Abstract
Cytotoxic T lymphocytes (CTL) play an important role in the control and elimination of infection by West Nile virus (WNV), yet the class I human leukocyte antigen (HLA)-presented peptide epitopes that enable CTL recognition of WNV-infected cells remain uncharacterized. The goals of this work were first to discover the peptide epitopes that distinguish the class I HLA of WNV-infected cells and then to test the T cell reactivity of newly discovered WNV epitopes. To discover WNV-immune epitopes, class I HLA was harvested from WNV (NY99 strain)-infected and uninfected HeLa cells. Then peptide epitopes were eluted from affinity-purified HLA, and peptide epitopes from infected and uninfected cells were comparatively mapped by mass spectroscopy. Six virus-derived peptides from five different viral proteins (E, NS2b, NS3, NS4b, and NS5) were discovered as unique to HLA-A*0201 of infected cells, demonstrating that the peptides sampled by class I HLA are distributed widely throughout the WNV proteome. When tested with CTL from infected individuals, one dominant WNV target was apparent, two epitopes were subdominant, and three demonstrated little CTL reactivity. Finally, a sequence comparison of these epitopes with the hundreds of viral isolates shows that HLA-A*0201 presents epitopes derived from conserved regions of the virus. Detection and recovery from WNV infection are therefore functions of the ability of class I HLA molecules to reveal conserved WNV epitopes to an intact cellular immune system that subsequently recognizes infected cells.
Keywords: epitope hierarchy, human leukocyte antigen, immunodominance, major histocompatibility complex, mass spectrometry
West Nile virus (WNV) is a single-stranded positive sense RNA flavivirus that is related to other human pathogens such as yellow fever virus and dengue fever virus. In nature, WNV exists in an enzootic cycle between birds and mosquitoes, with other species like horses and humans acting as incidental terminal hosts (1, 2). In humans, WNV causes a nonspecific febrile illness with rare cases of fatal encephalitis (3). Like other flaviviruses, WNV translates its genome into a polyprotein of ≈3,400 aa that is proteolytically cleaved into three structural proteins and seven nonstructural proteins (4). In contrast to other RNA viruses, such as HIV and influenza, WNV exhibits a high degree of sequence conservation in its natural reservoir (5). Such sequence conservation makes WNV a promising target with regard to targeting humoral and cellular immune responses to conserved epitopes.
WNV elicits a strong immune response from innate and adaptive branches of the immune system (6). With regard to adaptive immunity, WNV infection results in the generation of neutralizing antibodies that can protect mice from lethal infection when given passively (7–9), whereas mice deficient for cytotoxic T lymphocytes (CTL) or class I major histocompatibility complex (MHC) exhibit increased viral burdens and increased mortality (10–13). A key component of the cellular antiviral immune response is the antigen-specific interaction of T cell receptors and the class I MHC–peptide complex. Human MHC class I, or the human leukocyte antigens (HLA), are a complex consisting of a polymorphic α chain, β2-microglobulin, and a peptide ligand of 8–13 residues. Class I HLA molecules sample peptides that are representative of the intracellular proteome and present their peptide cargo on the cell surface. During viral infection, the proteome of the infected cell is altered as are the peptides presented to CTL by class I HLA. Although CTL recognize infected cells, the WNV epitopes presented by class I HLA have not been reported. The goals of this work were to identify peptide epitopes unique to infected cells and to assess T cell recognition of these epitopes.
To discover WNV immune epitopes, class I HLA molecules were gathered from infected and uninfected cells; then peptide epitopes from infected and uninfected cells were comparatively mapped, and peptides unique to infected cells were sequenced (14). Using this direct epitope discovery approach, we identified viral-encoded epitopes uniquely presented by HLA-A*0201 molecules of WNV-infected cells. We found A*0201 to be extremely effective in its interaction with WNV: highly conserved, high-affinity amino acid sequences dispersed throughout the viral proteome were sampled by class I HLA-A*0201. The discovered WNV epitopes were then tested for cellular reactivity in an IFN-γ ELISPOT assay in WNV-positive HLA-A*0201 patients. A hierarchy emerged whereby the conserved high-affinity epitopes were dominant cellular immune targets. These data represent a characterization of class I HLA-presented WNV epitopes and indicate that cellular immune responses focus on a conserved subset of the available WNV epitopes.
