Abstract
Cilia are present on nearly all cell types in mammals and perform remarkably diverse functions. However, the mechanisms underlying ciliogenesis are unclear. Here, we cloned a previously uncharacterized highly conserved gene, stumpy, located on mouse chromosome 7. Stumpy was ubiquitously expressed, and conditional loss in mouse resulted in complete penetrance of perinatal hydrocephalus (HC) and severe polycystic kidney disease (PKD). We found that cilia in stumpy mutant brain and kidney cells were absent or markedly deformed, resulting in defective flow of cerebrospinal fluid. Stumpy colocalized with ciliary basal bodies, physically interacted with γ-tubulin, and was present along ciliary axonemes, suggesting that stumpy plays a role in ciliary axoneme extension. Therefore, stumpy is essential for ciliogenesis and may be involved in the pathogenesis of human congenital malformations such as HC and PKD.
Keywords: brain, cilia, kidney, intraflagellar transport protein
Cilia are evolutionarily conserved organelles and are generally classified as primary (sensory, nonmotile) or motile [e.g., those expressed by ependymal cells in the brain that generate force necessary for cerebrospinal fluid (CSF) flow] (1, 2). The loss or defective function of cilia has been implicated in diseases such as obesity, diabetes, situs inversus, hydrocephalus (HC), and polycystic kidney disease (PKD) (3–8). The mechanisms regulating growth and stability of mammalian cilia during development are poorly understood. In attempting to explore the function of the transforming growth factor β1 gene (tgfb1) during development (9, 10), we established a conditional deficient mouse by inserting loxP sites 5′ of the promoter region of tgfb1 and 3′ of tgfb1 exon 1 (within the first intron). Importantly, during the generation of these mice, there were no known or predicted genes in close physical proximity to the tgfb1 promoter region. However, a hypothetical gene (ENSMUSG00000063439) was later predicted in this region encoding cDNA sequence BC028440 (GenBank accession no. NM_172148) (11). We have termed this hypothetical gene stumpy as a result of experimental evidence presented here showing a key role of stumpy in the growth of primary and motile cilia.
Results
Establishment of Conditional Stumpy Mutant Mice and Expression Analysis.
We deleted this genomic region in the brain by crossing floxed mice to nestin-cre deleter transgenic animals and determined deletion efficiency by PCR analysis of the floxed (but not deleted), deleted, or wild-type alleles (Fig. 1 A and Materials and Methods). The deleted allele was most obviously detected in DNA prepared from brains of fl/fl mice but was undetectable in control +/+ animals (Fig. 1B). Remarkably, stumpy mRNA expression was dramatically reduced from 41.0- to 84.7-fold by Affymetrix GeneChip, whereas, unexpectedly, tgfb1 mRNA expression was modestly (but significantly) increased by 1.8- to 3.0-fold in fl/fl vs. +/fl mice, raising the possibility of a stumpy-tgfb1 read-through transcript created by the deleted allele (Fig. 1C). Three approaches were taken to test this hypothesis: donor splice/recipient bioinformatics, RT-PCR, and Northern blot. Bioinformatics predicted a chimeric in-frame readthrough transcript encoded by the deleted allele, comprised of exons 2–3 of stumpy and exons 2–7 of tgfb1. Complementary DNA was prepared from postnatal (P) day 12 brains of +/+, +/fl, or fl/fl mice, and RT-PCR was performed by using various primer combinations, which revealed the presence of a readthrough transcript in +/fl and fl/fl mice (Fig. 1D). Sequencing of this chimeric stumpy-tgfb1 PCR product showed an in-frame transcript, and Northern blot analysis (Fig. 1E) further supported this conclusion. Therefore, we focused our efforts on characterizing the adjacent gene to determine whether this gene might underlie phenotypic defects present in these targeted mutant mice.
Fig. 1.
Generation of stumpy mutant mice and gene expression analysis. (A) Diagram showing insertion of loxP sites flanking exon 4 of stumpy and exon 1 of tgfb1. Positions of primers a–d used for RT-PCR or Northern blot probe are shown. (B) Multiplex PCR designed to detect floxed (but not deleted), deleted, or wild-type alleles at P0 from floxed but not deleted (+/+) or homozygous deleted (fl/fl) mice. (C) Affymetrix GeneChip results showing mRNA expression of stumpy or tgfb1 in P0 and P12 brains from fl/fl vs. heterozygous (+/fl) mice. (D) RT-PCR using primer pairs shown in A. (E) Northern blot using a probe amplified from primers c and d shown in A reveals tgfb1 or a chimeric stumpy-tgfb1 in-frame read-through transcript in brains from fl/fl vs. +/fl mice. (F) Q-PCR analysis of stumpy relative to hprt1 in mouse organs and cells. (G) RNA in situ hybridization with stumpy α-sense or sense (control) probes in wild-type C57BL/6 mouse brains. Higher-magnification images (Lower, scale bar denotes 50 μm, calculated for Lower Left and Lower Right) reveal expression in cells lining the 4v, the S. aq, Lv, and in C. Plexus (cp). (H) Brain homogenates from P12 +/+ or fl/fl mice were Western blotted with stumpy antibody (Upper). Note the ≈19-kDa band corresponding to the predicted molecular mass for stumpy. * shows a possible stumpy degraded product, and actin is shown Lower as a loading control.
