Abstract
Adipogenesis involves cell proliferation and differentiation, both of which have been shown to be regulated by micro (mi)RNA. During mouse preadipocyte 3T3L1 cell differentiation, we found that miR-17-92, a miRNA cluster that promotes cell proliferation in various cancers, was significantly up-regulated at the clonal expansion stage of adipocyte differentiation. Stable transfection of 3T3L1 cells with miR-17-92 resulted in accelerated differentiation and increased triglyceride accumulation after hormonal stimulation. By using a luciferase reporter assay, we demonstrated that miR-17-92 directly targeted the 3′ UTR region of Rb2/p130, accounting for subsequently reduced Rb2/p130 mRNA and protein quantities at the stage of clonal expansion. siRNA-mediated knock-down of Rb2/p130 at the same stage of clonal expansion recapitulated the phenotype of overexpression of miR-17-92 in the stably transfected 3T3L1 cells. These data indicate that miR-17-92 promotes adipocyte differentiation by targeting and negatively regulating Rb2/p130.
Given the global obesity epidemic, deciphering the mechanisms underlying adipocyte differentiation in mammals poses an important challenge. The mouse 3T3L1 preadipocyte cell line has been an ideal in vitro system for unraveling the molecular events of adipocyte differentiation (1, 2). The entire process of 3T3L1 preadipocyte cell differentiation involves several cellular stages. First, proliferating preadipocytes become growth-arrested by contact inhibition, which can be reversed by hormonal induction and reentry into the cell cycle. After several rounds of clonal expansion, the cells again become quiescent with the initiation of coordinate transcriptional activation of adipocyte genes such as C/Ebpα, Ap2, Lpl, Srebp, and Pparγ. Expression of these genes leads to terminal adipocyte differentiation (3).
Among the above cellular programs, clonal expansion is one of the key events taking place in early adipogenesis (4, 5). Blocking cell cycle reentry with a DNA synthesis inhibitor prevents adipocyte differentiation, suggesting that an active clonal expansion is required for the differentiation process (4, 6). Clonal expansion is critically regulated by retinoblastoma family (pRB) genes through interactions with the E2F transcription factors. A significant role of two members of the pRB family p130 and p107 in early adipocyte differentiation is suggested by the dramatic change in their expression within the first 24 h (i.e., clonal expansion) of hormonal induction. In growth-arrested preadipocytes, there are significant levels of p130 with little or no detectable p107. However, this trend is completely reversed at 24 h, with a significant decrease in p130 accompanied by a marked increase in p107. This dramatic regulation has been designated as the p130:p107 switch (6). The functional significance of the p130:p107 switch has now been demonstrated by disrupting the p130 expression pattern during the mitotic clonal expansion phase, resulting in complete inhibition of adipocyte differentiation (7).
In addition to the understanding of the classic molecular signaling involved in protein-coding genes, recent studies indicate that micro (mi)RNAs also play critical roles in adipocyte differentiation. Esau et al. (8) showed that inhibition of up-regulated miRNA (miR)-143 in human differentiating adipocytes effectively reduced the level of triglyceride accumulation as well as the expression level of adipocyte specific genes, which ultimately abolished adipocyte differentiation. miRNAs have been shown to play key roles in other cell differentiation events, including embryonic stem cell differentiation (9), myogenesis (10), angiogenesis and hematopoiesis (11), and neurogenesis (12). miRNAs are also actively involved in cell proliferation. One example is the miR-17-19 cluster, which comprises seven miRNAs (miR-17-5p, miR-17-3p, miR-18, miR-19a, miR-20, miR-19b, and miR-92-1) and resides in intron 3 of the C13orf 25 gene at 13q31.3 (13). This miR-17-92 cluster is frequently amplified and substantially overexpressed in B-cell lymphomas and lung cancers, and promotes tumor growth in human and mouse cell models (14, 15). The experimental data suggest that the growth stimulatory effect of miR-17-92 cluster is at least partially attributable to the rebalance between cell proliferation and apoptosis through interacting with E2F and pRB families (16)
In an effort to understand the functional significance of miRNAs during adipocyte differentiation, we used 3T3L1 preadipocyte cells to screen miRNA expression over seven time points after hormonal induction. Surprisingly, we found all five members of the miR-17-92 cluster queried were markedly up-regulated after hormonal stimulation and peaked at the clonal expansion stage. This distinctive expression pattern suggests that the miR-17-92 cluster may also regulate adipocyte differentiation. In this report, we describe the expression profiles of miR-17-92 and demonstrate that overexpression of miR-17-92 can accelerate adipocyte differentiation through targeting the tumor suppressor Rb2/p130.
