Abstract
HSP90 is a protein chaperone particularly important in the maturation of a diverse set of proteins that regulate key steps in a multitude of biological processes. Alterations in HSP90 function produce altered phenotypes at low penetrance in natural populations. Previous work has shown that at least some of these phenotypes are due to genetic variation that remains phenotypically cryptic until it is revealed by the impairment of HSP90 function. Exposure of such “buffered” genetic polymorphisms can also be accomplished by environmental stress, linking the appearance of new phenotypes to defects in protein homeostasis. Should such polymorphisms be widespread, natural selection may be more effective at producing phenotypic change in suboptimal environments. In evaluating this hypothesis, a key unknown factor is the frequency with which HSP90-buffered polymorphisms occur in natural populations. Here, we present Arabidopsis thaliana populations suitable for genetic mapping that have constitutively reduced HSP90 levels. We employ quantitative genetic techniques to examine the HSP90-dependent polymorphisms affecting a host of plastic plant life-history traits. Our results demonstrate that HSP90-dependent natural variation is present at high frequencies in A. thaliana, with an expectation that at least one HSP90-dependent polymorphism will affect nearly every quantitative trait in progeny of two different wild lines. Hence, HSP90 is likely to occupy a central position in the translation of genotypic variation into phenotypic differences.
Keywords: cryptic variation, molecular chaperone, morphological evolution
Several recent studies have suggested that the molecular chaperone HSP90 may facilitate rapid evolutionary change (1–4). Morphological diversity is dramatically enhanced upon reduction of HSP90 function in natural isolates of both the fruit fly Drosophila melanogaster (1) and the mustard plant Arabidopsis thaliana (2). In both organisms, HSP90-dependent altered morphologies differed between lines of different genetic background, suggesting that preexisting cryptic HSP90-dependent genetic variation had become phenotypically expressed. Rutherford and Lindquist (1) determined that the initially low penetrance of these variant traits could be increased by artificial selection and indeed driven to near fixation. Similar experiments revealed that other HSP90-dependent traits in D. melanogaster had a combined genetic and epigenetic origin (3). Importantly, many of these line-specific phenotypes could be revealed by modest environmental change in the absence of deliberate manipulation of HSP90 function (1). Taken together, these findings led to the proposal that HSP90 may play a key role in the transduction of environmental change to the phenotypic exposure of genetic differences, thereby acting as a capacitor of evolutionary change.
HSP90 is an essential, strictly conserved eukaryotic protein chaperone. Molecular chaperones are a class of proteins involved in altering the folding trajectories of a variety of other polypeptides (5). Unlike either generalist chaperones or highly substrate-specific chaperones, HSP90 interacts with a moderately sized group of substrates that are both functionally and structurally diverse (6–9). Most importantly, many HSP90 substrates (client proteins) have an inherent conformational flexibility or instability. This property is common to many signal transducers. HSP90's function in the folding of such proteins is illustrated by its interaction with certain mammalian hormone receptors, such as the glucocorticoid receptor (10, 11). HSP90 maintains such unstable receptors in a signaling competent state; inactive, but ready to be activated upon the acquisition of the proper upstream signal. After the signal, such as hormone binding, is received, substrates typically undergo a conformational change, leading to downstream signal propagation (5, 7). Importantly, other proteins may become HSP90 clients upon acquiring a genetic polymorphism that interferes with protein stability (12).
Due to the diverse activities of its substrates, HSP90 is a central, highly connected node in pathways regulating processes as diverse as cell growth, cell division, development, apoptosis, and environmental responses. Notably, although some HSP90 activity is essential in all eukarya, chaperone levels can be modulated significantly without clear deleterious effects for most individuals under optimal growth conditions (1, 4, 13–15). Under stress, however, the requirement for HSP90 increases because of compromised protein homeostasis, which may result in the exposure of underlying genetic polymorphisms (16). As inherently logical as such a proposal may be, as long as the underlying polymorphisms affected by HSP90 levels in genetically diverse populations remain unknown, such mechanistic hypotheses cannot be tested.
In the companion article (17), we have shown that naturally occurring HSP90-dependent variation can be genetically mapped in A. thaliana, using an early seedling trait as a model. These experiments decreased HSP90 activity with a light-sensitive, small-molecule inhibitor, an approach that is not suitable for the analysis of most plant traits. Here, we report the development of genetic mapping populations with constitutively reduced HSP90 levels and assess the frequency of natural HSP90-buffered polymorphisms across a variety of life-history traits in A. thaliana. We demonstrate that, in this model plant, HSP90-buffered variation is of global importance to variation in quantitative traits. These studies lay the foundation to assess both the mechanistic basis and the evolutionary potential of HSP90-dependent genetic variation.