We initially characterized stumpy expression by quantitative real-time RT-PCR (Q-PCR, normalized to hprt1 levels) by using cDNA prepared from organs of adult C57BL/6 mice, cultured Neuro-2a (N2a) neuroblastoma cells, or primary mouse microglia. Stumpy expression was widespread, with highest levels in mouse thymus and skeletal muscle, but we also detected expression in brain, N2a cells, and microglia (Fig. 1F). Using RNA in situ hybridization, we observed stumpy transcripts throughout the entire early postnatal brain, including cells lining the ventricles and Sylvian aqueduct (S. aq), and choroid plexus (C. Plexus) cells (Fig. 1G). To characterize stumpy mRNA, we then performed 5′- and 3′-RACE using cDNA prepared from adult C57BL/6 mouse brain, kidney, or liver. Interestingly, 5′-RACE revealed a noncoding exon 1 [supporting information (SI) Fig. 5 A and B], and 3′-RACE allowed for characterization of the 3′-UTR of stumpy mRNA (SI Fig. 5C). Bioinformatics analysis of stumpy mRNA (GenBank accession no. BC028440) revealed high conservation between vertebrate and invertebrate species (SI Fig. 6). SI Fig. 7A shows the complete coding sequence for stumpy (determined from RACE and cloning/sequencing analyses) with predicted translation indicated below (corresponding to hypothetical protein LOC232987, GenBank accession no. NP_742160) (11). Notably, protein family (Pfam) analysis revealed a B9/C2 calcium/lipid-binding region (Pfam references PF07162 and PF00168) spanning amino acids 4–106 (SI Fig. 7B). B9 is a highly conserved domain of unknown function present in 41 known or predicted eukaryotic proteins in both vertebrates and invertebrates (SI Fig. 8). The B9 domain is present in the ciliary proteome database (12), and a protein containing this domain, MKS1, has recently been linked to ciliogenesis (13). To confirm stumpy deficiency at the protein level, we generated a stumpy antibody and probed brain homogenates from +/+ and fl/fl mice. We noted the presence of a ≈19-kDa band, corresponding to the predicted molecular mass of stumpy, which was nearly absent in conditional stumpy deficient mice (Fig. 1H).
Perinatal HC and PKD in Conditional Stumpy Mutant Mice.
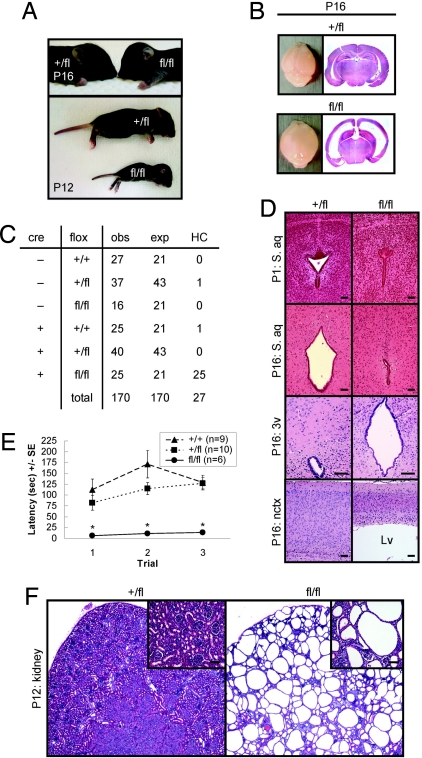
Upon gross anatomical observation, it was readily apparent that homozygous fl/fl stumpy mutant mice had a “domed” head that became prominent from P12 to P16, and was not observed in heterozygous (+/fl) animals (Fig. 2A). This phenotype suggested the presence of HC, a condition that results from cerebral ventricle dilation secondary to altered flow of CSF (14). Consistently, we found that fl/fl mice displayed an enlarged, balloon-like and translucent neocortex (nctx) by P16 (Fig. 2B). H&E staining of coronal brain sections revealed pronounced ventricular enlargement in fl/fl mice (Fig. 2B). Also at this age, bodies of stumpy fl/fl mice weighed significantly less than littermate controls, whereas their brains weighed more (likely because of increased CSF volume) (n = 7 for +/+, n = 10 for +/fl, and n = 6 for fl/fl; mean weights in grams ± SE, brain: 0.39 ± 0.02, 0.39 ± 0.02, 0.52 ± 0.04, P < 0.001; body: 7.59 ± 0.15, 7.14 ± 0.74, 4.34 ± 0.63, P < 0.001). Crosses of stumpy mutant mice heterozygous for the floxed allele and either positive (cre +, +/fl) or negative (cre−, +/fl) for the nestin-cre transgene yielded numbers for each genotype of observed (obs) offspring that did not significantly differ from expected (exp) Mendelian frequencies (χ(5)2 = 5.48, P = 0.36) (Fig. 2C), suggesting that fl/fl mice were not being lost due to embryonic lethality. Notably, perinatal HC occurred with complete penetrance in homozygous stumpy mutant mice (fl/fl, designated cre +, fl/fl in Fig. 2C). All of the hydrocephalic animals in this study died by the time of weaning (P21). Two control pups developed the disease, which may represent spontaneous incidence of hydrocephaly in C57BL/6 mice (<1%) (15). We used an important control mouse group in our analyses (+/fl mice, heterozygous for the nestin-cre transgene) to rule out cre-mediated effects in fl/fl mice (heterozygous for the nestin-cre transgene and homozygous for the floxed allele), given that mice homozygous for this transgene develop microencephaly and hydrocephaly (16). We did not observe any incidence of HC in +/fl mice.