Results
miRNAs Are Critical for Adipocyte Differentiation.
Before we tried to identify critically important miRNAs for adipocyte differentiation, we functionally tested transfection efficiency by siRNA-mediated knockdown of PPARγ, a master regulator of adipocyte differentiation. In 3T3L1 cells in which the expression of PPARγ protein was reduced 78% by siRNA targeting, differentiation was absent (Fig. 1 A, B, and I), confirming the utility of this approach. Then, we addressed whether miRNAs are important for adipocyte differentiation by siRNA-mediated suppression of Drosha, an essential enzyme for miRNA maturation. Results showed that transfection of Drosha-targeted siRNA 2 days before hormonal induction suppressed 80% of Drosha expression and effectively abolished adipocyte differentiation (Fig. 1 C, D, and I); the effectiveness significantly decreased 1 day before hormonal induction, although a 4.2-fold reduction of Drosha expression was achieved by siRNA-mediated knockdown (Fig. 1 E, F, and I), and no obvious effect was observed 1 day after induction (Fig. 1 G–I). These data suggest that miRNAs are critical at the early stage of adipocyte differentiation.
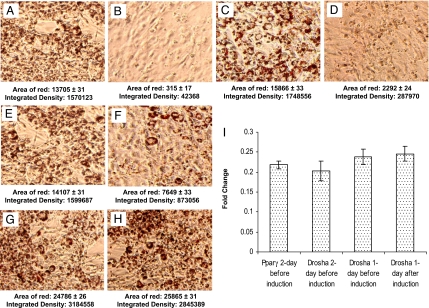
Fig. 1.
Representative oil red O staining of 3T3L1 cells after 7 days differentiation under varied experimental conditions. A, C, E, and G are negative siRNA controls (siCONTROL Non-Targeting siRNA Pool purchased from Dharmacon) for B, D, F, and H, respectively. Transfection of Pparγ siRNA 2 days before hormonal induction abolished adipocyte differentiation (B). Transfection of Drosha siRNA 2 days before hormonal induction effectively blocked adipocyte differentiation (D). Transfection of the same Drosha siRNA 1 day before induction had a limited effect on blocking differentiation (F), whereas transfection 1 day after hormonal induction had no effect on adipocyte differentiation (H). The area of red droplets and their integrated density were measured for each image by using IPLabs software 4.0 from BD Biosciences, which quantitatively reflects the degree of adipocyte differentiation. QRT-PCR analyses confirmed effective siRNA knockdown of Pparγ and Drosha, which led to a >4-fold reduction of their mRNA levels compared with the control siRNA transfections (I).
miR-17-92 Cluster Is Significantly Up-Regulated During Adipocyte Differentiation.
To identify potential miRNAs that regulate adipocyte differentiation, we systematically profiled miRNA at 2, 4, 8, and 12 h and 1, 2, and 6 days after hormonal induction. These data showed 74–219 miRNAs were differentially expressed (≥2-fold) over these time periods. The complete lists for these seven sets of data are available upon request. A striking observation was that every member of the miR-17-92 cluster (miR-17-5p, miR-17-3p, miR-18, miR-19b, and miR-20) was up-regulated at each time point (Fig. 2 A and B). Two-way ANOVA showed that expression levels of all members, except for miR-19, were significantly higher than noninduced controls at all time points, except for 8 h. Expression levels over times exhibited two peaks at 4 h and 1 day after induction. We independently confirmed the differential expression of miR-17-5p and miR-18 at 1, 2, and 6 days after induction by quantitative (Q)RT-PCR (Fig. 2C).
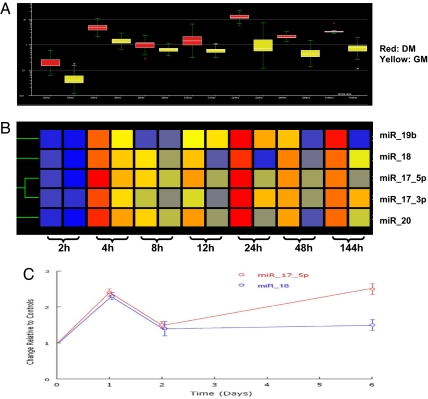
Fig. 2.