Results
Recombinant Inbred Population Construction.
We have previously described the construction of A. thaliana (Columbia) with a constitutive reduction in HSP90 levels (13). This reduction is caused by the stable genomic insertion of a vector producing double-stranded HSP90 RNA, which targets the four cytosolic HSP90 isoforms by an RNA interference (RNAi) mechanism. The phenotypic consequences and genome-wide expression differences resulting from the constitutive reduction of HSP90 levels have been previously quantified in the standard inbred Columbia genetic background (13).
To determine the effect of this constitutive HSP90 reduction on the translation of genotype to phenotype, we constructed a recombinant inbred line (RIL) set with constitutively reduced HSP90 levels [supporting information (SI) Fig. 3]. We crossed a Columbia (Col-0) HSP90-RNAi line with ≈68% of wild-type cytosolic HSP90 protein levels (13) to a wild-type Landsberg erecta (Ler-2) line. RILs were created by seven generations of single-seed descent, with continuous selection for the HSP90-RNAi transgene. A control RIL set was created in a similar manner, except that the Col-0 parent was a control line containing the same integrated transgene backbone without any HSP90 sequence (13).
HSP90 Reduction in RILs.
We verified that the HSP90-RNAi RILs were reduced in HSP90 levels by RT-PCR of the inducible HSP90–1 isoform, as described in ref. 13. Analysis of 11 RILs from each set revealed a significant decrease, 1.6 cycles on average, in HSP90 levels in the HSP90-RNAi RIL population (Table 1; P ≪ 0.001 by ANOVA). This reduction is virtually identical to that reported for the parental HSP90-RNAi line in ref. 13. One RIL in the control set was an outlier. It had HSP90 levels within the range of HSP90-reduced RILs. No other overlap occurred between the sets.
Table 1.
HSP90-RNAi RILs have constitutively reduced HSP90 levels
| RIL | Line cycle time | Average cycle time | P-value |
|---|---|---|---|
| HSP90-RNAi 1 | 19.09 | 20.48 | 6.7 × 10−11 |
| HSP90-RNAi 2 | 20.04 | 20.48 | 6.7 × 10−11 |
| HSP90-RNAi 3 | 20.88 | 20.48 | 6.7 × 10−11 |
| HSP90-RNAi 4 | 21.11 | 20.48 | 6.7 × 10−11 |
| HSP90-RNAi 5 | 21.26 | 20.48 | 6.7 × 10−11 |
| Control 501 | 18.57 | 18.6 | 6.7 × 10−11 |
| Control 502 | 18.45 | 18.6 | 6.7 × 10−11 |
| Control 503 | 17.76 | 18.6 | 6.7 × 10−11 |
| Control 506 | 20.10 | 18.6 | 6.7 × 10−11 |
| Control 507 | 18.02 | 18.6 | 6.7 × 10−11 |
| HSP90-RNAi 13 | 18.60 | 17.91 | 6.0 × 10−8 |
| HSP90-RNAi 26 | 18.15 | 17.91 | 6.0 × 10−8 |
| HSP90-RNAi 39 | 17.58 | 17.91 | 6.0 × 10−8 |
| HSP90-RNAi 62 | 17.99 | 17.91 | 6.0 × 10−8 |
| HSP90-RNAi 89 | 17.40 | 17.91 | 6.0 × 10−8 |
| HSP90-RNAi 102 | 17.71 | 17.91 | 6.0 × 10−8 |
| Control 512 | 16.62 | 16.63 | 6.0 × 10−8 |
| Control 523 | 16.99 | 16.63 | 6.0 × 10−8 |
| Control 545 | 17.20 | 16.63 | 6.0 × 10−8 |
| Control 556 | 16.65 | 16.63 | 6.0 × 10−8 |
| Control 573 | 16.04 | 16.63 | 6.0 × 10−8 |
| Control 584 | 16.29 | 16.63 | 6.0 × 10−8 |
RT-PCR cycle times using primers recognizing HSP90–1 are presented. Upper and lower sets represent independent experiments (see Materials and Methods). Overall HSP90-RNAi vs. control P-values by ANOVA.
Genetic Mapping.
Both RIL sets were genotyped by an array hybridization method (18) using 240 insertion/deletion markers that are polymorphic between Ler and Col. The markers are distributed in a genome-wide fashion, with an average spacing of 0.56 Mb/marker. Only four gaps of >2 Mb exist between markers; these contain the centromeres of chromosomes 1, 3, 4, and 5.
