Abstract
In the Fluid Mosaic Model for biological membrane structure, proposed by Singer and Nicolson in 1972, the lipid bilayer is represented as a neutral two-dimensional solvent in which the proteins of the membrane are dispersed and distributed randomly. The model portrays the membrane as dominated by a membrane lipid bilayer, directly exposed to the aqueous environment, and only occasionally interrupted by transmembrane proteins. This view is reproduced in virtually every textbook in biochemistry and cell biology, yet some critical features have yet to be closely examined, including the key parameter of the relative occupancy of protein and lipid at the center of a natural membrane. Here we show that the area occupied by protein and lipid at the center of the human red blood cell (RBC) plasma membrane is at least ≈23% protein and less than ≈77% lipid. This measurement is in close agreement with previous estimates for the RBC plasma membrane and the recently published measurements for the synaptic vesicle. Given that transmembrane proteins are surrounded by phospholipids that are perturbed by their presence, the occupancy by protein of more than ≈20% of the RBC plasma membrane and the synaptic vesicle plasma membrane implies that natural membrane bilayers may be more rigid and less fluid than has been thought for the past several decades, and that studies of pure lipid bilayers do not fully reveal the properties of lipids in membranes. Thus, it appears to be the case that membranes may be more mosaic than fluid, with little unperturbed phospholipid bilayer.
Keywords: fluid mosaic model, membrane lipid bilayer
The protein-to-lipid ratio has often been measured, but such measures do not address the question we raise here, because the proteins usually extend well beyond the bilayer and often occupy surface area—the finding from the recent modeling of the synaptic vesicle is that there is little lipid exposed, being covered by extrabilayer protein domains (1). There is great variability in the measured protein-to-lipid ratios, from myelin, with ≈20% protein, to the inner mitochondrial membrane at 76% protein; the plasma membranes of most animal cells are ≈50% protein. Nonetheless, with the exception of the purple membrane from Halobacterium salinarium, there is little accurate information concerning the area occupancy of protein and lipid at the center of natural membranes (2, 3).
Results
We measured the area occupancy of protein and lipid at the center of the human RBC plasma membrane, a much-studied membrane with direct relevance to other cell types. RBC plasma membranes were “shaved” by using a combination of high-pH washes with 10 mM NaOH and proteolytic digestion with proteinase K to remove loops and domains extending beyond the lipid characteristic groups of the membrane bilayer surface. The basic assumption in conducting the membrane “shaving” experiment is that the membranes should be resistant to proteolysis of the transmembrane regions, allowing determination of their abundance. Several key ideas support the use of membrane shaving to characterize the area occupancy at the center of natural membranes. The first is the observation that the dominant transmembrane structural element in membrane proteins is the α-helix (4). The second idea is that individual α-helices remain stable as transbilayer elements, or independently folded domains (4, 5). This assumption is supported by a number of studies that have shown that fragments of membrane proteins in which the loops between helices are disrupted or absent can retain the ability to self-assemble in a membrane (4). Experiments have shown that the bacteriorhodopsin molecule can be cut in several loops and reassembled (6, 7), and related experiments on other proteins have demonstrated that coexpression of fragments cut in loop regions results in functional proteins inserted into membranes (see table in ref. 4). Another supporting argument is that the transmembrane peptides will be either predominantly or entirely hydrophobic. Thus, there will be no driving force for the peptides to flip out of the membrane bilayer into a horizontal orientation or into the solvent. Finally, a transmembrane orientation is particularly favorable if the length of the peptide roughly matches the thickness of the membrane. Therefore, the process of membrane shaving is expected to reduce most of the proteins contained within the membrane bilayer to their transmembrane helices. There will be a few helices that are, in fact, unstable, some proteins that bind at the surface of the bilayer, and regions of structure that arise from apparent reentry in folding (such as the partial TMs in aquaporin); each of these would be removed in our treatment. Thus, the remaining helices are expected to be a lower bound for the amount of protein contained within the lipid bilayer.