Fig. 2.
PTvHC communicating HC and polycystic kidney disease in stumpy mutant mice. (A) Photographs of representative +fl and fl/fl mice (Upper) showing classic “domed head” of hydrocephalic fl/fl mice at P16. By P12, fl/fl mice consistently appear runted (Lower). (B) Whole brains isolated from +/fl and fl/fl mice (Left), and low-magnification photomicrographs of H&E-stained coronal brain sections (Right) showing pronounced venticulomegaly. (C) Nestin-cre-positive mice bearing one floxed allele (cre +, +/fl) were crossed with nestin-cre-negative –, +/fl mice, and all deficient offspring (cre +, fl/fl) manifested perinatal hydrocephaly. An additional 20/104 hydrocephalic cre +, fl/fl mice were identified from cre +, +/fl x cre−, fl/fl breedings. (D) Coronal H&E-stained sections show complete stenosis of the S. aq in fl/fl brains (Right). An enlarged 3v and thinner neocortex (nctx) are also observed in fl/fl brains (Right). (Scale bars, 100 μm.) (E) Motor function in wild-type (+/+, n = 9), +/fl (n = 10), and fl/fl mice (n = 6) was assayed by accelerating roto-rod, and data are represented as means ± SE of time to fall (latency, in seconds) for each of three consecutive trials. *, P < 0.001 for fl/fl vs. +/fl or +/+. (F) H&E-stained sections from +/fl or fl/fl P12 kidneys show evidence of polycystic kidney disease. High-magnification Insets are shown in the upper right corners. (Scale bars, 20 μm.)
Congenital HC in mice (and in humans) can be attributed to blockage of CSF flow, a failure in CSF absorption, or overproduction of CSF (15). Histological analysis of brains from +/fl vs. fl/fl mice at P1 (but not at P0) revealed stenosis of the mesencephalic S. aq (which joins the third and fourth ventricles) in fl/fl mice before the onset of ventriculomegaly (Fig. 2D). By P16, the S. aq remained stenotic in fl/fl mice and was accompanied by marked third ventricle (3v) and lateral ventricle (Lv) enlargement, producing a thinner neocortex (Fig. 2D). These pathological changes were consistent with a form of communicating HC known as perinatal triventricular HC (PTvHC) characterized by another group in axonemal dynein heavy chain gene mdnah5 mutant mice (3). PTvHC in fl/fl mice was associated with tonic seizures and severe motor dysfunction (data not shown), and behavioral assay of motor function at P16 revealed significant (P < 0.001) impairment in fl/fl mice in an accelerating rotating rod apparatus (Fig. 2E). In addition to efficiently recombining cre/loxP in the central nervous system, nestin-cre transgenic mice also delete floxed alleles in the kidney (17). Accordingly, we noted stumpy deletion in the kidney (Fig. 1B), which prompted us to examine kidney sections by H&E histochemistry. Strikingly, all of the fl/fl mice examined had prominent evidence of PKD (Fig. 2F). Confocal microscopy with lectins lotus tetragonolobus agglutinin and dolichos biflorus agglutinin revealed that both proximal and distal segments of the nephron were cystic in fl/fl mice (data not shown).
Stumpy Conditional Mutant Mice Lack Functional Intact Cilia.