This genotyping array has the advantage of providing high throughput analysis of markers whose position is known in the sequenced Col-0 genome, allowing for facile correlation of physical and genetic maps. Maps of the genotyped RILs are presented in SI Table 3 and SI Fig. 4. The RIL construction procedure selected for regions of the genome that are linked to the inserted construct. Therefore, the HSP90-reduced RIL population should show significant segregation distortion, biased toward Col-0, throughout chromosome 5, the site of the RNAi transgene insertion (SI Fig. 4; P < 0.01, χ2 test). This distortion should, and did, increase to 100% Col-0 at the location of the transgene (34 cM). Similarly, the control population showed segregation distortion on chromosome 1 from 0 to 56 cM, as the control vector insertion was located at 25 cM (SI Fig. 4).
HSP90-Responsive QTL Affecting Hypocotyl Length in the Dark.
A previously created Col × Ler RIL population (19) has been demonstrated to harbor several HSP90-dependent QTL affecting the hypocotyl length of dark-grown seedlings (17). These experiments used geldanamycin (GDA), a highly specific HSP90 inhibitor (20). To determine whether similar results could be obtained by genetic reduction of HSP90, we measured the hypocotyl elongation in the dark for 76 RILs of each of the HSP90-reduced and control sets.
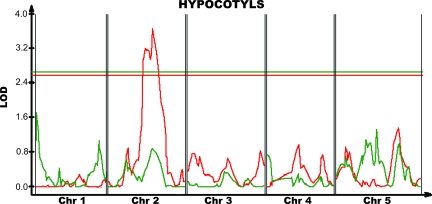
One significant dark hypocotyl length QTL was discovered in the HSP90-reduced population, whereas no significant QTL were found in the control set (Fig. 1 and SI Fig. 5). This QTL, on chromosome 2, is significantly different between the RIL sets and is HSP90-responsive (P = 0.05 by nonparametric epistasis test; see Materials and Methods). The QTL is responsible for 20% of the phenotypic variance and an additive effect of 0.90 mm of hypocotyl length.
Fig. 1.
QTL affecting dark hypocotyl length. Red, HSP90-reduced set; green, control set. QTL significance (interval map) is in LOD; a horizontal line of the appropriate color denotes a genome-wide α = 0.05 significance threshold.
This HSP90-responsive QTL is in a very similar position—≈10 cM distal to the erecta locus—to a QTL observed in previous experiments using GDA to inhibit HSP90 (17). A colocalizing QTL has also been found in the CVI × Ler RIL set (17). The other HSP90-responsive QTL found in the previous study with Col × Ler RILs were not observed with the genetic reduction of HSP90 levels. However, a region on chromosome 4, which colocalizes with one of these previously detected HSP90-dependent QTL, is highly significant for epistasis between HSP90 state and genotype (P = 0.003, nonparametric epistasis test). That is, although neither the control nor the HSP90-reduced QTL are themselves significant, the predicted additive effects are opposite in sign and significantly different.
HSP90-Responsive QTL Affecting Life-History Traits.
Hypocotyl and root elongation of dark-grown seedlings were the sole traits assessed in the accompanying study investigating HSP90-responsive QTL in A. thaliana (17) because the light sensitivity of GDA restricted us to early phenotypes in the dark. The construction of the genetically HSP90-reduced RIL set allowed us to expand the analysis of the effects of HSP90 reduction to any quantitative trait. Therefore, to assess the overall frequency of HSP90-responsive QTL, we measured a variety of life-history traits in the HSP90-reduced and control RIL sets, including flowering, size, and fitness traits. Because many of these traits are correlated (SI Table 4), principal components were also analyzed. The first four principal components each accounted for more than 10% of the total variance (37%, 22%, 13%, and 10%, respectively), so we approximate that this experiment measured the equivalent of four independent traits. Further quantitative genetic parameters are presented in SI Tables 5 and 6.
Consistent with previously reported results (13), the HSP90-reduced population tended to flower slightly later than the control population (assessed by total leaf number; plants stop producing leaves at flowering, P = 0.0017, Wilcoxon test). Also consistent with a previously published hypothesis (16), the HSP90-reduced population displayed significantly increased developmental noise (P = 0.0065, Wilcoxon test). Here, developmental noise was measured for each plant by the coefficient of variation, using the length of the stem between the first five siliques.