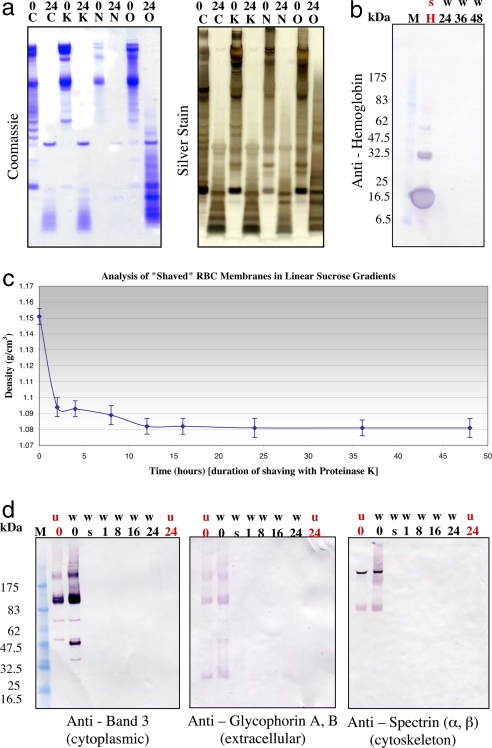
As a method to measure the completion of membrane shaving, the progressive proteolytic digestion of RBC plasma membrane open ghosts with 3.0 mg/ml proteinase K and 5 mM phenylmethylsulfonyl fluoride (PMSF) as the protease inhibitor was monitored as a function of digestion time. A series of three different types of membrane washes [1 M KCl (K), 10 mM NaOH (N), and 100 mM Na2CO3 (O)] were tested to remove peripherally associated proteins from the extracellular and cytoplasmic surfaces of the RBC open ghost membranes before conducting proteolytic digestion experiments (Fig. 1a). The 10 mM NaOH wash was the most effective and efficient wash medium (Fig. 1a). The procedure of using a 10 mM NaOH wash before shaving with 3.0 mg/ml proteinase K in 10 mM NaOH for 24 h also resulted in the elimination of contamination from hemoglobin (Fig. 1b). By using buoyant density measurements obtained from equilibrium centrifugation in linear sucrose gradients, the time progression of membrane shaving was extended to 48 h of digestion with proteinase K to establish completion of the shaving (Fig. 1c). A buoyant density plateau was reached by 24 h (Fig. 1c). By repeating the buoyant density analysis for each time point a total of 10 times, the density of untreated RBC ghost membranes was determined to be 1.15 ± 0.004 g/cm3 (Fig. 1c) and the density of the fully shaved RBC plasma membrane was found to be 1.081 ± 0.006 g/cm3 (Fig. 1c).
Fig. 1.
Time progression of membrane shaving. (a) Membrane washes. RBC open ghost membranes were washed in 1 M KCl (K), 10 mM NaOH (N), or 100 mM Na2CO3 (O), and then shaved with 3.0 mg/ml proteinase K in the respective wash buffer for 24 h (C, control, membranes were not washed). The membrane samples were then run on 1D SDS/PAGE gels and subjected to standard Coomassie and silver staining procedures. 0, no shaving with proteinase K; 24, membranes were shaved with 3.0 mg/ml proteinase K for 24 h. (b) Elimination of contamination from hemoglobin. RBC open ghost membranes washed (w) in 10 mM NaOH were shaved with 3.0 mg/ml proteinase K in 10 mM NaOH for various lengths of time up to 48 h. The membrane samples were then run on 1D SDS/PAGE gels and subjected to standard immunoblot analysis by using anti-hemoglobin (Sigma, H4890). Lane M, markers; lane H, hemoglobin supernatant (positive control for hemoglobin). (c) Time progression of membrane shaving by using buoyant density measurements from equilibrium centrifugation in linear sucrose gradients. RBC plasma membrane open ghosts were progressively shaved by proteolytic digestion with 3.0 mg/ml proteinase K. The proteolytic digestion was stopped with 5 mM PMSF as a function of time. At each time point, buoyant density measurements were made by using equilibrium centrifugation in continuous, linear sucrose gradients. Membranes were loaded on 5–45% continuous, linear sucrose gradients made by using the BioComp Gradient Master and spun at 215,578 × g for 24 h. Gradients were then fractionated by using the BioComp Gradient Fractionator. The density of sucrose at which the sample band of membranes was found by refractometry (equal to a buoyant density measurement for the membrane itself) was measured by using a temperature-controlled, small-volume, automatic refractometer (Rudolph Research Analytical, J57). The density at each time point was measured independently a total of ten times. (d) Time progression of membrane shaving by using immunoblot analysis. RBC open ghost membranes washed (w) in 10 mM NaOH and unwashed (u) RBC open ghost membranes were shaved with 3.0 mg/ml proteinase K in 10 mM NaOH for various lengths of time up to 24 h. 0, control, no proteinase K; s, shaved for 5 s. The membrane samples were then run on 1D SDS/PAGE gels and subjected to standard immunoblot analysis. Three different antibodies were analyzed in the immunoblot analysis including anti-band 3 (Sigma B9277) (cytoplasmic), anti-glycophorins A and B (Sigma G7650) (extracellular), and anti-spectrin (α and β) (Sigma S3396) (cytoskeleton). Lane M, markers.
