely determined. The concentration of functionalized PEG polymers on the AFM tip is such that on average one peptide chain binds to the tip. The attachment exhibits long-time stability, so that up to 2,000 desorption experiments can be recorded with the same peptide probe molecule. Force–extension traces are obtained from the deflection piezopath signal as described elsewhere (21). They show no dependency on the pulling velocity in the typical range for AFM experiments of 0.1–10 μm/s. The traces are taken at least at two different positions on the hydrogen-terminated diamond with an MFP-3D from Asylum Research.
The contour length of a C16 peptide in the fully extended conformation, including the PEG linker, amounts to 259 nm: 210 nm stems from the spider silk protein, which consists of a total of 575 aa, i.e., 16 × 35 plus an additional 15 aa from the nonrepetitive region and using a length of 0.365 nm per amino acid. The remaining 49 nm originates from the 6-kDa PEG linker molecule, which consists of ≈136 -CH2CH2O- monomers with a contour length of ≈0.36 nm per monomer in the all-trans conformation, which is the predominant conformation at these elevated forces. The polydispersity of the PEO linker is 1.13. The mean of the measured maximal plateau length of the C16 peptide construct in our experiments is 236 nm, which should be close to the actual extension of the peptide chain at the plateau force of ≈58 pN. This is indeed ≈91% of the fully extended contour length of 259 nm. The difference is brought about by conformational chain fluctuations, and the stretching found here is in rough agreement with previously reported values of ≈85% stretching of hydrophilic peptide chains at a force of 60 pN (22).
Simulation Methods.
Molecular dynamics simulations at fixed particle number N, constant ambient pressure P = 1 bar, and at different temperatures T are performed by using the Gromacs simulation package (23, 24). For the hydrophobic diamond, a slab of extension up to 6 nm × 3 nm × 2 nm with a fully hydrogen-terminated (100) surface in the z direction is built. For the hydrophilic diamond, every other surface hydrogen is replaced by a hydroxyl group with partial charges adjusted to those in serine (C, 0.266 e; O, −0.674 e; H, 0.408 e). All other partial atomic charges in the diamond are set to zero. Up to 3,000 single point charge water molecules are added above and below the diamond slab, filling the simulation box of approximate size 6 nm × 3 nm × 7 nm. For comparison, we also model a SAM consisting of a 6 × 8 grid of C20H42 molecules with a lattice constant that represents the spacing on a gold (111) surface, yielding a 30° tilt angle of the alkane strands.
One repetitive unit of the C16 motif of spider silk contains 35 aa, which is too long for an efficient simulation with explicit solvent, therefore we split it into three pieces: the first fragment, C16-1, contains the residues 1–10, GSSAAAAAAA, the second, C16-2, the residues 11–26, ASGPGGYGPENQGPSG, and the third, C16-3 contains the residues 21–35, NQGPSGPGGYGPGGP (see SI Fig. 7; for amino acid abbreviations see ref. 25). The negative charge of glutamic acid in fragment 2 is compensated by a sodium ion, which corresponds to a Na+ concentration of 25 mmol/liter in the simulation system. The force field parameters for the peptide and the surface atoms are taken from the Gromos96 version 53A6 force field (26). Initially, the peptide is placed in the simulation cell above the surface before the cell is filled with water. To equilibrate the system, we first perform an energy minimization of the simulation system, followed by a 5-ps NVT relaxation and a 100-ps NPzAT relaxation. During this initial relaxation, the peptide adsorbs readily onto the surface. Simulations of 5–20 ns are done employing the Berendsen scheme (27) with semiisotropic pressure coupling. The particle-mesh Ewald method (28) is used for the periodic treatment of coulombic interactions, and bonds to hydrogen atoms are constrained by using LINCS (29). The center of mass translation and the overall rotational motion are removed at every time step.
In simulations mimicking AFM experiments, denoted as process I, in which the peptide is pulled vertically away from the surface, a harmonic restraint potential with spring constant k = 166 pN/nm is applied on the z coordinate of the center of mass of the first residue of the peptide, leaving the lateral coordinate unperturbed. The center of the restraint potential is moved with constant velocity v0 in the z direction away from the surface. The pulling force F is calculated during the simulation from the extension of the harmonic spring. In the beginning of the simulations the AFM tip potential is located at a height of approximately z = 0.6 nm, which is measured with respect to the first layer of carbon atoms (oxygen atoms for the OH-terminated diamond surface), and pulling is done until the peptide is completely desorbed. On H-terminated diamond, simulations for all three fragments are performed with pulling rates of v0 = 10 m/s, 1 m/s, and 0.1 m/s. On the OH-terminated diamond and on the SAM surface, simulations are performed only for the third fragment, with pulling rates of v0 = 1 m/s and 0.1 m/s. Every simulation is repeated at least once with a different starting conformation. In addition to these dynamic pulling simulations, we simulate every system with a static restraining potential kept at a constant elevation for 8 ns; here we start from configurations obtained within the dynamic simulations at v0 = 1 m/s pulling rate. We carefully check for equilibration by starting with different initial conditions and making sure that the same final state is obtained. The pulling force is averaged over the last 4 ns in these static simulations. Note that in contrast to the hydrophobic substrates, for the hydrophilic diamond surface different starting configurations do not equilibrate to the same final structure, and equilibrium plateau forces are inaccessible in simulations. The simulations are done for tip elevations in the range from complete adsorption to complete desorption with a step size of 0.2 nm. Most simulations are done at a temperature of 300 K. For fragment 3 on H-terminated diamond, we repeat static simulations for temperatures of 280 K and 320 K. The results are shown in Table 1.