Expression profiles of five members of the miR-17-92 cluster over seven time points with and without hormonal induction. (A) Box plots showing the data distribution of the miR-17-92 cluster over time. The horizontal lines in the boxes indicate the median values; the lower and upper edges of the boxes indicate the 25th and 75th percentiles of the datasets, respectively; the whiskers indicate the minimum and maximum values (or 1.5 times the interquartile range). The red- and yellow-colored boxes represent cells grown in differentiation and growth medium, respectively. (B) Cluster analysis for all five members of miR-17-92 cluster available on Invitrogen miRNA chips. The red color shows relatively abundant expression of the same miRNA compared with other time points, whereas the blue color indicates a low expression relative to others. Cluster analyses were performed with GeneSpring GX 7.3.1 by using the default settings. (C) The relative expression levels of miR-17-5p and miR-18 at days 1, 2, and 6 after differentiation, as detected by QRT-PCR. The red line represents miR-17-5p, and the blue line denotes miR-18. The data shown are means and SDs of three replicates.
miR-17-92 Cluster Accelerates Adipocyte Differentiation.
Given these striking observations, we hypothesized that the up-regulation of the miR-17-92 cluster has a functional significance in adipocyte differentiation. To test this hypothesis, we established a 3T3L1 cell line with stable transfection of miR-17-92 cluster in an murine stem cell virus (MSCV) vector; a vector-only stable transfectant served as control for these studies. QRT-PCR analysis showed that enhanced expression of miR-17-92 resulted in 2.8- and 2.7-fold increases in expression of miR-17-5p and miR-18, respectively, compared with controls with vector alone, and 2.5- and 1.4-fold increases compared with nontransfected 3T3L1 cells (Fig. 3A). There was no evident change in 3T3L1 cells in which there was overexpression of the miR-17-92 cluster without hormonal induction (data not shown). However, when hormonally induced, the cells transfected with miR-17-92 cluster differentiated into adipocytes significantly faster than vector alone (Fig. 3B). The accelerated differentiation was accompanied by the increased mRNA levels of several key transcription factors considered to be adipocyte-specific markers. As depicted in Fig. 3C, Pparγ, Ap2, and Lpl mRNA expressions were significantly increased in the cells transfected with miR-17-92 compared with controls (vector alone) during the progression of adipocyte differentiation. The expression levels of C/Ebpα and Srebp were also significantly elevated at 2 and 3 days after hormonal induction (data not shown).
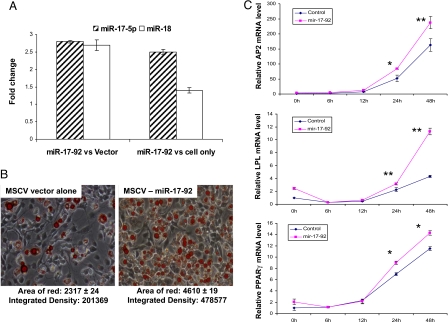
Fig. 3.
Overexpression of miR-17-92 cluster accelerates adipocyte differentiation. (A) Enhanced expression of the miR-17-92 cluster resulted in an elevated expression of miR-17-5p and miR-18, as shown by QRT-PCR. RNA was extracted from stably transfected 3T3L1 cells with either MSCV-miR-17-92 construct or MSCV alone without hormonal induction, respectively. The empty and cross-hatched bars represent miR-18 and miR-17-5p, respectively. The left two bars are the expression ratios of measured miRNAs between cells with MSCV-miR-17-92 and the MSCV vector alone, and the right two bars are the expression ratios of measured miRNAs between cells harboring the MSCV-miR-17-92 construct and 3T3L1 cells only. (B) Overexpression of miR-17-92 resulted in a >2-fold increase of adipocyte differentiation (Right) compared with vector control (Left), as shown by oil red O staining and measured by the area of red and integrated density 7 days after hormonal induction. (C) QRT-PCR analyses of key adipogenic transcription factors during 3T3L1 adipocyte differentiation. The expression levels of Pparγ, Ap2, and Lpl were significantly increased in the cells transfected with miR-17-92 construct compared with vector alone at 24 and 48 h. The data shown are means and SDs of three replicates. **, P < 0.01; *, P < 0.05.
Search for Targets of miR-17-92 Cluster.
As a first step to understand mechanisms underlying the actions of the miR-17-92 cluster, we focused initially on identifying the potential targets of the miR-17-92 cluster. Because the miR-17-92 cluster was up-regulated during differentiation, we expected that the expression of its targets should be concomitantly down-regulated. Therefore, we compared gene expression profiles between baseline (no induction control) and 1 day of hormonal induction by using Affymetrix mouse 430 2.0 arrays. We identified 323 genes that were down-regulated >1.5-fold after induction and were also predicted to be potential targets of the miR-17-92 cluster (a list of the genes is available upon request). Among these genes, Rb2/p130 was arguably the most relevant because several members of miR-17-92 cluster, including miR-17-5p, miR-18, and miR-20, can target the 3′ UTR of Rb2/p130; additionally, previous studies have indicated that decreased expression of Rb2/p130 within the first day of hormonal stimulation is critically important for adipocyte differentiation (7).
miR-17-92 Cluster Directly Targets Rb2/p130.