The membrane shaving process was also monitored as a function of time by using immunoblot analysis (Fig. 1d) including anti-band 3 (Sigma B9277) (cytoplasmic), anti-glycophorin A and B (Sigma G7650) (extracellular), and anti-spectrin (α and β) (Sigma S3396) (cytoskeleton). These antibodies were used to monitor the shaving completion of human RBC membrane protein components on the cytoplasmic surface, the extracellular surface, and the erythrocyte cytoskeleton. Band 3 is estimated to account for 26% of the amount of total RBC ghost surface proteins, whereas glycophorin A is estimated to account for 1.6% and glycophorin B to account for 0.2% of the amount of total RBC ghost surface proteins (8). Spectrin is estimated to account for 25% of the amount of total RBC ghost surface proteins and 75% of the cytoskeletal mass (8). The immunoblot analysis for band 3, glycophorins A and B, and spectrin (α and β) revealed that by 24 h of shaving with proteinase K, a significant amount of extramembranous material had been successfully removed from the cytoplasmic surface, the extracellular surface, and the cytoskeleton of the RBC open ghost membrane samples (Fig. 1d).
The area occupancy of protein and lipid at the center of the human RBC plasma membrane was measured by using two different biochemical techniques: (i) a compositional analysis and (ii) a densitometric analysis. For the compositional analysis, dry weight measurements of RBC open ghost plasma membranes, fully shaved RBC plasma membranes (DWmembrane), lipid-extracted membrane protein from fully shaved RBC plasma membranes (DWprotein), total membrane lipid from RBC open ghost plasma membranes, and total lipid from fully shaved RBC plasma membranes (DWlipid) were determined (Table 1). Based on the dry weight measurements reported in Table 1, the area occupancy of the RBC plasma membrane was determined to be 20 ± 3% protein and 79 ± 5% lipid.
Table 1.
Dry weight measurements of RBC plasma membranes
| Sample | Dry weight, mg/1010 RBCs |
|---|---|
| RBC open ghost plasma membrane | 11.6 ± 0.1 |
| Fully shaved RBC plasma membrane (DWmembrane) | 6.5 ± 0.1 |
| Lipid-extracted membrane protein from fully shaved RBC plasma membranes (DWprotein) | 1.3 ± 0.2 |
| Total membrane lipid from RBC open ghost plasma membranes | 5.1 ± 0.3 |
| Total membrane lipid from fully shaved RBC plasma membranes (DWlipid) | 5.1 ± 0.3 |
Dry weight measurements for RBC open ghost plasma membranes, fully shaved RBC plasma membranes, lipid-extracted membrane protein from fully shaved RBC plasma membranes, total membrane lipid from RBC open ghost plasma membranes, and total membrane lipid from fully shaved RBC plasma membranes were determined by using standard methods. All dry weight measurements were repeated a total of 10 times for each of the five different types of samples.
A much more accurate method proved to be buoyant density measurements with use of equilibrium centrifugation in continuous, linear sucrose gradients. Buoyant density measurements were made of RBC open ghost plasma membranes, fully shaved RBC plasma membranes (Dmembrane), lipid-extracted membrane protein from fully shaved RBC plasma membranes (Dprotein), total membrane lipid from RBC open ghost plasma membranes, and total lipid from fully shaved RBC plasma membranes (Dlipid) (Table 2). Based on the buoyant density measurements reported in Table 2, the area occupancy of the RBC plasma membrane was determined based on the following equation, assuming additivity: Dmembrane = xDlipid + (1 − x)Dprotein, where x is the fractional area occupied by protein. The buoyant density measurements yielded an area occupancy of 23 ± 0.003% protein and 77 ± 0.003% lipid.
Table 2.