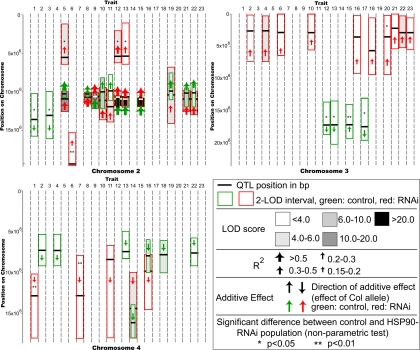
Significant QTL are depicted in Fig. 2 and SI Fig. 6, with details in SI Table 7. A major effect QTL was detected surrounding the ERECTA locus in several traits, including leaf length, plant height, and seed mass. This result is expected because ERECTA is known to pleiotropically affect many traits (21). Similarly, QTL for traits relating to flowering time were detected around the known natural variation at the FLC locus (22). Neither of these effects were HSP90-dependent. The first principal component appears to be dominated by the effects of ERECTA, the second by FLC. Overall, nine independent regions affected the traits mapped in the control set, while nine also appeared in the HSP90-reduced set. Some of these regions overlapped, whereas others did not.
Fig. 2.
Significant QTL for chromosomes 2–4. Red, HSP90-RNAi set; green, control set. Stars denote significantly HSP90-responsive QTL. Black bar, most significant location for QTL; box, 2-LOD interval. QTL significance is in shades of gray. Additive effect by direction of arrow, up signifies that Col-0 allele increases the trait mean. Trait variance explained by QTL by size of arrow. Traits: 1, days from germination to flowering; 2, rosette leaf number; 3, cauline leaf number; 4, total leaf number; 5, longest leaf length; 6, longest leaf width; 7, longest leaf depth; 8, longest leaf petiole length; 9, longest leaf petiole length:longest leaf length; 10, longest leaf surface area; 11, longest leaf volume; 12, internode mean; 13, plant height; 14, days actively flowering; 15, growth per day; 16, primary stem fertile siliques; 17, primary stem infertile siliques; 18, primary stem total siliques; 19, secondary stems from rosette; 20, secondary stems from primary stem; 21, dry mass of aerial tissue; 22, total seed weight; 23, total seed weight:dry mass.
Of the 11 independent regions with significant QTL, 9 displayed significant HSP90-buffering effects, defined as epistasis between construct and genotype, for at least one trait (nonparametric epistasis test, P < 0.05 after multiple-comparison correction). Six of these occurred on chromosomes 2, 3, and 4, which are unaffected by the segregation distortion inherent in the populations. In summary, assuming that we analyzed four independent traits, we detect 0.5 HSP90-dependent QTL per chromosome per trait.
Confirmation of Flowering Time QTL in Near-Isogenic Lines (NILs).
To confirm the statistical association of phenotype and genotype in QTL analysis, one commonly examines the trait in NILs. NILs are isogenic for most of the genome except for a small region of interest that segregates between different NILs. We focused on an HSP90-dependent total leaf number QTL on chromosome 4, because flowering time, strongly correlated to total leaf number, is a biologically important, well studied trait in A. thaliana and a QTL had not been previously reported in this region. We created NILs in a Col-0 genomic background except for two-thirds of chromosome 4, which was either homozygous Col-0 or Ler-2. Control and HSP90-RNAi lines were created for both genotypes. Because total leaf number, a flowering time correlate, had shown a significant HSP90-responsive effect in this region, we measured this trait in the NILs (Table 2). The genotype-dependent effect of HSP90 on flowering time was confirmed. Whereas control Col-0 plants had, on average, 1.4 more leaves than control Ler-2 plants, reduction of HSP90 caused Col-0 plants to have two fewer leaves than Ler-2 plants (HSP90 interaction effect, P = 0.0003, nonparametric epistasis test).
Table 2.
Confirmation of an HSP90-responsive QTL in NILs
| Genotype at 10.8 Mb | Mean total leaf number |
||
|---|---|---|---|
| Control | HSP90-reduced | Response | |
| Col | 19.3 | 16.8* | −2.5** |
| Ler | 17.9 | 18.8 | +0.9 |
*, P < 0.05;
**, P < 0.01 (by t-test).
Discussion
We have vastly expanded the utility of A. thaliana to characterize the phenomenon of HSP90-buffered genetic variation. The presented Col-0 × Ler RIL population with a constitutive genetic reduction in HSP90 levels dramatically broadens the traits and alleles whose phenotypic interactions with HSP90 may be assessed. The work presented here is thus of critical importance to determining the mechanism of HSP90 buffering and the associated potential evolutionary consequences.