We hypothesized that HC in conditional stumpy mutant mice might be due to a defect in the ependymal cell layer that regulates CSF flow. High magnification of H&E-stained brain sections from fl/fl vs. +/fl mice revealed that ependymal cells in the 3v appeared dysmorphic but apparently intact (Fig. 3A). More notably, we observed that cilia on ependymal cells lining the S. aq and the cerebral ventricles (3v is shown, similar results were observed in the Lv and 4v) were nearly absent in fl/fl mice (Fig. 3A). This observation suggests that stumpy may be implicated in ciliogenesis. Indeed, stumpy fl/fl mutants had dramatically fewer motile cilia (as detected by antibody against acetyl α-tubulin, a major structural component of ciliary axonemes) (18–20) in the foramen of Monroe (F. Monroe) and in the fourth ventricle (4v; similar findings were made in the Lv and 3v) (Fig. 3B). Ciliary axonemes that were present in fl/fl mice had a “stumpy-like” appearance. Interestingly, ciliary rootlets (at the ciliary base, detected by rootletin antibody) (21) appeared normal in cells lining the CSF system of +/fl and fl/fl mice (Fig. 3B). Similar results were observed for primary cilia on C. Plexus cells; specifically, acetyl α-tubulin antibody-positive ciliary axonemes were almost completely absent, whereas ciliary basal bodies (bb, another anchoring component of cilia, detected by γ-tubulin antibody) (22) or rootletin-positivity was comparable between P4 fl/fl and +/fl brains (Fig. 3B). Electron microscopy analysis revealed numerous intact bb structures in the S. aq of P0 fl/fl mice; however, cells lining the ventricles either completely lacked proper ciliary axonemal extensions, or had markedly deformed structures (Fig. 3C). Although +/fl mice displayed robust cilia, serial sectioning confirmed the lack of axonemal protrusion from bb structures in the fl/fl mouse (Fig. 3C). Recent evidence has shown that defective cilia lead to kidney cyst formation (6), leading us to examine cilia on kidney tubule cells. We noted a pattern of results similar to what we observed in the brain: ciliary axonemes in fl/fl mice were either completely absent or, when present, often had a “stumpy” appearance (Fig. 3D).
Fig. 3.
Ciliogenesis defects in stumpy mutant mice. (A) P16 +/fl or fl/fl H&E-stained sections showing the S. aq (Upper) and the ependyma of 3v (Lower). Arrowheads indicate cilia. (Scale bars, 20 μm.) (B) Vibratome brain sections from the F. Monroe (Top) or cryosections from 4v or the C. Plexus from +/fl (Left) or fl/fl mice (Right) at P4 were immunostained for acetylated α-tubulin (against the ciliary axoneme), rootletin (against ciliary rootlets), and/or γ-tubulin (recognizes ciliary bb) (colors for each antibody are indicated), and nuclear counterstained with DAPI (blue). Images were captured by using a Zeiss ApoTome-equipped fluorescence microscope (Top) or a confocal microscope (all other images). Arrowheads in the C. Plexus show ciliary axonemes. (Scale bars, 10 μm.) (C) Transmission electron micrographs of cells lining the S. aq are shown from P0 +/fl (Left) or fl/fl mice (Right and Bottom). Higher magnifications of Insets (Top) are shown below in Middle (Middle Insets show additional examples). Note the presence of bb structures in both +/fl and fl/fl mice, whereas fl/fl cells either do not have ciliary axonemes (c) or they appear deformed. Bottom show serial EM sections (0–420 nm) through bb structures of the boxed region, demonstrating the absence of c structures in a mutant S. aq cell. [Scale bars: Top, 2 μm; Middle and Bottom, 0.5 μm.] (D) Confocal microscopy was performed on kidneys isolated from P12 +/fl or fl/fl mice after α-tubulin immunostaining. Arrowheads indicate cilia. (Scale bars, 20 μm.)
We tested whether cilia beating frequency was altered in fl/fl mice vs. +/fl littermate controls by using an electrophysiological technique. Although generally difficult to detect ciliary movement in the S. aq of fl/fl mice (i.e., we observed a flat line), likely due to the widespread loss of cilia, beating frequency of remaining cilia was not significantly different in fl/+ vs. fl/fl mice (number, mean ± SD for fl/+ vs. fl/fl mice: n = 5, 12.8 ± 2.2 Hz vs. n = 3, 10.9 ± 1.2 Hz, P > 0.05) (SI Fig. 9). The presence of few cilia may be due to incomplete deletion in a small fraction of conditional stumpy mutant cells, and suggests that stumpy may be required more for growth (ciliogenesis) rather than function per se. Finally, we assayed CSF flow in +/fl vs. fl/fl mice by acutely injecting India Ink (as a tracer) into the CSF system of live brain slices and monitored flow by real-time video microscopy. This analysis revealed a profound defect in CSF flow in fl/fl mice (SI Movie 1). Thus, stumpy appears to be essential for cilia outgrowth, the lack of which results in failure of CSF flow.
Defective Ciliogenesis in Stumpy Conditional Mutant Mice Is tgfb1 Independent.