Results
Production and Purification of HLA-A*0201-Specific Peptide Pools from WNV-Infected Cells.
The first objective of this work was to understand the number and nature of class I HLA-presented peptide epitopes unique to WNV-infected cells. Secreted class I HLA proteins (sHLA) were used to produce a sufficient quantity of peptides for comparative proteomics. To confirm that one set of peptides in the comparative analysis were derived from infected cells, we tested for viral RNA in the cells and in the sHLA-containing culture supernatant. Florescence in situ hybridization (FISH) and real-time PCR (RT-PCR) confirmed the presence of viral RNA in the cells and supernatant, respectively. Viral RNA was detected 12 h after infection in the cells and in the supernatant (data not shown). Optimally infected cell supernatant from days 0.5 to 5.5 after infection were pooled for affinity chromatography purification. Infected and uninfected sHLA purified was 9.1 and 11 mg, respectively, as determined by ELISA.
Comparative Ion Mapping Identifies Six WNV-Derived Peptides.
Pools of peptides eluted from class I HLA molecules contain thousands of different peptides. To reduce the complexity of the peptides before comparative analysis, peptides were separated by reverse-phase HPLC (RP-HPLC) into fractions containing ≈200–250 peptides each (Fig. 1A). Similar HPLC elution patterns for uninfected and infected peptide pools were obtained (Fig. 1A). MS ion maps were generated for corresponding infected and uninfected HPLC fractions. These ion maps were compared to identify ions unique to infected fractions. The vast majority of ions were shared in the infected and uninfected MS ion maps (Fig. 1B). Tandem mass spectrometry (MS/MS) sequence analysis of shared ions in the infected and uninfected ion maps produced like sequences (data not shown), confirming the alignment of HPLC fractions and overlap in the peptide preparations. Fig. 1B shows an ion map generated from HPLC fraction 63, where ion 524.8 was identified as unique to the WNV-infected peptide pools. MS/MS fragmentation was performed on ions unique to infected cells and individual peptide sequences were determined (Fig. 1C). MS/MS fragmentation at the same location in the corresponding uninfected fraction (and neighboring uninfected fractions) confirmed the absence of this peptide in uninfected cells. Fragmentation patterns between the newly discovered WNV-derived peptides were compared with synthetic peptide patterns to ensure sequence identity. Fig. 1C shows the MS/MS fragmentation pattern of ion 524.8 (identified as WNV-derived SLF9 peptide) overlaid with a synthetic peptide demonstrating a correct sequence assignment. Finally, WNV-derived peptides were assayed for their binding affinity to HLA-A*0201. Fig. 1D demonstrates that SLF9 is a high-affinity HLA-A*0201 ligand with an IC50 of 204 nM.
Fig. 1.
Direct epitope discovery of WNV epitope SLF9. (A) RP-HPLC profile of HLA-A*0201 peptides from uninfected (blue) and WNV-infected (red) cells. (B) MS ion spectra of HPLC fraction 63 from uninfected (blue) and infected (red) cells. MS data were recorded for 300 scans. (C) Overlay of the MS/MS fragmentation pattern of ion 528.4 in the infected HPLC fraction 63 (identified as the SLF9 peptide; red) with the MS/MS fragmentation pattern for the SLF9 synthetic peptide (blue). (D) Florescence polarization competitive binding assay with the SLF9 synthetic peptide and HLA-A*0201.