The new HSP90-reduced RIL population alleviates the drawbacks of relying on GDA to assess HSP90-buffered genetic variation in A. thaliana. We demonstrate that genetic reduction of HSP90 levels can have comparable effects to inhibition with GDA through the common assessment of hypocotyl length. The major HSP90-dependent QTL previously detected with GDA, along with one other region of significant interaction, was also detected by comparison of the HSP90-reduced and control RIL sets. Other HSP90-dependent QTL previously identified with GDA may not have been detected in this study for two reasons. First, the RNAi lines reduce the level of HSP90, but GDA blocks its chaperone cycle by preventing ATP hydrolysis. The two methods might affect the activities of certain client proteins in different ways. Second, the overall decrease in HSP90 activity may have been higher in GDA experiments than the minor reduction achieved with the RNAi construct (≈68% of wild-type activity remaining). Thus, the two different methods likely lead to different levels of functional HSP90 and the expression of slightly different subsets of revealed variation.
Our survey of life-history traits revealed at least six QTL with significant HSP90-dependent effects on the three chromosomes that were unaffected by linkage to the insertions that were selected for during RIL construction. Notably, no HSP90-responsive QTL were found to colocalize with the HSP90 locus itself; all HSP90-responsive QTL reported here represent the effects that reduced HSP90 levels have on other loci. Due to correlations between some traits, we measured approximately four that were independent. The accompanying study (17) on hypocotyl and root elongation detected eight HSP90-dependent QTL in two independent traits across 2.5 genetically independent RIL sets. Our rate of discovery of 0.5 HSP90-dependent QTL per trait per chromosome per cross is quite similar to the corresponding value of 0.32 obtained in the accompanying study. These values are based on several extremely conservative assumptions. First, we only consider QTL that had both a significant mean effect in either the control or HSP90-RNAi set and that also had a significant HSP90 dependence. Second, we only score QTL as being independent loci if their positions are statistically significantly different. Third, we assume overlapping QTL in different traits to be pleiotropic effects of the same underlying genetic difference. Therefore, because our positional resolution is low, we almost certainly underestimate the number of underlying HSP90-dependent polymorphisms.
We also demonstrate that normal HSP90 levels affect phenotypic variation in another way: suppressing intraindividual developmental noise. Previously, we had shown that HSP90 activity increases interindividual developmental stability in A. thaliana populations (2). Here, we used the coefficient of variation of the first five internodes as a proxy for intraindividual noise, or fluctuating asymmetry. The observed increase in noise upon HSP90 reduction conflicts with several results from D. melanogaster (23, 24). It might be that the greater environmental responsiveness of A. thaliana and the decreased natural canalization of the examined trait may result in greater power to detect such effects in our system.
With these results, the characteristics of the effects of HSP90 on the map between genotype and phenotype come into much better focus. The RIL set and genetic approaches detailed herein provide an excellent foundation for complete mechanistic and ecological dissection of this phenomenon. We demonstrate that HSP90-responsive natural genetic variation can be observed in A. thaliana at such a frequency that nearly every trait could be expected to be affected. This result strengthens the plausibility of previous suggestions that HSP90 might play an important role in enhancing the rate of evolutionary change. Moreover, we propose that far more genetic variation may be available to alter phenotype than quantitative genetic studies generally suggest.
Materials and Methods
Recombinant Inbred Population Construction.
Constitutively HSP90-reduced line RNAi-A1 and control line Control-2 from ref. 13 had been backcrossed twice to wild-type Col-0, followed by one generation of self-propagation and selection on kanamycin for insert homozygosity. Ler-2 (CS8581) pollen was crossed to these lines. Single F1 progeny were self-propagated.
One hundred sixty kanamycin-resistant F2 seedlings were selected, and the resulting lines were subjected to single-seed descent through the F8 generation. For F2 and subsequent generations through the F6, kanamycin-resistant seedlings were selected on soil by aerosol spray of 100 μg/ml kanamycin every 3 days for the first 12 days after germination.
All generations were grown in thoroughly wetted 50% Metro-Mix 200:50% Pro-Mix BX soil with 32 pots per flat and one pot per RIL. Four to six seeds were initially planted in each pot and stratified on soil for 4 days at 4°C, followed by growth in a greenhouse environment (≈22°C with 80 μmol·m−2·s−1 cool-white light in a long-day light cycle). Humidity domes were removed after 10 days. After 15 days, all seedlings were eliminated from each pot except for the kanamycin-resistant plant closest to the center. Lines containing different constructs were grown in separate flats until through the F6. Single F7 plants of all lines were grown at the same time in a randomized design without kanamycin selection to minimize maternal environmental differences between the F8 seeds, which were used for phenotypic analysis. Lines that did not yield a resistant plant among the seeds planted or that failed to germinate or set seed were not replaced.
HSP90 Reduction in RILs.