To examine whether the miR-17-19 cluster can regulate the expression of Rb2/p130, we measured Rb2/p130 mRNA and protein quantities in 3T3L1 cells stably transfected with miR-17-92 cluster or vector alone by QRT-PCR and Western blotting, respectively. As shown in Fig. 4A, Rb2/p130 mRNA in cells overexpressing miR-17-92 was lower than that in control cells by 1.5-, 2.7-fold, and 2.0-fold at 0, 12, and 24 h after induction, respectively. Similarly, Rb2/p130 protein was visibly lower at baseline and nearly undetectable at 24 h in the miR-17-92 cluster-transfected cells (Fig. 4B). These data are consistent with the hypothesis that Rb2/p130 is a direct target of miR-17-92 cluster in 3T3L1 cells. To test this hypothesis, a standard reporter assay in which the miR-17-5p predicted target sequence in the Rb2/p130 3′ UTR was inserted downstream of the firefly luciferase gene. These studies were performed in MES13 cells, in which the level of endogenous miR-17-92 expression is 50 times lower than that of 3T3L1 cells (data not shown). There was a significant reduction in luciferase activity when chemically synthesized miR-17-5p was transfected into MES13 cells. The degree of the reduction in luciferase activity depended on the quantity of synthetic mir-17-5p (Fig. 4C). These data provide evidence that Rb2/p130 is a direct target of the miR-17-19 cluster during adipocyte differentiation.
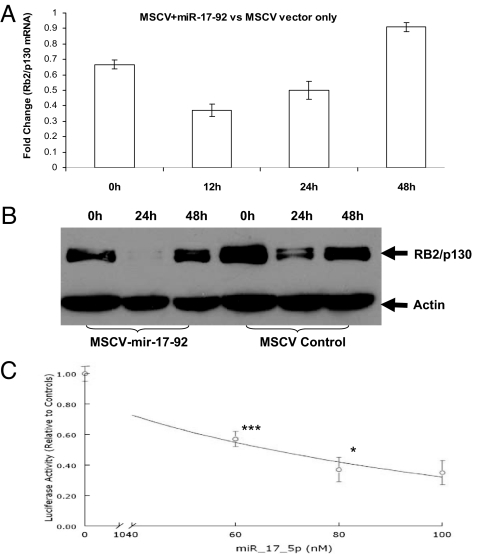
Fig. 4.
Rb2/p130 is a direct target of miR-17-92. (A) Rb2/p130 mRNA in the cells transfected with miR-17-92 was lower than that in the control cells at different time points of hormonal induction. (B) Rb2/p130 protein in the cells transfected with miR-17-92 was markedly reduced compared with the cells transfected with empty MSCV vector. (C) Luciferase activities were decreased in MES13 cells in a dose-dependent fashion by transfection of different concentration of synthetic miR-17-5p. There was no difference in luciferase activities between the cells harboring the pGL3-Rb2/p130 construct and the cells harboring the pGL3 control construct before cotransfection. The luciferase activities were decreased with the increase of synthetic miR-17-92 concentrations. ***, P < 0.001 between 0 and 60 nM; *, P < 0.05 between 60 and 80 nM. There was no significant difference between 80 and 100 nM, which could be attributable to the saturation of synthetic miR-17-5p at 80 nM.
siRNA-Mediated Knockdown of Rb2/p130 Recapitulates the Differentiation Phenotype of Overexpression of miR-17-92.
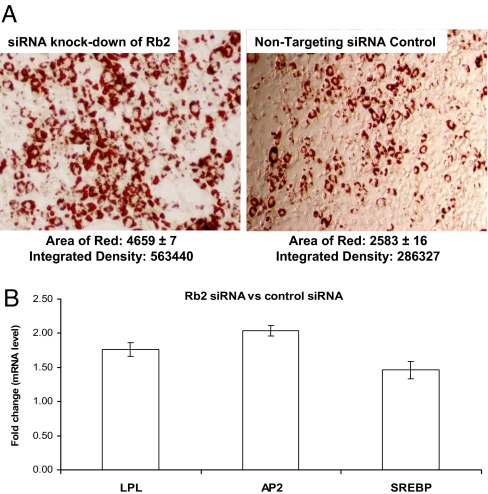
To establish a functional link between the promotion of adipocyte differentiation and suppression of Rb2/p130 expression by miR-17-92, we transfected siRNA against Rb2/p130 into the 3T3L1 cells 6 h before hormonal induction, which led to a 3.3-fold reduction of Rb2/p130 expression 24 h after hormonal stimulation (data not shown). Consistent with the role of miR-17-92 in accelerating differentiation by repressing Rb2/p130, siRNA-mediated knockdown of Rb2/p130 enhanced adipocyte differentiation (Fig. 5A) and up-regulated several adipocyte-specific markers, such as Ap2, Lpl, and Srebp (Fig. 5B).