By using this direct comparative approach, six HLA-A*0201 WNV-derived peptides were identified from five different viral proteins (E, NS2b, NS3, NS4b, and NS5) (Fig. 2 and Table 1). Five peptides were nonamers and one a decamer. Three of the peptides (RLD10, SLF9, and SLT9) had a common A*0201 P2 leucine anchor residue, peptides YTM9 and ATW9 had a threonine P2 anchor, and SVG9 had a valine at P2. At their C terminus, A*0201 peptide ligands prefer a leucine or valine anchor, and only SLT9 (C-terminal alanine) did not have a common A*0201 C-terminal anchor. These putative WNV epitopes fit the reported A*0201-binding motif. When tested for binding to HLA-A*0201, these six WNV peptides were found to be high-affinity binders with SLF9, SVG9, and YTM9 being the strongest binders, having an IC50 of 204, 247, and 291 nM, respectively (Table 1). The other peptides, SLT9, ATW9, and RLD10, had comparatively lower affinities, with IC50 values of 503, 780, and 847 nM, respectively. The fluorescence polarization assay used to assess A*0201 binding categorizes peptides with IC50 values <5,000 nM as high affinity (15). In summary, we see that HLA-A*0201 presents six high-affinity peptides derived from five different proteins throughout the WNV proteome (Fig. 2).
Fig. 2.
Location of A*0201 WNV peptide epitopes on the WNV polyprotein. C, nucleocapsid; M, membrane; E, envelope glycoprotein; NS, nonstructural.
Table 1.
Identified WNV-derived peptides
| Peptide | Sequence† | Protein | Location† | IC50, nM |
|---|---|---|---|---|
| SVG9 | SVGGVFTSV | Env | 430–438 | 247 |
| RLD10 | RLDDDGNFQL | NS2b | 78–87 | 847 |
| YTM9 | YTMDGEYRL | NS3 | 518–526 | 291 |
| SLF9 | SLFGQRIEV | NS4b | 68–76 | 204 |
| SLT9 | SLTSINVQA | NS4b | 15–23 | 503 |
| ATW9 | ATWAENIQV | NS5 | 862–870 | 780 |
†WNV NY99 polyprotein accession no. AAF20092.
CTL from Infected Individuals Recognize WNV Peptides.
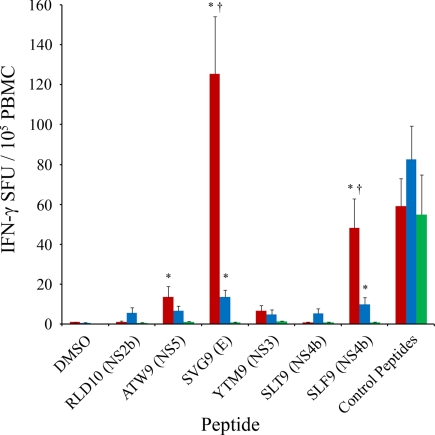
These WNV peptide epitopes were tested for recognition by CTL from HLA-A*0201 WNV-infected individuals. Peripheral blood mononuclear cells (PBMCs) from WNV-infected patients were tested in ELISPOT assays with the six identified WNV peptides. To assess CTL responsiveness over time, we tested three sets of CTL from HLA-A*0201 individuals: (i) the early set, PBMCs isolated from individuals who had recently recovered from infection (mean, 26.9 days after infection); (ii) the late set, PBMCs isolated from individuals ≈1 year after infection; and (iii) PBMCs from WNV-negative healthy controls. In both the WNV early and late infection sets, PBMCs responded strongly to SVG9 [mean = early, 125.3; late, 13.4 spot-forming units (SFU) per 105 PBMCs] and SLF9 (mean = early, 48.0; late, 9.8 SFU per 105 PBMCs) with a moderate but significant response in the early set to ATW9 (mean = 13.6 SFU per 105 PBMCs) compared with healthy controls. The WNV peptide epitopes RLD10, YTM9, and SLT9, as a group, demonstrated no significant activity above the healthy controls in the both early and late sets (Fig. 3). These data demonstrate an epitope hierarchy whereby SVG9>SLF9>ATW9>RLD10-SLT9-YTM9.
Fig. 3.
CTL reactivity of identified WNV epitopes. Shown are IFN-γ ELISPOT assays with WNV peptides and PBMC isolated early (red) and late (blue) after infection from WNV-positive individuals and healthy controls (green). *, significant increase from healthy controls; †, significant increase from the late set (blue). Significance was determined by Kruskal–Wallis one-way ANOVA followed by Dunn's method; P < 0.05.