Plant growth and RT-PCR was performed as in ref. 13. Four independent experiments were performed with HSP90-RNAi RILs 1, 2, 3, 4, and 5 and control RILs 501, 502, 503, 506, and 507. One experiment was performed to assay HSP90-RNAi RILs 13, 26, 39, 62, 89, and 102 and control RILs 512, 523, 545, 556, 573, and 584. Each experiment was analyzed with three RT-PCR reactions. Least-squares means for each RIL and for comparing HSP90-RNAi and control sets were calculated from a linear regression model including construct state, RIL nested within construct state, and experiment for the lines with multiple experiments. The two groups of RILs were modeled separately. RIL means were compared by Tukey's honestly significant difference (HSD) post hoc test and the effect of construct by ANOVA.
Genetic Mapping.
The A. thaliana indel array (18) was used to genotype 86 HSP90-reduced RILs and 93 control RILs at 240 insertion/deletion markers polymorphic between Col-0 and Ler. DNA was isolated from pooled tissue from several F8 seedlings per line. DNA preparation, array layout, hybridization, and data analysis were as described in ref. 18. Markers whose genotype could not be assigned with a posterior confidence greater than 95% were scored as missing data. A further four HSP90-reduced and three control RILs were genotyped by PCR at 10 insertion/deletions per chromosome. Markers whose genotype could not be assigned at >95% confidence (18) were treated as missing data.
Genetic maps for the RIL populations were calculated with JoinMap 4, using the regression algorithm and Haldane's mapping function (25). Only the HSP90-reduced population was used for chromosome 1, and only the control population was used for chromosome 5. Both populations were combined for chromosomes 2, 3, and 4. The total genetic distance of 405 cM is similar to the 427 cM calculated for an independent Col × Ler RIL population (26).
Hypocotyl and Root Length Analysis.
Surface-sterilized seeds were plated on germination (GM) medium (27). The medium contained 0.056% DMSO for consistency with prior similar experiments (2, 17). Plates were wrapped in foil and cold-treated at 4°C for 72 h to synchronize germination. The foil was removed and the plates were incubated at 22°C in 150 μmol/m2·sec continuous cool-white light for 2 h to increase germination frequency. The plates were then rewrapped in foil, incubated at 22°C in the dark, and photographed after 7 days. Hypocotyl and root lengths were measured on digital images with Scion Image (Scion Corporation).
Seventy-six Hsp90-reduced RILs and an equal number of control RILs were included in this experiment. Twenty seeds were plated per line. One seed from each of 10 different RILs was sown on each plate, with Hsp90-reduced and control seeds alternated spatially. The same block of 10 RILs was used for 20 plates, with seed of each RIL twice occupying each of the 10 growth positions on the plates.
Mean hypocotyl and root values were used for QTL analysis by interval mapping with QTL Cartographer v.2.5. Significance thresholds were determined by 300 permutations of the data. The effect of Hsp90 on significant QTL was determined by the nonparametric epistasis test presented below.
Life-History Trait Analysis.
Seventy-six lines of the HSP90-reduced and control RIL sets were used for genetic analysis of a variety of life-history traits. These lines were selected from among those mapped to maximize recombination breakpoint diversity with MapPop (28). F8 RIL seed of all lines had been grown in a randomized design and harvested at the same time. All lines were planted in six fully randomized blocks with one plant of each line per block. The soil was thoroughly wetted Pro-Mix BX, supplemented with 7 lbs/yd3 Osmocote 14:14:14 extended-release fertilizer. Eighteen pots were placed in a checkboard pattern within each flat, such that plants only had diagonal neighbors and did not overlap with adjoining pots. Five to 10 seeds were planted in each pot and were stratified at 4°C for 4 days. The flats were subsequently moved to the greenhouse for growth at ≈22°C with 80 μmol·m−2·s−1 cool-white light in a long-day light cycle. Humidity domes were removed after 10 days, and all plants were discarded except for the seedling closest to the center of each pot. All flats were rotated daily between growth racks to minimize the effect of microenvironments within the greenhouse. Each flat was bottom-watered overnight once per week. Orthene (acephate) insecticide (0.1 g/liter) was included in the water. Plants were removed to drying flats at the cessation of flowering, with a final watering 1 week after flowering cessation. The plants were then allowed to dry for at least 1 month before harvest. Drying flats were not rotated.
The following traits were scored: flowering date; cauline and rosette leaf number; longest leaf length, width, depth, and petiole length; length of the first five internodes on the main stem; cessation of flowering date; plant height; number of fertile and infertile siliques on the primary stem; number of secondary stems emerging from the rosette and primary stem; dry plant mass; and total seed mass. The flowering date was defined as the day the primary inflorescence reached 1 cm in height. Longest leaf measurements were conducted 1 week after flowering; internode measurements were conducted at least 2 weeks after flowering. Dry mass was measured after separating seeds.