Fig. 5.
siRNA-mediated knockdown of Rb2/p130 accelerates adipocyte differentiation. (A) siRNA knockdown of Rb2/p130 accelerated adipocyte differentiation (Right) compared with control siRNA (Left), as shown by oil red O staining and measured by the area of red and integrated density 7 days after hormonal induction. (B) QRT-PCR analyses of key adipocyte markers during 3T3L1 adipocyte differentiation. The y axis represents the mRNA expression ratio of Ap2, Lpl, and Srebp between the cells transfected with siRNA against Rb2/p130 and the cell transfected with control siRNA at 48 h after induction. The siRNA transfection was performed 6 h before hormonal induction. The data shown are mean ratios and SDs of three replicates.
Discussion
The present study establishes for the first time the important role played by miR-17-92 in adipogenesis. After the observation of elevated expression of miR-17-92 cluster members during adipocyte differentiation, we demonstrated that: (i) overexpression of miR-17-92 is associated with the accelerated adipocyte differentiation; (ii) miR-17-92 directly targets and down-regulates the expression of Rb2/p130; and (iii) siRNA knockdown of Rb2/p130 recapitulates the differentiation phenotype created by overexpression of miR-17-92. Thus, we conclude that miRNA-17-92 accelerates adipocyte differentiation by negatively regulating the key cell cycle regulator and tumor suppressor, Rb2/p130.
Before terminal differentiation of 3T3L1 preadipocytes, growth-arrested preadipocytes must undergo clonal expansion through cell cycle events before undergoing terminal differentiation. In this study, we observed two elevated expression peaks of miR-17-92 members during the clonal expansion. The first was at 4 h, corresponding to the G1 stage, when RNAs and proteins are actively synthesized and the G1 checkpoint is activated to ensure that everything is ready for DNA synthesis. This is a critical stage to initiate and/or prepare for reentering the cell cycle. The second expression peak was at 24 h, coinciding with the end of clonal expansion. In contrast, the expression of miR-17-92 in control cells (without hormonal induction) remained relatively constant after an initial moderate elevation at 2 h. These differential expression patterns suggest that miR-17-92 may modulate adipocyte differentiation by regulating the reentry and exit of cell cycle. This is consistent with the data obtained by siRNA-mediated Drosha inhibition, which showed an essential role for miRNAs at the clonal expansion stage of adipocyte differentiation.
To evaluate the functional significance of the distinctive expression pattern of miR-17-92 cluster members during the clonal expansion stage of adipocyte differentiation, we established a stably transfected 3T3L1 cell line overexpressing miR-17-92 cluster genes. In agreement with expression data, enhanced expression of miR-17-92 had profound effects on adipocyte differentiation. However, up-regulation of miR-17-92 cannot trigger adipocyte differentiation without hormonal induction, which suggests that miR-17-92 is a necessary adjunct to hormonal signals to initiate differentiation.
In an effort to find target genes of miR-17-92, gene expression profiles were compiled during adipocyte differentiation. Among the numbers of down-regulated genes 1 day after the onset of hormonal induction, we focused on Rb2/p130 because it has an established role in adipogenesis and its 3′ UTR has several targeting sites for miR-17-5p, miR-18, and miR-20. In differentiating 3T3L1 cells, we observed reduced Rb2/p130 mRNA levels, as well as concomitantly decreased protein levels, when miR-17-92 cluster was overexpressed within the first 24 h of differentiation, thereby confirming in silico predictions that Rb2/p130 would represent a target gene of the miR-17-92 cluster. Subsequent luciferase assays provided evidence that miR-17-5p directly targets the 3′ UTR of Rb2/p130 mRNA. Clearly, Rb2/p130 is unlikely the only target of miR-17-92, because the members of the miR-17-92 cluster have a number of other potential targets that were also observed to be down-regulated during adipocyte differentiation.