Process II describes the transformation of a surface-adsorbed peptide to the relaxed bulk conformation. This transformation is accomplished by applying two restraint potentials at each end of the peptide chain. The first restraint potential brings the peptide chain into a stretched state where one end is still adsorbed on the surface. This is the final state of process I. Then the second restraint potential is turned on and moves the other peptide end away from the surface. This path is sampled in discrete steps of 0.2 nm, and every step consists of an 8-ns simulation with fixed spring positions, averaging the force during the last 4 ns. In the final state of process II, both ends of the peptide chain are confined at a certain distance from the surface, with no restriction on the lateral position and on the distance between them.
For both processes, the free energy is obtained as the integral over the force along the pulling path. The internal energy U and its contributions are obtained as time averages during the last 4 ns of static simulations, the constant restraint potential is not included in U. The energy differences for process I are calculated by linear regression over different separations from the surface. For process II, the energy in the totally detached state is used.
We determine the contact angle θ from the interfacial tensions of the solid–water (γsl), solid–vapor (γsv), and vapor–water (γvl) interfaces by the Young equation cos θ = (γsv − γsl)/γvl. The surface tensions are obtained from 4-ns simulations as the difference between the pressure tensor components normal and parallel to the interface.
Supplementary Material
ACKNOWLEDGMENTS.
Discussions with Andreas Bausch are acknowledged. This work was supported by Deutsche Forschungsgemeinschaft Grants NE 10/4 and HU997/2, the German Excellence Initiative via the Nanosystems Initiative Munich, and the Elite Netzwerk Bayern (CompInt). M.G. acknowledges support by the Stiftung Industrieforschung.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707879105/DC1.
References
- 1.Blokzijl W, Engberts JBFN. Angew Chem Int Ed Engl. 1993;32:1545–1579. [Google Scholar]
- 2.Hummer G, Garde S, Garcia AE, Paulaitis ME, Pratt LR. J Phys Chem B. 1998;102:10469–10482. [Google Scholar]
- 3.Meyer EE, Rosenberg KJ, Israelachvili J. Proc Natl Acad Sci USA. 2006;103:15739–15746. doi: 10.1073/pnas.0606422103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christenson HK, Claesson PM. Adv Colloid Interface Sci. 2001;91:391–436. [Google Scholar]
- 5.Carambassis A, Jonker LC, Attard P. Phys Rev Lett. 1998;80:5357–5360. [Google Scholar]
- 6.Podgornik R, Parsegian VA. Chem Phys. 1991;154:477–483. [Google Scholar]
- 7.Wallqvist A, Berne BJ. J Phys Chem. 1995;99:2893–2899. [Google Scholar]
- 8.Hayashi T, Pertsin AJ, Grunze M. J Chem Phys. 2002;117:6271–6280. [Google Scholar]
- 9.Chandler D. Nature. 2005;437:640–647. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- 10.Mountain RD, Thirumalai D. Proc Natl Acad Sci USA. 1998;95:8436–8440. doi: 10.1073/pnas.95.15.8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sotomayor M, Schulten K. Science. 2007;316:1144–1148. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
- 12.Schneider SW, Nuschele S, Wixforth A, Gorzelanny C, Alexander-Katz A, Netz RR, Schneider MF. Proc Natl Acad Sci USA. 2007;104:7899–7903. doi: 10.1073/pnas.0608422104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Härtl A, Garrido JA, Nowy S, Zimmermann R, Werner C, Horinek D, Netz RR, Stutzmann M. J Am Chem Soc. 2007;129:1287–1292. doi: 10.1021/ja066543b. [DOI] [PubMed] [Google Scholar]
- 14.Huemmerich D, Helsen CW, Quedzuweit S, Oschmann J, Rudolph R, Scheibel T. Biochemistry. 2004;43:13604–13612. doi: 10.1021/bi048983q. [DOI] [PubMed] [Google Scholar]
- 15.Châtellier X, Senden TJ, Joanny J-F, di Meglio J-M. Europhys Lett. 1998;41:303–308. [Google Scholar]
- 16.Hanke F, Livadaru L, Kreuzer HJ. Europhys Lett. 2005;69:242–248. [Google Scholar]
- 17.Serr A, Netz RR. Europhys Lett. 2006;73:292–298. [Google Scholar]
- 18.Singh N, Husson SM. Langmuir. 2006;22:8443–8451. doi: 10.1021/la0533765. [DOI] [PubMed] [Google Scholar]
- 19.Exler JH, Hummerich D, Scheibel T. Angew Chem Int Ed. 2007;46:3559–3562. doi: 10.1002/anie.200604718. [DOI] [PubMed] [Google Scholar]
- 20.Hinterdorfer P, Baumgartner W, Gruber H, Schilcher K, Schindler H. Proc Natl Acad Sci USA. 1996;93:3477–3481. doi: 10.1073/pnas.93.8.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hugel T, Seitz M. Macromol Rapid Comm. 2001;22:989–1016. [Google Scholar]
- 22.Neuert G, Hugel T, Netz RR, Gaub HE. Macromolecules. 2006;39:789–797. [Google Scholar]
- 23.Lindahl E, Hess B, van der Spoel D. J Mol Model. 2001;7:306–317. [Google Scholar]
- 24.Berendsen HJC, van der Spoel D, van Drunen R. Comp Phys Commun. 1995;91:43–56. [Google Scholar]
- 25.Stryer L. Biochemistry. 4th Ed. New York: Freeman; 1995. p. 23. [Google Scholar]
- 26.Scott WRP, Hünenberger PH, Tironi IG, Mark AE, Billeter SR, Fennen J, Todd AE, Huber T, Krüger P, van Gunsteren WF. J Phys Chem A. 1999;103:3596–3607. [Google Scholar]
- 27.Berendsen HJC, Postma JPM, van Gunsteren WF, di Nola A, Haak JR. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 28.Darden T, York D, Pedersen L. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 29.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.