Statistical Analysis.
Analysis of data were performed with JMP5.0 (SAS Institute). A multiple linear regression model was used to estimate the least-squares mean of each trait for each RIL. Flat, position within the flat, construct, cytoplasm, construct × cytoplasm interaction, and RIL were included in the model, with RIL nested within construct × cytoplasm. All measured traits were analyzed, along with the following composite traits: longest leaf length:longest leaf petiole length, longest leaf length × width (approximates surface area), longest leaf length × width × depth (approximates aerial volume occupied by the leaf), inflorescence growth per day, siliques produced per day, and seed weight:dry mass. Mean internode distance and the coefficient of variation (CV) of internode distance for each plant was calculated before modeling. To improve normality, rosette leaf number, total leaf number, longest leaf length × width × depth, internode CV, dry mass, and total seed weight were log-transformed before analysis, and the number of infertile siliques was probit-transformed. Broad-sense heritabilities were estimated with the same model with RIL considered as a random effect.
Because many measured characters are highly correlated, principal-component analysis was used to examine independent life-history trajectories. Least-squares means per genotype of the traits days to flower; total leaf number (log transformed); longest leaf length, width, depth, and petiole length; plant height; days flowering; total siliques on the primary stem; dry mass (log transformed); and total seed mass (log transformed) were transformed to standard normal distributions and included in the principal-component analysis because these traits have the potential to be developmentally independent. Principal components that explained greater than 10% of the total variance were treated as traits in further analyses.
Least-squares means and principal components were used for QTL analysis by interval mapping with QTL Cartographer (28). One thousand permutations per trait determined that LOD 2.7 was appropriate as a genome-wide significance threshold of α = 0.05 for all traits. Estimates of additive effects and the percentage of variation explained by each QTL were calculated with QTL Cartographer.
Because the several regions of segregation distortion may cause difficulties for epistasis tests that assume traits to be normally distributed, the effect of HSP90 on significant QTL was determined by the following highly conservative nonparametric epistasis test. For a particular trait, sort all HSP90-reduced RILs in ascending trait value and replace the trait values by rank order. Repeat separately for the control RIL set. At the marker to be tested, divide each RIL set into Col-0 and Ler-2 genotypic classes. Denote the ranks of HSP90-reduced lines with Col-0 genotype and Ler-2 genotype as RC and RL respectively. Similarly, define CC and CL for the control lines. The test statistic is
Significance is determined by comparison with a distribution of all possible such test statistics given the number of lines in all four genotypic classes. For large sample sizes with greater than five lines in each class, this distribution is well approximated by the normal distribution:
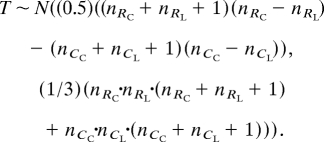 |
All P-values reported are two-tailed.
Near-Isogenic Lines.
RIL102, containing the HSP90-RNAi construct, was crossed to Col-0 (CS60000), followed by self-propagation of a single F1 plant. F2 plants were genotyped by PCR at insertion/deletion markers (18) at 1.4, 7.4, 11.6, and 17.5 Mb on chromosome 3 and at 10.8 and 14.6 Mb on chromosome 4. The presence of the HSP90-RNAi construct was assessed by segregation tests using the tightly linked kanamycin-resistance marker. Five lines were selected that were of Col-0 genotype at all tested loci, three of which were homozygous for the presence of the HSP90-RNAi construct and two of which were homozygous for its absence. Five further lines were chosen that were of Col-0 genotype at the chromosome 3 markers and Ler-2 genotype at the chromosome 4 markers. Two of these were homozygous for the presence of the HSP90-RNAi construct and three were homozygous for its absence. F3 seeds from these plants were used for experiments.
Nine plants of each of the four genotypes were grown in each of six replicate flats, with approximately equal division between lines of the same genotype. Planting, seedling selection, growth conditions, and measurement were performed as described above. Least-squares effects of genotype were derived from a linear regression model containing flat, position within the flat (corner/edge/interior), genotype, HSP90-RNAi construct presence/absence, and interaction between the HSP90 state and genotype.
Supplementary Material
ACKNOWLEDGMENTS.
T.A.S. was a Howard Hughes Medical Institute predoctoral fellow. T.A.S. and S.L. were supported by the Howard Hughes Medical Institute and the G. Harold and Leila Y. Mathers Foundation. N.S., H.N.L., E. W., K.S., K.M., H.W., S.U., and C.Q. were supported by National Institute of General Medical Sciences National Centers for Systems Biology Grant GM068763 and a Bauer Fellowship (to C.Q.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712210105/DC1.