Buoyant density measurements of RBC plasma membranes
| Sample | Density, g/cm3 |
|---|---|
| RBC open ghost plasma membrane | 1.15 ± 0.004 |
| Fully shaved RBC plasma membrane (Dmembrane) | 1.081 ± 0.006 |
| Lipid-extracted membrane protein from fully shaved RBC plasma membranes (Dprotein) | 1.281 ± 0.003 |
| Total membrane lipid from RBC open ghost plasma membranes | 1.022 ± 0.004 |
| Total membrane lipid from fully shaved RBC plasma membranes (Dlipid) | 1.022 ± 0.004 |
Buoyant density measurements for RBC open ghost plasma membranes, fully shaved RBC plasma membranes, lipid-extracted membrane protein from fully shaved RBC plasma membranes, total membrane lipid from RBC open ghost plasma membranes, and total membrane lipid from fully shaved RBC plasma membranes were determined by using standard methods. All buoyant density measurements were repeated a total of 60 times for each of the five different types of samples.
Discussion
The measured value of at least ≈23% of the RBC plasma membrane area occupied by protein transmembrane regions is in close agreement with previous estimates (9, 10), and is significantly higher than that contemplated in most general models for membrane structure. If we add the perturbation by proteins bound at the surfaces of the bilayer and the influence of protein masses that may have been located in the bilayer but unstable to digestion, there may be little lipid that is in the state of thickness and dynamic motion present in a pure bilayer with the same lipid composition as the red blood cell membrane.
Methods
Purification of Human Red Blood Cells (RBCs) from Whole Blood.
Fresh whole human blood (tested negative for HBV, HCV, HIV 1&2, HTLV 1&2, and syphilis) with sodium heparin was obtained from Bioreclamation and stored at 4°C. RBCs were purified within 48 h of withdrawal from human donors. Human RBCs were initially isolated by centrifugation at 600 × g for 20 min at 4°C. After removing the plasma and buffy layers by careful suction, the RBC pellet was resuspended in cold isotonic buffer (145 mM NaCl, 5 mM KCl, and 5 mM Hepes, pH 7.4) by trituration and then recentrifuged at 2,000 × g for 10 min at 4°C. The RBCs were washed with the isotonic buffer a total of four times to completely remove the plasma and buffy layers. To remove white blood cells from the RBC suspensions, the samples were passed through white blood cell filters (Plasmodipur, Euro-diagnostica B.V.) a total of two times. At each step in the purification process, the upper RBC layer was removed. The final preparation of washed RBCs was resuspended in cold isotonic buffer at pH 7.4 to an approximate hematocrit of 50% and stored at 4°C. All RBC samples were stored at 4°C and used for analysis within 24 h of purification.
Purification of Human RBC Open Ghosts.
Packed RBCs (1 ml) were lysed by trituration and stirring on ice for 10 min in 100 ml of hypotonic lysing solution containing 15 mM KCl, 0.01 mM EDTA, 1 mM EGTA, and 5 mM Hepes, pH 6.0, at 4°C to reduce premature, spontaneous resealing of the erythrocyte ghosts. The membranes were then centrifuged at 12,000 × g for 10 min at 4°C. After discarding the supernatant, the membranes were then washed in 100 ml of hypotonic lysing solution without EGTA by trituration and recentrifuged at 12,000 × g for 10 min at 4°C. Once again, after discarding the supernatant, the membranes were washed in 100 ml of EGTA-free hypotonic lysing solution containing 2 mM Mg2+ by trituration and recentrifuged at 12,000 × g for 10 min at 4°C. After removing the supernatant a final time, the erythrocyte ghost membranes were concentrated in 4 ml of the EGTA-free lysing solution containing 2 mM Mg2+ by centrifugation at 12,000 × g for 10 min at 4°C. This method has been shown to dilute most cytosolic components of RBCs by 4-million-fold (11).
RBC Plasma Membrane Washes.
Purified RBC open ghost membranes (300 μl) were resuspended in 10 mM NaOH (30 ml) at 4°C. The membrane suspensions were then placed on a nutator and were rotated at medium speed at 4°C for 30 min. Washed membrane samples were then pelleted at 12,000 × g for 10 min and resuspended in fresh 10 mM NaOH (1 ml) at 4°C. The washed membrane samples were then immediately subjected to proteolytic digestion with proteinase K.
Proteolytic Digestion of RBC Plasma Membranes.