Conditional stumpy mutants lack exon 4 of stumpy and exon 1 of tgfb1 (Fig. 1), resulting in generation of a chimeric, in-frame stumpy-tgfb1 readthrough mRNA transcript. To determine whether deficiency in tgfb1 exon 1 contributed to the ciliogenesis defect and HC observed in fl/fl mice, we crossed heterozygous (+/fl) stumpy mutant mice with another mouse that is heterozygous for egfp knocked-in to exon 1 of tgfb1, resulting in a null tgfb1 frame-shift mutation (progeny heterozygous for both alleles are designated +/fl/tgfb1egfpKI+; see SI Fig. 10A for a diagram). These mice did not manifest HC, and had normal ciliogenesis in ependymal cells lining the ventricles and the C. Plexus (SI Fig. 10A). Alternatively, the defects in conditional stumpy mutants could be owed to a gain of (dys) function of the stumpy-tgfb1 chimera. To test this, we cloned the stumpy complete coding sequence into an adenovirus with a bicistronic EGFP reporter (pAdTrack) (23). We then intracerebroventricularly (i.c.v.) injected embryonic day 14.5 fl/fl embryos with adenovirus and analyzed their brains at P12. The adenovirus predominantly infected C. Plexus cells (SI Fig. 10B). Remarkably, we observed that EGFP-positive cells clearly had multiple detectable α-tubulin-positive ciliary axonemes, whereas surrounding EGFP-negative cells lacked these structures, or they appeared “stumpy” (SI Fig. 10B). Finally, we evaluated mice conditionally deficient in brain TGF-β receptor II (required for TGF-β signaling, obtained by crossing TGF-β receptor II floxed mice with nestin-cre transgenics), and these animals appear normal without incidence of hydrocephaly (data not shown). Together, these data suggest that neither tgfb1 (signaling) deficiency nor stumpy-tgfb1 chimerism underlies the phenotype, and that stumpy deficiency is responsible for both the cilogenesis defect and HC.
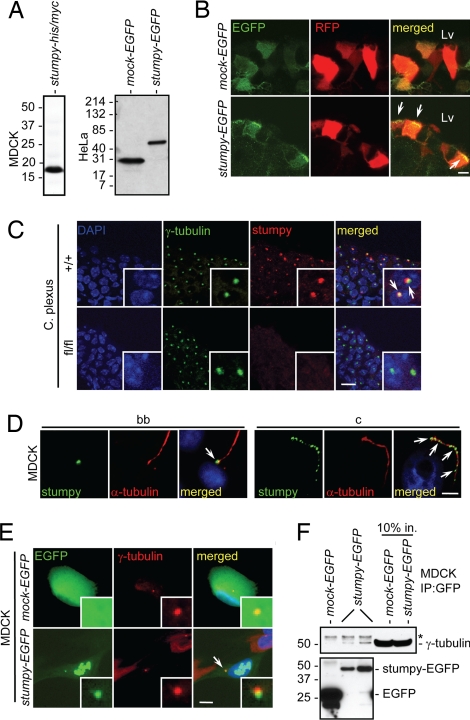
Stumpy Localizes to the Ciliary-bb and Physically Interacts with γ-Tubulin.
To gain insight into the function and localization of stumpy, we first generated either poly-histidine/poly-myc (his/myc) or EGFP-tagged stumpy constructs and transiently transfected canine kidney epithelial (MDCK) or human cervical cancer (HeLa) cells. Western blots of transfectants with myc or GFP antibodies revealed that stumpy protein was ≈19 kDa (Fig. 4A), consistent with Western blot of mouse brain homogenates with stumpy antibody (Fig. 1H). We then coelectroporated mouse fetus brain Lvs with red fluorescence protein (RFP) and mock-EGFP or stumpy-EGFP constructs and imaged Lv ependymal cells by confocal microscopy. We observed that stumpy-EGFP (but not mock-EGFP) localized to the apical surface of RFP-positive ependymal cells, reminiscent of ciliary bb structures (Fig. 4B). To directly assess whether stumpy localized to the bb, we immunostained brains from +/+ or fl/fl mice with γ-tubulin and stumpy antibodies. We found colocalization of these proteins in ependymal cells (data not shown) and in C. Plexus (Fig. 4C). Consistently, MDCK cells showed stumpy signal at the base of α-tubulin-positive ciliary axonemes by immunostaining (Fig. 4D). Further, we observed a punctate pattern of stumpy immunoreactivity along ciliary axonemes (Fig. 4D). To further confirm γ-tubulin colocalization and to test for putative physical interaction between stumpy and γ-tubulin, we generated MDCK cell lines stably expressing stumpy-EGFP or mock-EGFP constructs and performed immunofluorescence microscopy and coimmunoprecipitation assays. Stumpy colocalized with γ-tubulin, and could also be observed in euchromatic regions of the nucleus (Fig. 4E). Further, pull-down with GFP antibody coimmunoprecipitated γ-tubulin in stumpy-EGFP but not mock-EGFP MDCK transfectants (Fig. 4F). These results suggest that stumpy is a critical component of the ciliary bb and axoneme.