The peaks of miR-17-92 expression, which coincided with the time of reentry and exit of the cell cycle, suggest that the molecular mechanism behind the acceleration of adipocyte differentiation by miR-17-92 is linked to the clonal expansion. As previously shown, the reentry into the cell cycle is a key event for initiating adipocyte differentiation. This event depends on the activation of the pRB–E2F pathway that controls the G1-to-S transition of the cell cycle. In this pathway, E2F transcription factors control the expression of genes involved in cell cycle progression, whereas pRB family members regulate E2F activities through complex formation (17). Specifically, Rb2/p130 can form heterodimers with E2F4 and E2F5 (18), leading to repression of target genes (19). Based on our experimental data [showing that (i) the peaks of miR-17-92 expression coincided with the time of reentry and exit of the cell cycle; (ii) miR-17-92 directly targeted Rb2/130; and (iii) overexpression of miR-17-92 significantly suppressed both Rb2/p130 mRNA and protein within the first 24 h], we propose the following model: after hormonal induction, the expression of miR-17-92 increases, resulting in a decrease of Rb2/p130. Insufficient quantities of Rb2/p130 with which to dimerize lead to an increase of free E2F4 and E2F5. These then activate E2F target genes, triggering reentry into the cell cycle. After 1 day of hormonal stimulation, the expression of miR-17-92 reached a peak and then started to decrease, which corresponds to increased complex formation between Rb2/p130 and E2F. Loss of activities of E2F silences E2F-responsive genes, leading to the induction of cell cycle exit and terminal differentiation. This model not only explains our experimental observations but is also consistent with previously published data, i.e., inactivation of Rb2/p130 enables clonal expansion, whereas growth arrest after this expansion phase requires active Rb2/p130, which positively influence terminal differentiation (20).
In summary, the current study represents, to our knowledge, the first demonstration that miR-17-92 promotes adipocyte differentiation by directly targeting Rb2/p130. Overexpression of miR-17-92 cannot trigger adipocyte differentiation without hormonal induction. Thus, miR-17-92 is necessary, but not sufficient, to initiate differentiation. The results described here provide an experimental framework for further functional dissection of this miRNA cluster and its targets to fully delineate its role in adipocyte differentiation.
Materials and Methods
3T3L1 Cell Culture and Differentiation.
3T3-L1 cells were obtained from American Type Culture Collection and were cultured to confluence in DMEM containing 10% FCS by changing the medium every 2 days. Two days after cell confluence, differentiation was initiated by adding differentiation medium (5 μM methylisobutylxanthine, 0.25 μM dexamethasone, and 1 μg/ml insulin in DMEM containing 10% FBS). Methylisobutylxanthine and dexamethasone were removed after 2 days, but insulin (1 μg/ml) was maintained for an additional 2 days. Thereafter, cells were grown in DMEM containing 10% FBS by replacing the media every 2 days.
Pparγ, Drosha, and Rb2/p130 Gene Knockdown.
The siRNAs for Pparγ, Drosha, and Rb2/p130 knockdown were purchased from Dharmacon. There are four individual siRNA sequences for each gene. The Pparγ siRNA target sequences are CGAAGAACCAUCCGAUUGA, ACCCAAUGGUUGCUGAUUA, UCACAAUGCCAUCAGGUUU, and CGACAUGAAUUCCUUAAUG. The Drosha siRNA sequences are UGGAAGGAGUUACGCUUUA, GCCAAAUACGGAUCGGCAA, UGUGUAAAGUGAUUCGAUU, and GGAUGGAAUUUCUGGGCGA. The Rb2/p130 siRNA sequences are GCGAUGAUCUGGUCAAUUC, GGGACAGAAUUAGAGAUAA, GGCUAUCGCUGACAGAUUG, and GUACAGUUAUCUCAAAGUC. The negative control siRNA used in the experiments was siCONTROL Non-Targeting siRNA Pool (Dharmacon). The siRNAs were transfected into the cell with DharmaFECT-3 transfection reagent (Dharmacon) according to the instructions of the manufacturer. Briefly, 3T3L1 cells were cultured, and differentiation was induced under the same conditions as described above. At day −2, −1, and 1 after hormonal induction, the cells were transfected separately with each Pparγ and Drosha siRNA at 40 nM with 2 μl of DharmaFECT-3 transfection reagent. Knockdowns of Pparγ and Drosha were confirmed at the mRNA level by QRT-PCR. The Rb2/p130 siRNA transfection was performed 6 h before hormonal induction. The knockdown of Rb2/p130 was confirmed at both mRNA and protein levels by QRT-PCR and Western blot.
Oil Red O Staining.