Although the early and late groups demonstrate a CTL response to WNV epitopes, a significant decrease in the magnitude of the CTL response is observed at the latter time point. For CTL that respond to the SVG9 peptide, there is a 9.3-fold decrease in average SFU from early to late, and for the SLF9 peptide there is a 4.9-fold decrease, suggesting a reduction in the number of CTL available to respond to specific WNV peptides over time (Fig. 3). The magnitude of CTL recall needed to confer immunity to subsequent infection is unknown. These data demonstrate a hierarchical CTL response to these newly discovered HLA-A*0201-restricted epitopes in both early and late populations, with a reduction in response at a later time after infection.
Heterogeneity Among Responses to HLA-A*0201-Restricted WNV Epitopes.
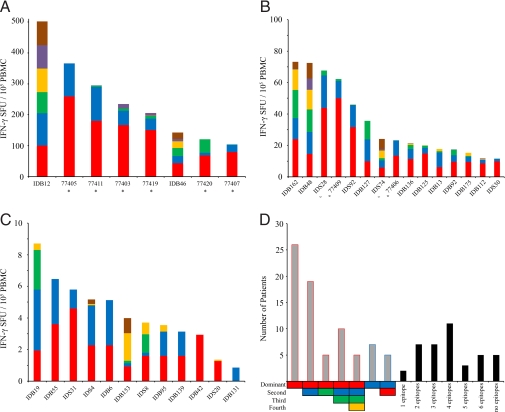
Having identified common trends in epitope hierarchy and response strength (early and late; Fig. 3), we then analyzed CTL response heterogeneity in three ways: (i) the magnitude of each individual's response to each epitope; (ii) each individual's epitope hierarchy; and (iii) the total number of epitopes recognized by each individual's CTL (Fig. 4). Responders were categorized as high responders [>100 SFU per 105 PBMCs (Fig. 4A)], medium responders [10–100 SFU per 105 PBMCs (Fig. 4B)], and low responders [<10 SFU per 105 PBMCs (Fig. 4C)]. Eight individuals, six early and two late, were high responders (Fig. 4A). Fifteen medium responders were identified, seven of which followed the SVG9>SLF9>ATW9 hierarchy (Fig. 4B). Twelve low responders were identified, and although a hierarchy was not clear in many of these weak responders, responses to SVG9 and SLF9 predominated (Fig. 4C). Five infected individuals did not respond to WNV epitopes above background, although two of these nonresponders did not recognize the positive control peptides (data not shown); only 3 of 40 infected A*0201 individuals with measurable CTL responses failed to recognize the peptides discovered here.
Fig. 4.
Heterogeneity among CTL responses. Patient-specific responses to WNV epitopes SVG9 (red), SLF9 (blue), ATW9 (green), RLD10 (yellow), YTM9 (purple), and SLT9 (brown). (A) High responders, >100 SFU. (B) Moderate responders, 1–100 SFU. (C) Low responders, <10 SFU. Histograms represent the average number of SFU per patient minus the healthy control (plus 2× SEM) and DMSO control. *, patient is from the early group. (D) (Right) Number of patients with a specific epitope hierarchy. (Left) Number of epitopes to which each patient responded. Colored boxes, WNV epitopes as stated.
Heterogeneity was observed in immune epitope hierarchies. (Fig. 4D Left). The largest grouping of donors (26 in total) responded to SVG9 (red) as the dominant epitope. Of the 26 donors that predominantly recognized SVG9, 19 recognized SLF9 (blue), and 5 recognized ATW9 (green) as a secondary epitope. Of the 19 donors that responded to SVG9 followed by SLF9, most (10 of 19) responded to ATW9 as their third epitope. Five of the donors that responded to SVG9>SLF9>ATW9 responded to RLD10 (yellow) as their fourth. For 7 donors, the dominant epitope was SLF9 of which 5 reacted to SVG9 as their second most dominant epitope. Finally, donors were grouped by the number of epitopes to which they responded (Fig. 4D Right). Few donors (2 of 40) responded to only one epitope with four epitopes most commonly recognized (11 of 40 donors).
Viral Sequence Homology of the Epitopes.