Fig. 4.
Stumpy is an essential component of the ciliary bb and axoneme. (A) MDCK (Left) or HeLa (Right) cells were transiently transfected with stumpy-his/myc, mock-EGFP, or stumpy-EGFP plasmids. After 48 h, cell lysates were Western blotted with anti-myc or GFP antibodies. (B) Embryonic day 15 fetuses were coelectroporated with plasmids encoding RFP and mock-EGFP or stumpy-EGFP. RFP+ ependymal cells along Lv were imaged at P10. Arrowheads show apical localization of stumpy-EGFP compared with diffuse EGFP signal in Mock-transfected cells. (Scale bar, 5 μm.) (C) C. Plexus from P8 +/+ or fl/fl mice was immunostained for γ-tubulin (green signal) and stumpy (red signal) (arrowheads show colocalization; high-magnification Insets are in the bottom right). (Scale bar, 20 μm.) (D) Differentiated MDCK cells were immunostained for stumpy (green signal) or α-tubulin (red signal) (merged images are shown to the right). (Scale bar, 5 μm.) Note stumpy signal at the bb (Left, see arrowhead) and ciliary axoneme (c) (Right; see arrowheads). MDCK cells were stably transfected with mock-EGFP or stumpy-EGFP plasmids (green signal), and (E) immunostained with γ-tubulin (red signal) (merged images are shown to the right; arrowhead shows colocalized signals; high-magnification Insets are in the bottom right) (scale bar, 5 μm) or (F) cell lysates were immunoprecipitated with GFP antibody and Western blotted with γ-tubulin (Upper) or GFP antibody (Lower). Ten percent input controls are shown to the right, and * indicates rabbit Ig heavy chain. For C–E, DAPI was used as a nuclear counterstain (blue signal). Images were captured by using a confocal microscope (B and C), a Zeiss ApoTome-equipped fluorescence microscope (D), or an epifluorescence microscope (E).
Discussion
Here, we present evidence that stumpy mutant mice have a profound defect in ciliogenesis associated with complete penetrance of PTvHC and PKD. A causal relationship between disturbed CSF flow homeostasis (brought on by dysfunctional cilia) and HC has been suggested by others (3–5). This may be due to poor CSF flow resulting from dysfunction of motile cilia on ependymal cells or failure of primary cilia on C. Plexus cells, which are thought to be important regulators of CSF production (4). Ciliogenesis has been suggested to be calcium-dependent (24), and it is noteworthy that another B9/C2 calcium/lipid-binding domain-containing protein, MKS1, has recently been localized to the ciliary bb (25). Mutation in this gene has recently been linked to Meckel Syndrome, a severe fetal developmental disorder hallmarked by occipital meningoencephalocele, cystic kidney dysplasia, fibrotic changes of the liver, and polydactyly (13). Stumpy appears to have a general role in ciliogenesis, because we show that stumpy mutant mice also have PKD and striking loss of cilia on nephron epithelial cells. Interestingly, our results show that the nestin-cre deleter mice are able to recombine in the kidney when crossed with stumpy floxed mice, consistent with recent reports showing nestin expression in human kidney and recombination of kidney cells in nestin-cre mice (17, 26). This should be a consideration for the neuroscientist who is interested in only recombining brain cells and is considering using nestin-cre deleter mice. Consistent with our findings in mammals, another group has shown that knocking down the stumpy ortholog in paramecium using RNA interference results in loss of cilia stability (27).
Interestingly, stumpy is atypical in that it is located in tight physical proximity to the tgfb1 gene, and conditional stumpy mutant mice lack stumpy exon 4 and tgfb1 exon 1. To determine how tgfb1 deficiency impacted the phenotype of stumpy mutant mice, we took two approaches: (i) crossed heterozygous stumpy mutant mice with mice heterozygous for a null tgfb1 allele, and (ii) i.c.v. delivered stumpy cDNA in pAdTrack adenovirus into fetuses of stumpy mutant mice or control littermates. Progeny from the crossing strategy homozygously deficient for tgfb1 but heterozygous for stumpy were essentially normal. When we reintroduced stumpy into brains of conditional stumpy mutant mice by adenoviral delivery, we noted that infected C. Plexus cells had intact cilia, whereas neighboring noninfected cells did not. Taken together, these results support that stumpy deficiency as opposed to loss of tgfb1 is responsible for defective ciliogenesis.