On purifying erythrocyte plasma membranes from human RBC ghosts and washing the membrane samples with 10 mM NaOH, the RBC ghost membranes were shaved by using proteinase K, a serine protease with broad cleavage specificity (12–15). Proteinase K is active over the pH range 7.5–12.0 but is most often used in the pH range 7.5–9.0 (14, 15). The activity of the enzyme is at a maximum at 37°C, but the activity is >80% of its maximum between 20°C and 60°C (14, 15). Washed RBC ghost membranes were pelleted in a microcentrifuge at 12,000 × g at room temperature for 10 min. The supernatant was then removed and discarded and the pellet was resuspended in 10 mM NaOH. Proteinase K was added to the membrane suspension at a final concentration of 3.0 mg/ml per 100 μl of washed membranes for each shaving reaction and the samples were left on a nutator at medium speed for varying durations of time (up to 48 h) at room temperature. The digestions with proteinase K were conducted at room temperature rather than at 37°C where protease activity is maximized to reduce the amount of resealing of the erythrocyte open ghost membranes that occurs completely within 6 min at 37°C (16, 17). At designated time points, PMSF, a protease inhibitor specific for proteinase K, was added at a final concentration of 5 mM and the samples were left on a nutator at medium speed for 30 min to be sure that all of the proteinase K activity was inhibited. The membranes were then pelleted in a microcentrifuge at 12,000 × g at room temperature for 10 min. The supernatant was then removed and discarded and the pellet was resuspended in 10 mM NaOH once again. This washing procedure of the fully washed and shaved RBC open ghost membranes was repeated two more times for a total of four washes. In the final round, the membrane samples were resuspended in buffer K (15 mM KCl, 5 mM Hepes, 2 mM Mg2+, 0.01 mM EDTA, pH 8.0). To be sure that proteinase K and PMSF were removed from the membrane samples with the series of four wash steps, samples of the supernatants and membrane pellets from each wash step were run on 1D SDS/PAGE gels and silver-stained. By the fourth wash step, all of the proteinase K and PMSF were removed from the membrane pellet.
One-Dimensional Gel Electrophoresis of RBC Plasma Membranes.
Membranes before and after washes with 10 mM NaOH and proteolytic digestion with proteinase K were analyzed by SDS/PAGE by using precast 4–12% NuPAGE Bis-Tris gels (Invitrogen). This particular gel system was chosen because it provides a much lower pH environment than traditional SDS/PAGE systems. The advantages of the lower pH include sharper band resolution and higher accuracy of results. NuPAGE Mes running buffer was used in all gel electrophoresis experiments (Invitrogen). Membrane samples were loaded onto the gel after heat denaturation at 70°C for 10 min according to the Invitrogen protocol for NuPAGE gels. Protein bands were visualized by using standard Coomassie blue and silver staining (SilverQuest Silver Staining Kit from Invitrogen) techniques.
Immunoblot Analysis of RBC Plasma Membranes.
The membrane shaving process was also monitored as a function of time by using immunoblot analysis as a means to assess the degree of shaving completion. The aim was to follow key structural elements: extracellular and intracellular digestion targets, as well as key cytoskeletal components. Standard immunoblot procedures were used for all samples by using the following antibody combinations:
Monoclonal Anti-Glycophorin A and B (Mouse) (Sigma G7650) (primary antibody at 1:800) and Anti-Mouse IgG (Fc specific)–Alkaline Phosphatase Antibody (Goat) (Sigma A1418) (secondary antibody at 1:20,000). This antibody recognizes an epitope located in the extracellular domain of glycophorin A and glycophorin B, within amino acid residues 1–26 (the fragment that is MN-unrelated and identical in glycophorins A and B) (18, 19).
Monoclonal Anti-Band 3 Antibody (Mouse) (Sigma B9277) (primary antibody at 1:5,000) and Anti-Mouse IgG (Fc specific)–Alkaline Phosphatase Antibody (Goat) (Sigma A1418) (secondary antibody at 1:20,000). This antibody recognizes an epitope located on the cytoplasmic N terminus of band 3 (20). Three other groups of bands localized by this antibody are in the regions of 55–60 kDa, 38–42 kDa, and 21–26 kDa (20). The 20- and 40-kDa fragments are formed by cleavage on the cytoplasmic side (20). Even though the 60-kDa fragment is transmembrane protein, its cleavage site is available at the external surface of the erythrocyte (20).
Monoclonal Anti-Spectrin (α and β) Antibody (Mouse) (Sigma S3396) (primary antibody at 1:500) and Anti-Mouse IgG (Fc specific)–Alkaline Phosphatase Antibody (Goat) (Sigma A1418) (secondary antibody at 1:20,000). This antibody localizes the 220-kDa (β) and 240-kDa (α) bands of human erythrocyte spectrin, the most abundant protein in the erythrocyte cytoskeleton (Sigma Product Information Sheet, no. S3396). In addition, a number of polypeptides in the region of 170, 150, 85, and 45 kDa react with this antibody (Sigma Product Information Sheet, no. S3396).