References
- 1.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 2.Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 3.Sollars V, et al. Evidence for an epigenetic mechanism by which Hsp90 acts as a capacitor for morphological evolution. Nat Genet. 2003;33:70–74. doi: 10.1038/ng1067. [DOI] [PubMed] [Google Scholar]
- 4.Cowen LE, Lindquist S. Hsp90 potentiates the rapid evolution of new traits: Drug resistance in diverse fungi. Science. 2005;309:2185–2189. doi: 10.1126/science.1118370. [DOI] [PubMed] [Google Scholar]
- 5.Wegele H, Muller L, Buchner J. Hsp70 and Hsp90—A relay team for protein folding. Rev Physiol Biochem Pharmacol. 2004;151:1–44. doi: 10.1007/s10254-003-0021-1. [DOI] [PubMed] [Google Scholar]
- 6.Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med (Maywood) 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 7.Picard D. Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci. 2002;59:1640–1648. doi: 10.1007/PL00012491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young JC, Moarefi I, Hartl FU. Hsp90: A specialized but essential protein-folding tool. J Cell Biol. 2001;154:267–273. doi: 10.1083/jcb.200104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 10.Picard D, et al. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature. 1990;348:166–168. doi: 10.1038/348166a0. [DOI] [PubMed] [Google Scholar]
- 11.Pratt WB, Morishima Y, Murphy M, Harrell M. Chaperoning of glucocorticoid receptors. Handb Exp Pharmacol. 2006:111–138. doi: 10.1007/3-540-29717-0_5. [DOI] [PubMed] [Google Scholar]
- 12.Citri A, et al. Drug-induced ubiquitylation and degradation of ErbB receptor tyrosine kinases: Implications for cancer therapy. EMBO J. 2002;21:2407–2417. doi: 10.1093/emboj/21.10.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sangster TA, et al. Phenotypic diversity and altered environmental plasticity in Arabidopsis thaliana with reduced Hsp90 levels. PLoS ONE. 2007;2:e648. doi: 10.1371/journal.pone.0000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borkovich KA, Farrelly FW, Finkelstein DB, Taulien J, Lindquist S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeyati PL, Bancewicz RM, Maule J, van Heyningen V. Hsp90 selectively modulates phenotype in vertebrate development. PLoS Genet. 2007;3:e43. doi: 10.1371/journal.pgen.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sangster TA, Lindquist S, Queitsch C. Under cover: Causes, effects and implications of Hsp90-mediated genetic capacitance. BioEssays. 2004;26:348–362. doi: 10.1002/bies.20020. [DOI] [PubMed] [Google Scholar]
- 17.Sangster TA, et al. HSP90 affects the expression of genetic variation and developmental stability in quantitative traits. Proc Natl Acad Sci USA. 2008;105:2963–2968. doi: 10.1073/pnas.0712200105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salathia N, et al. Indel arrays: An affordable alternative for genotyping. Plant J. 2007;51:727–737. doi: 10.1111/j.1365-313X.2007.03194.x. [DOI] [PubMed] [Google Scholar]
- 19.Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- 20.Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: Essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci USA. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torii KU, et al. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milton CC, Batterham P, McKenzie JA, Hoffmann AA. Effect of E (sev) and Su(Raf) Hsp83 mutants and trans-heterozygotes on bristle trait means and variation in Drosophila melanogaster. Genetics. 2005;171:119–130. doi: 10.1534/genetics.104.038463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milton CC, Huynh B, Batterham P, Rutherford SL, Hoffmann AA. Quantitative trait symmetry independent of Hsp90 buffering: Distinct modes of genetic canalization and developmental stability. Proc Natl Acad Sci USA. 2003;100:13396–13401. doi: 10.1073/pnas.1835613100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stam P. Construction of integrated genetic-linkage maps by means of a new computer package—Joinmap. Plant J. 1993;3:739–744. [Google Scholar]
- 26.Alonso-Blanco C, et al. Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 1998;14:259–271. doi: 10.1046/j.1365-313x.1998.00115.x. [DOI] [PubMed] [Google Scholar]
- 27.Queitsch C, Hong SW, Vierling E, Lindquist S. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell. 2000;12:479–492. doi: 10.1105/tpc.12.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vision TJ, Brown DG, Shmoys DB, Durrett RT, Tanksley SD. Selective mapping: A strategy for optimizing the construction of high-density linkage maps. Genetics. 2000;155:407–420. doi: 10.1093/genetics/155.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.