Cells were washed twice with PBS and fixed with 10% formalin for 60 min at room temperature. After fixation, cells were stained with filtered oil red O solution (stock solution: 3 mg/ml in isopropanol; working solution: 60% oil red O stock solution and 40% distilled water) for 2 h at room temperature. Cells were then washed with water to remove unbound dye, visualized by light microscopy, and photographed. Oil red O stain was quantified by using IPLabs software 4.0 (BD Biosciences). After correction of background, the color images were segmented by using the red channel intensity. The area of red droplets, as well as their integrated density, was measured to quantitatively indicate the degree of cell differentiation.
Generation of Stable Clones.
pMSCV-mir-17-92 and pMSCV negative control constructs, which contains a PGK–puromycin–IRES–GFP (PIG) cassette, were kind gifts from Lin He (14). The miR-17-92 cluster sequence was PCR-amplified and cloned into MSCVpuro–IRES–GFP vector through multiclone site (21). pMSCV–mir-17-92 and pMSCV negative control constructs were transfected into 3T3 L1 cells by Lipofectamine 2000 (Invitrogen). The stably transfected cell lines were established by puromycin selection (2 μg/ml) in DMEM, which were then confirmed by fluorescence microscopy (GFP marker) and by QRT-PCR.
miRNA Microarray/Gene Array Hybridization and Data Analysis.
Total RNAs were isolated by TRIzol (Invitrogen) from 3T3L1 cell cultures growing either on growth medium or differentiation medium at 2, 4, 8,12, 24, 48, and 144 h after hormonal induction. miRNAs were enriched by using a PureLink miRNA isolation kit (Invitrogen). The labeled miRNAs were hybridized to a Ncode MultiSpecies miRNA Microarray (Invitrogen) according to the instructions of the manufacturer. Data analyses were performed by using GeneSpring GX 7.3.1. All mouse 430 2.0 gene array hybridizations were performed at the Functional Genomics Facility (University of Chicago). The target preparation protocol followed the GeneChip expression analysis manual (Affymetrix). The arrays were washed and stained with streptavidin phycoerythrin in Fluidics Station 450 by using the Affymetrix GeneChip protocol and scanned by using the Affymetrix GeneChip 7G scanner (all from Affymetrix). Data analyses were performed by using DNA-Chip Analyzer 1.3 (22). The invariant set approach was used for data normalization. The thresholds for selecting significant genes were set at a relative difference of >1.5-fold, an absolute difference of >100 signal intensity, and P < 0.05.
Quantitative Real-Time PCR Analyses.
miRNA mmu–mir-17-5p and mmu–mir-18 were analyzed by a TaqMan MicroRNA assay kit (Applied Biosystems). The sno234 gene was used as an internal control for normalization. An SYBR Green-based detection system was used for QRT-PCR analysis (Applied Biosystems) with the following primers: Rb2 forward, AGGTCATGCCACCTCAAAAC; Rb2 reverse, TCTCGAATAGCCGCCTTCTA; Rb1 forward, TCACCACGCCTGTAGCTTCA; Rb1 reverse, CCAGCGACGATGCTCTGTAA; Pparγ forward, AAGAGCTGACCCAATGGTTG; Pparγ reverse, ACCCTTGCATCCTTCACAAG; Drosha forward, GGACCATCACGAAGGACACT; Drosha reverse, GATGTACAGCGCTGCGATAA; SREBP1 forward, GATCAAAGAGGAGCCAGTGC; SREBP1 reverse, TAGATGGTGGCTGCTGAGTG; Ap2 forward, TCACCTGGAAGACAGCTCCT; Ap2 reverse, AATCCCCATTTACGCTGATG; Lpl forward, GGGCTCTGCCTGAGTTGTAG; and Lpl reverse, CCATCCTCAGTCCCAGAAAA. Expression levels were normalized to Gapdh mRNA with the following l primers: Gapdh forward, AACTTTGGCATTGTGGAAGG; and Gapdh reverse, ACACATTGGGGGTAGGAACA. All QRT-PCR measurements were performed by using a 7900 HT Fast Real Time PCR system (Applied Biosystems).
Western Blotting.
3T3L1 cells were lysed in radioimmunoprecipitation assay buffer (150 mM NaCl, 1.0% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris, pH 8.0) with protease inhibitor mixture (Roche Applied Science). Protein concentrations were determined with BCA protein assay kit (Pierce). Proteins were separated on 10% SDS/PAGE gels under reducing conditions and electroblotted onto a PVDF membrane. The membranes were probed with primary (Rb2/p130 antibody; BD Biosciences) and secondary antibodies as detailed previously (23). The membranes were reprobed with anti-actin antibody (Sigma) for data normalization.
Luciferase Assay.