Anti-Human Hemoglobin (Rabbit) (Sigma H4890) (primary antibody at 1:50) and Anti-Rabbit IgG (whole molecule)–Alkaline Phosphatase Antibody (Goat) (Sigma A3812) (secondary antibody at 1:40,000). This hemoglobin antiserum was determined to be specific for hemoglobin and it showed strong reactivity with human hemoglobin at 1 mg/ml (Sigma Product Information Sheet, no. H4890).
Characterization of Membrane Shaving by Proteolytic Digestion by Buoyant Density Measurements from Equilibrium Centrifugation in Linear Sucrose Gradients.
As an alternative method to follow the membrane shaving process, the progressive proteolytic digestion of RBC plasma membrane ghosts was monitored by using buoyant density measurements obtained from equilibrium centrifugation in linear sucrose gradients. RBC plasma membrane open ghosts were progressively shaved by proteolytic digestion with 3.0 mg/ml proteinase K. The proteolytic digestion was stopped with 5 mM PMSF as a function of time. At each time point, buoyant density measurements were made by using equilibrium centrifugation in sucrose gradients. Membranes were loaded on 5–45% continuous, linear sucrose gradients made by using the BioComp Gradient Master and spun at 215,578 × g for 24 h. Gradients were fractionated by using the BioComp Gradient Fractionator. The density of sucrose at which the sample band of membranes was found, by refractometry (equal to a buoyant density measurement for the membrane itself), was measured by using a temperature-controlled, small-volume, automatic refractometer (Rudolph Research Analytical, J57). By measuring the refractive index of the membrane sample in a particular sucrose gradient fraction, the buoyant density of this sample was determined by using a simple conversion based on published tables (ICUMSA SPS-3). By using the aforementioned technique, buoyant density measurements were obtained for RBC plasma membrane open ghosts as a function of proteolytic digestion time.
Lipid Extraction of RBC Open Ghosts and Fully Shaved RBC Plasma Membranes.
Lipids were extracted from RBC open ghosts and fully shaved RBC plasma membranes by using chloroform/isopropyl alcohol 7:11 (vol/vol) according to the Rose–Oklander method of lipid extraction (21). This method was chosen over the well known Folch extraction procedure because the Folch extraction is plagued by significant contamination with heme (22). The Rose–Oklander method of lipid extraction was also chosen as the preferred method because it results in full phospholipid recovery (21). Reagent grade chloroform (redistilled) and isopropyl alcohol were used in all lipid extraction experiments. RBCs were purified from whole human blood according to the methods described earlier. RBC open ghost plasma membranes were then purified according to the methods described earlier, and the lipid extraction procedures detailed below were carried out on this sample. Fully shaved RBC plasma membranes were also subjected to lipid extraction according to the Rose–Oklander method of lipid extraction. Packed RBC open ghosts or fully shaved RBC plasma membranes (1 ml) were pipetted by using a siliconized serological pipette into a 20-ml tube with a teflon-lined screw cap. One milliliter of distilled water was added to the suspension of packed RBC open ghosts (or fully shaved RBC plasma membranes) and the contents of the tube were mixed with a vortex and allowed to stand for 15 min. Isopropyl alcohol (11 ml) was then added to the RBC open ghost membrane (or fully shaved RBC plasma membrane) suspension slowly with continuous mixing. The RBC membrane samples gradually turned dark and clumped together. After 1 h with occasional mixing, 7.0 ml of chloroform was added to the RBC open ghost (or fully shaved RBC plasma membrane) suspension and mixed. At the end of another hour, the tube containing the RBC open ghost (or fully shaved RBC plasma membrane) suspension was centrifuged at 500 × g for 30 min. The supernatant was removed and stored at 4°C. The pellet, representing the lipid-extracted membrane protein from RBC open ghosts or fully shaved RBC plasma membranes was carefully separated from the supernatant, and the Rose–Oklander extraction procedure was repeated two more times to remove the lipid bound tightly to the membrane protein. After the third and final extraction, the lipid-extracted membrane protein was lyophilized for 12–24 h, separate from the lipid material. The extracted RBC membrane lipid from all three rounds of the Rose–Oklander extraction procedure was pooled together and lyophilized for 12–24 h. All lipid extractions were carried out at 4°C and at room temperature. No differences in extraction efficiencies were observed at the two different temperatures. Particular care was taken in controlling both the pellet volume and the water volume so that the water volume did not exceed the pellet volume under any circumstances. Under these conditions, no differences in lipid extraction between RBC open ghosts or fully shaved RBC plasma membranes were observed.