Stumpy is critical for the proper elongation of cilia. Although the precise biochemical nature of its function requires further investigation, the presence of stumpy within ciliary axonemes suggests it could associate with intraflagellar transport proteins (IFTs), large proteins that orchestrate bidirectional movement of substances within cilia and are required for steady-state assembly of tubulin onto the distal end of the ciliary axoneme (2). Our observation of stumpy localization at the ciliary bb further suggests an IFT role for this molecule. Strikingly, we find complete penetrance of both PTvHC and PKD in stumpy mutant mice, suggesting a general role for stumpy in mammalian ciliogenesis. We hypothesize that mutations in the highly conserved human homolog of this gene (MGC4093, located on chromosome 19 in close proximity to tgfb1, GenBank accession no. NM_030578) may be linked to congenital HC, PKD, and/or other developmental defects associated with abnormal cilia growth in humans. Such findings would serve to further highlight the role of previously underappreciated mammalian cilia in both physiological and disease processes.
Materials and Methods
Mice.
Conditional stumpy mutant mice were established by inserting one loxP site in the intronic region just upstream of the last exon of stumpy and another loxP sequence in the intronic region between exons 1 and 2 of tgfb1 (28). Additional mice were established with an egfp sequence “knocked-in” in place of exon 1 of tgfb1 (designated tgfb1egfpKI+), resulting in a null frame-shift mutation in tgfb1 (M.O.L and R.A.F., unpublished work). Both of these mice were bred with transgenic nestin-cre mice [B6.Cg-Tg(Nes-cre)1Kln/J] (29) obtained from The Jackson Laboratory. We examined >300 mice over a 3-year period in this study. All of the mice used in this study were fully backcrossed onto a C57BL/6 background.
Histochemistry and Light Microscopy.
Postnatal mice (from P0 to P16) were perfused with ice-cold PBS, and brains and kidneys were rapidly isolated. Brains were dissected down the midline (for sagittal sectioning) or at the level of the anterior commissure (for coronal sectioning) using a mouse brain slicer (World Precision Instruments); kidneys were dissected down the midline. Cerebral pieces were immersion-fixed in 4% PFA overnight at 4°C. For H&E histochemistry, cerebral pieces were routinely embedded in paraffin for microtome sectioning at 8-μm thickness, applied to Superfrost Plus Gold slides (Fisher Scientific), deparaffinized in xylene, hydrated in a graded series of ethanols, and stained with H&E Y (Sigma) according to standard practice. Stained sections were dehydrated and mounted in Permount (Fisher Scientific), air-dried, and viewed by using an automated Olympus BX-61 microscope with attached Olympus MagnaFire CCD imaging system. See SI Text for more methodological details.
Supplementary Material
ACKNOWLEDGMENTS.
We thank T. Li (Harvard University, Boston) for providing mouse rootletin antibody; F. J. Sim and S. A. Goldman (University of Rochester, Rochester, NY) for providing pAdTrack reagents and technical assistance; M. Pappy, Y. Morozov, S. Mane, and S. T. Kim for assistance with experiments; S. Somlo for helpful discussions; and F. Manzo for assistance with manuscript preparation. This work was supported in part by a National Institutes of Health/National Research Service Award/National Institute on Aging postdoctoral fellowship (T.T.), an Epilepsy Foundation of America postdoctoral fellowship (M.R.S.), a National Institutes of Health K01 award (M.O.L.), a National Institutes of Health/National Institute on Aging “Pathway to Independence” award (1K99AG029726, to T.T.), and by grants from the U.S. Public Health Service and the Kavli Institute (to P.R.). R.A.F. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712385105/DC1.