The pGL3 vector was modified by insertion of a puromycin fragment into the plasmid between KpnI and XhoI sites. The 3′ UTR sequence of Rb2/p130 was generated by PCR with the following primers: ATATCTAGAGTGTCCAGGAGGAAACTGTC (forward) and ATATCTAGATGCCACTACCACAAATGGAAG (reverse). The PCR product was digested with XbaI and then inserted into pGL3-Puro vector at the downstream of luciferase gene. The correct clone of pGL3-puro-Rb2–3UTR plasmid was transfected into a mouse mesangial cell line, MES13, growing on DMEM plus 10% FBS. The stable cell line harboring the plasmid was selected by using 1 μg/ml puromycin in DMEM. Mmu–mir-17-5p mimic (Dharmacon) was transfected into this cell at the concentration of 0, 60, 80, and 100 nM by using HiPerFect (Qiagen). Twenty-four hours after transfection, firefly luciferase activities were measured by using a luciferase assay kit (Promega) and were normalized to protein concentrations.
ACKNOWLEDGMENTS.
We thank Dr. Lin He (Cold Spring Harbor Laboratory, Watson School of Biological Sciences, Cold Spring Harbor, NY) for providing the MSCV-mir-17-92 construct. This work was supported in part by National Institutes of Health Cancer Research Center Grants R01DK055357 and R01HL085793.
Footnotes
The authors declare no conflict of interest.
References
- 1.MacDougald OA, Lane MD. Transcriptional regulation of gene expression during adipocyte differentiation. Annu Rev Biochem. 1995;64:345–373. doi: 10.1146/annurev.bi.64.070195.002021. [DOI] [PubMed] [Google Scholar]
- 2.Cowherd RM, Lyle RE, McGehee RE., Jr Molecular regulation of adipocyte differentiation. Cell Dev Biol. 1999;10:3–10. doi: 10.1006/scdb.1998.0276. [DOI] [PubMed] [Google Scholar]
- 3.Cornelius P, MacDougald OA, Lane MD. Regulation of adipocyte development. Annu Rev Nutr. 1994;14:99–129. doi: 10.1146/annurev.nu.14.070194.000531. [DOI] [PubMed] [Google Scholar]
- 4.Patel YM, Lane MD. Mitotic clonal expansion during preadipocyte differentiation: Calpain-mediated turnover of p27. J Biol Chem. 2000;275:17653–17660. doi: 10.1074/jbc.M910445199. [DOI] [PubMed] [Google Scholar]
- 5.Debril MB, Renaud JP, Fajas L, Auwerx J. Peroxisome proliferator-activated receptor gamma: From adipogenesis to carcinogenesis. J Mol Med. 2001;79:30–47. doi: 10.1677/jme.0.0270001. [DOI] [PubMed] [Google Scholar]
- 6.Richon V, Lyle RE, McGehee REJ. Regulation and expression of retinoblastoma proteins p107 and p130 during 3T3–L1 adipocyte differentiation. J Biol Chem. 1997;272:10117–10124. doi: 10.1074/jbc.272.15.10117. [DOI] [PubMed] [Google Scholar]
- 7.Prince AM, May JS, Burton GR, Lyle RE, McGehee RE., Jr Proteasomal degradation of retinoblastoma-related p130 during adipocyte differentiation. Biochem Biophys Res Commun. 2002;290:1066–1071. doi: 10.1006/bbrc.2001.6291. [DOI] [PubMed] [Google Scholar]
- 8.Esau C, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 9.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naguibneva I, et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8:278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- 11.Yang WJ, et al. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280:9330–9335. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- 12.Schratt GM, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 13.Ota A, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31–q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 14.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashita Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 16.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 17.Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- 18.Cobrinik D, Whyte P, Peeper DS, Jacks T, Weinberg RA. Cell cycle-specific association of E2F with the p130 E1A-binding protein. Genes Dev. 1993;7:2392–2404. doi: 10.1101/gad.7.12a.2392. [DOI] [PubMed] [Google Scholar]
- 19.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 20.Fajas L, Debril MB, Auwerx J. Peroxisome proliferator-activated receptor-gamma: From adipogenesis to carcinogenesis. J Mol Endocrinol. 2001;27:1–9. doi: 10.1677/jme.0.0270001. [DOI] [PubMed] [Google Scholar]
- 21.Hemann MT, et al. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat Genet. 2003;33:396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- 22.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, et al. Genetic network and pathway analysis of differentially expressed proteins during critical cellular events in fracture repair. J Cell Biochem. 2007;100:527–543. doi: 10.1002/jcb.21017. [DOI] [PubMed] [Google Scholar]