Dry Weight Analysis of RBC Plasma Membranes.
Total RBC plasma membrane lipid from RBC open ghosts and fully shaved RBC plasma membranes was prepared as described earlier. The total lipid from these two different samples was lyophilized and dried to constant weight under vacuum. The dry weight of the total lipid was then measured by using a precision microbalance. The lipid extraction and lipid dry weight measurements of both sample types (RBC open ghosts and fully shaved RBC plasma membranes) were repeated a total of 10 times. Lipid-extracted membrane protein from RBC open ghosts and fully shaved RBC plasma membranes was prepared as described earlier. The total protein from these two different samples was lyophilized and dried to constant weight under vacuum. The dry weight of the total lipid-extracted membrane protein was then measured by using a precision microbalance. The lipid extraction and protein dry weight measurements of both sample types (lipid-extracted membrane protein from RBC open ghosts and fully shaved RBC plasma membranes) were repeated a total of 10 times. The dry weight of purified RBC open ghost plasma membranes and fully shaved RBC plasma membranes were also measured a total of 10 times each by using the procedures described earlier.
Determination of the Buoyant Density of the Protein Component of Fully Shaved RBC Plasma Membranes.
Lipid-extracted membrane protein from fully shaved RBC plasma membranes was purified according to the methods presented previously. The lipid-extracted membrane protein was resuspended in a buffer containing 25 mM Hepes-KOH and 5 mM MgCl2 at pH 7.4 (same buffer as the shaved RBC plasma membrane and the extracted RBC lipid). The buoyant density of the total lipid-extracted membrane protein from fully shaved RBC plasma membranes was measured according to the methods detailed earlier. The only modification to these methods was that a series of density gradients (20–60%, 30–60%, and 40–60% sucrose) were used in combination to determine the precise protein density measurement. The buoyant density of the lipid-extracted membrane protein from “fully” shaved RBC plasma membranes was measured in six separate sucrose gradients in a total of 10 experimental trials for a total of 60 measurements of protein density.
Determination of the Lipid Density of RBC Open Ghost Membranes and Fully Shaved RBC Plasma Membranes.
The total membrane lipid from RBC open ghost membranes and from fully shaved RBC plasma membranes was purified according to the methods presented earlier. Unilamellar lipid vesicles were made from the extracted membrane lipid from RBC open ghosts and from fully shaved RBC plasma membranes by extrusion. The dried lipid film from the total membrane lipid purification of RBC open ghost membranes and fully shaved RBC plasma membranes was rehydrated in buffer (25 mM Hepes-KOH, 5 mM MgCl2, pH 7.4) at 40°C, for a total of 2 h (with vortexing), and passed through 100-nm pores in a MiniExtruder (Avanti Polar Lipids) a total of 20 times. The buoyant density of the total lipid extracted from RBC open ghost membranes and from fully shaved RBC plasma membranes (in the form of unilamellar lipid vesicles) was measured according to the methods detailed earlier. The only modification to these methods was that a series of density gradients (5–20%, 5–30%, and 5–45% sucrose) were used in combination to determine the precise lipid density measurement. The buoyant density of the total membrane lipid from RBC open ghost membranes and from fully shaved RBC plasma membranes were each measured in six separate sucrose gradients in a total of 10 experimental trials for a total of 60 measurements of lipid density of each sample type.
ACKNOWLEDGMENTS.