References
- 1.Porter ME, Sale WS. The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J Cell Biol. 2000;151:F37–F42. doi: 10.1083/jcb.151.5.f37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davenport JR, Yoder BK. An incredible decade for the primary cilium: a look at a once-forgotten organelle. Am J Physiol. 2005;289:F1159–F1169. doi: 10.1152/ajprenal.00118.2005. [DOI] [PubMed] [Google Scholar]
- 3.Ibanez-Tallon I, et al. Dysfunction of axonemal dynein heavy chain Mdnah5 inhibits ependymal flow and reveals a novel mechanism for hydrocephalus formation. Hum Mol Genet. 2004;13:2133–2141. doi: 10.1093/hmg/ddh219. [DOI] [PubMed] [Google Scholar]
- 4.Banizs B, et al. Dysfunctional cilia lead to altered ependyma and choroid plexus function, and result in the formation of hydrocephalus. Development. 2005;132:5329–5339. doi: 10.1242/dev.02153. [DOI] [PubMed] [Google Scholar]
- 5.Sawamoto K, et al. New neurons follow the flow of cerebrospinal fluid in the adult brain. Science. 2006;311:629–632. doi: 10.1126/science.1119133. [DOI] [PubMed] [Google Scholar]
- 6.Yoder BK. Role of primary cilia in the pathogenesis of polycystic kidney disease. J Am Soc Nephrol. 2007;18:1381–1388. doi: 10.1681/ASN.2006111215. [DOI] [PubMed] [Google Scholar]
- 7.Davenport JR, et al. Disruption of intraflagellar transport in adult mice leads to obesity and slow-onset cystic kidney disease. Curr Biol. 2007;17:1586–1594. doi: 10.1016/j.cub.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ainsworth C. Cilia: tails of the unexpected. Nature. 2007;448:638–641. doi: 10.1038/448638a. [DOI] [PubMed] [Google Scholar]
- 9.Pintavorn P, Ballermann BJ. TGF-beta and the endothelium during immune injury. Kidney Int. 1997;51:1401–1412. doi: 10.1038/ki.1997.192. [DOI] [PubMed] [Google Scholar]
- 10.Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming Growth Factor-beta Regulation of Immune Responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 11.Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 12.Gherman A, Davis EE, Katsanis N. The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nat Genet. 2006;38:961–962. doi: 10.1038/ng0906-961. [DOI] [PubMed] [Google Scholar]
- 13.Kyttala M, et al. MKS1, encoding a component of the flagellar apparatus basal body proteome, is mutated in Meckel syndrome. Nat Genet. 2006;38:155–157. doi: 10.1038/ng1714. [DOI] [PubMed] [Google Scholar]
- 14.Bruni JE, Del Bigio MR, Cardoso ER, Persaud TV. Hereditary hydrocephalus in laboratory animals and humans. Exp Pathol. 1988;35:239–246. doi: 10.1016/s0232-1513(88)80094-x. [DOI] [PubMed] [Google Scholar]
- 15.The Jackson Laboratory. Hydrocephalus in Laboratory Mice. 2003 http://jaxmice.jax.org/library/notes/490f.html.
- 16.Forni PE, et al. High levels of Cre expression in neuronal progenitors cause defects in brain development leading to microencephaly and hydrocephaly. J Neurosci. 2006;26:9593–9602. doi: 10.1523/JNEUROSCI.2815-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubois NC, Hofmann D, Kaloulis K, Bishop JM, Trumpp A. Nestin-Cre transgenic mouse line Nes-Cre1 mediates highly efficient Cre/loxP mediated recombination in the nervous system, kidney, and somite-derived tissues. Genesis. 2006;44:355–360. doi: 10.1002/dvg.20226. [DOI] [PubMed] [Google Scholar]
- 18.Piperno G, LeDizet M, Chang XJ. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J Cell Biol. 1987;104:289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LeDizet M, Piperno G. Identification of an acetylation site of Chlamydomonas alpha-tubulin. Proc Natl Acad Sci USA. 1987;84:5720–5724. doi: 10.1073/pnas.84.16.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LeDizet M, Piperno G. Detection of acetylated alpha-tubulin by specific antibodies. Methods Enzymol. 1991;196:264–274. doi: 10.1016/0076-6879(91)96025-m. [DOI] [PubMed] [Google Scholar]
- 21.Yang J, et al. Rootletin, a novel coiled-coil protein, is a structural component of the ciliary rootlet. J Cell Biol. 2002;159:431–440. doi: 10.1083/jcb.200207153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oakley BR. Gamma-tubulin: the microtubule organizer? Trends Cell Biol. 1992;2:1–5. doi: 10.1016/0962-8924(92)90125-7. [DOI] [PubMed] [Google Scholar]
- 23.He TC, et al. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueno H, Gonda K, Takeda T, Numata O. Identification of elongation factor-1alpha as a Ca2+/calmodulin-binding protein in Tetrahymena cilia. Cell Motil Cytoskeleton. 2003;55:51–60. doi: 10.1002/cm.10111. [DOI] [PubMed] [Google Scholar]
- 25.Dawe HR, et al. The Meckel-Gruber Syndrome proteins MKS1 and meckelin interact and are required for primary cilium formation. Hum Mol Genet. 2007;16:173–186. doi: 10.1093/hmg/ddl459. [DOI] [PubMed] [Google Scholar]
- 26.Perry J, et al. The intermediate filament nestin is highly expressed in normal human podocytes and podocytes in glomerular disease. Pediatr Dev Pathol. 2007;10:369–382. doi: 10.2350/06-11-0193.1. [DOI] [PubMed] [Google Scholar]
- 27.Ponsard C, et al. Identification of ICIS-1, a new protein involved in cilia stability. Front Biosci. 2007;12:1661–1669. doi: 10.2741/2178. [DOI] [PubMed] [Google Scholar]
- 28.Li MO, Wan YY, Flavell RA. T cell-produced transforming growth factor-beta1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity. 2007;26:579–591. doi: 10.1016/j.immuni.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 29.Tronche F, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.