We thank the members of the D.M.E. laboratory for discussions. This work was supported in part by National Institutes of Health Grants GM070895 and GM073857 (to D.M.E.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Takamori S, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 2.Henderson R, Unwin PN. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975;257:28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- 3.Henderson R. The structure of the purple membrane from Halobacterium hallobium: Analysis of the X-ray diffraction pattern. J Mol Biol. 1975;93:123–138. doi: 10.1016/0022-2836(75)90123-0. [DOI] [PubMed] [Google Scholar]
- 4.Popot JL, Engelman DM. Helical membrane protein folding, stability, and evolution. Annu Rev Biochem. 2000;69:881–922. doi: 10.1146/annurev.biochem.69.1.881. [DOI] [PubMed] [Google Scholar]
- 5.Engelman DM, Steitz TA. The spontaneous insertion of proteins into and across membranes: The helical hairpin hypothesis. Cell. 1981;23:411–422. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- 6.Kataoka M, et al. Bacteriorhodopsin reconstituted from two individual helices and the complementary five-helix fragment is photoactive. Photochem Photobiol. 1992;56:895–901. doi: 10.1111/j.1751-1097.1992.tb09710.x. [DOI] [PubMed] [Google Scholar]
- 7.Marti T. Refolding of bacteriorhodopsin from expressed polypeptide fragments. J Biol Chem. 1998;273:9312–9322. doi: 10.1074/jbc.273.15.9312. [DOI] [PubMed] [Google Scholar]
- 8.Beutler E, Williams WJ. Williams Hematology. New York: McGraw–Hill; 2001. pp. xxvii–1941. [Google Scholar]
- 9.Engelman DM. Surface area per lipid molecule in the intact membrane of the human red cell. Nature. 1969;223:1279–1280. doi: 10.1038/2231279a0. [DOI] [PubMed] [Google Scholar]
- 10.Guidotti G. Membrane proteins. Annu Rev Biochem. 1972;41:731–752. doi: 10.1146/annurev.bi.41.070172.003503. [DOI] [PubMed] [Google Scholar]
- 11.James-Kracke MR. Calmodulin activation of the Ca2+ pump revealed by fluorescent chelator dyes in human red blood cell ghosts. J Gen Physiol. 1992;99:41–62. doi: 10.1085/jgp.99.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu CC, Yates JR., III The application of mass spectrometry to membrane proteomics. Nat Biotechnol. 2003;21:262–267. doi: 10.1038/nbt0303-262. [DOI] [PubMed] [Google Scholar]
- 13.Wu CC, MacCoss MJ, Howell KE, Yates JR., III A method for the comprehensive proteomic analysis of membrane proteins. Nat Biotechnol. 2003;21:532–538. doi: 10.1038/nbt819. [DOI] [PubMed] [Google Scholar]
- 14.Kraus E, Femfert U. Proteinase K from the mold Tritirachium album Limber. Specificity and mode of action. Hoppe Seylers Z Physiol Chem. 1976;357:937–947. doi: 10.1515/bchm2.1976.357.2.937. [DOI] [PubMed] [Google Scholar]
- 15.Ebeling W, et al. Proteinase K from Tritirachium album Limber. Eur J Biochem. 1974;47:91–97. doi: 10.1111/j.1432-1033.1974.tb03671.x. [DOI] [PubMed] [Google Scholar]
- 16.Steck TL, Kant JA. Preparation of impermeable ghosts and inside-out vesicles from human erythrocyte membranes. Methods Enzymol. 1974;31:172–180. doi: 10.1016/0076-6879(74)31019-1. [DOI] [PubMed] [Google Scholar]
- 17.Dodge JT, Mitchell C, Hanahan DJ. The preparation and chemical characteristics of hemoglobin-free ghosts of human erythrocytes. Arch Biochem Biophys. 1963;100:119–130. doi: 10.1016/0003-9861(63)90042-0. [DOI] [PubMed] [Google Scholar]
- 18.Telen MJ, Bolk TA. Human red cell antigens. IV. The abnormal sialoglycoprotein of Gerbich-negative red cells. Transfusion. 1987;27:309–314. doi: 10.1046/j.1537-2995.1987.27487264736.x. [DOI] [PubMed] [Google Scholar]
- 19.Telen MJ, Scearce RM, Haynes BF. Human erythrocyte antigens. III. Characterization of a panel of murine monoclonal antibodies that react with human erythrocyte and erythroid precursor membranes. Vox Sang. 1987;52:236–243. doi: 10.1111/j.1423-0410.1987.tb03035.x. [DOI] [PubMed] [Google Scholar]
- 20.Czerwinski M, et al. Degradation of the human erythrocyte membrane band 3 studied with monoclonal antibody directed against an epitope on the cytoplasmic fragment of band 3. Eur J Biochem. 1988;174:647–654. doi: 10.1111/j.1432-1033.1988.tb14147.x. [DOI] [PubMed] [Google Scholar]
- 21.Rose HG, Oklander M. Improved procedure for the extraction of lipids from human erythrocytes. J Lipid Res. 1965;6:428–431. [PubMed] [Google Scholar]
- 22.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]