We next examined sequence variability among reported WNV isolates to assess conservation and divergence for these newly discovered A*0201 peptide epitopes. Of the dominant epitopes, SVG9 was the most conserved, with 97.2% (600 of 617 sequences) encoding the epitope sequence we identified in the NY99 strain. Within SVG9, 1.6% (10 sequences) displayed a conservative valine to isoleucine substitution at P2. The SLF9 and ATW9 epitopes demonstrated 86.1% (118 of 137) and 86.4% (171 of 198) homology among reported WNV sequences. Variants of the SLF9 epitope altered 3 or 4 of the 9 residues in the epitope, whereas the most common variation of the ATW9 epitope (11.1%) was a glutamate to histidine at P8. The YTM9, SLT9, and RLD10 epitopes displayed 97.8% (134 of 137), 100% (137 of 137), and 97.1% (135 of 139) sequence homology with reported WNV isolates, respectively. These data show that the WNV peptide epitopes sampled by A*0201 are highly conserved among hundreds of viral isolates.
Discussion
Several experimental observations can be gleaned from the endogenously processed WNV epitopes reported here. First is that the WNV-encoded peptides clearly distinguish themselves by MS intensity, as shown in Fig. 1B; the WNV peptides found here were high-abundance peptides compared with numerous unchanged host ligands. Another observation is that the six viral peptides were derived from multiple proteins in the WNV proteome. We observed that the class I molecule HLA-A*0201 was able to sample epitopes from half of the viral proteins, both structural and nonstructural, during infection. These peptides were of high affinity for A*0201 and were conserved among numerous WNV isolates.
An immune hierarchy emerged when these viral epitopes were assayed by IFN-γ ELISPOT. As a group, three WNV epitopes were recognized by CTL significantly above background: SVG9>SLF9>ATW9 (Fig. 3A). Epitopes RLD10 and YTM9 displayed moderate reactivity, and SLT9 was the least immunogenic. Furthermore, there was a significant decrease in CTL reactivity to WNV peptides in blood collected from infected individuals at a later time point, indicating a loss of CTL specific for WNV epitopes over time. Individual immune responses were heterogeneous in their level of response, epitope hierarchy, and number of total epitopes recognized. Moreover, there were no statistically significant associations of age, gender, race, duration of infection, and severity of disease with level of responses (data not shown). Cumulatively, these data show that six WNV epitopes were presented by A*0201, that most individuals recognized a dominant target and an auxiliary epitope, and that a small group of patients responded to five to six epitopes. Until now, this type of immune response hierarchy has been impossible to envision because a thorough survey of epitopes presented for recognition by the cellular immune system had not been completed. Here, the direct identification of WNV epitopes and subsequent immunogenicity testing provides metrics for how HLA molecules reduce viral proteins into a handful of optimal CTL targets.
The epitope hierarchy presented coincides with that seen in mice. Two dominant epitopes have recently been identified in mice during WNV infection. An E glycoprotein-derived epitope is presented by the mouse class I MHC molecule H-2Kb, and an NS4b epitope is restricted to H-2Db (16, 17). In the Brien et al. work (16), the dominant H-2Db peptide epitope accounted for 50–70% of the reactivity, and the remaining CTL reactivity was dispersed among three other WNV peptide epitopes. These murine data fit the observations presented here in that we find a dominant SVG9 epitope along with subdominant SLF9 and ATW9 epitopes presented by A*0201. Moreover, these mouse studies reported a trend of dominant epitopes having the highest binding affinities, of having multiple viral proteins sampled, and for having epitope sequences conserved among numerous viral isolates (data not shown). The discovery of conserved/dominant WNV epitopes presented by human MHC molecules in this work is therefore consistent with observations in mice: dominant viral epitopes are presented by MHC class I and efficiently targeted by CTL.
One would be hard pressed to arrive at a more eloquent model of antigen presentation and cellular immune reactivity: multiple epitopes in half of the viral proteins are sampled by A*0201, a cellular immune response recognizes several of the available viral epitopes, and conserved epitopes of high affinity for A*0201 are preferentially targeted. These data are quite powerful for two reasons. First, understanding how the immune response successfully samples WNV epitopes provides a comparative model as we try to elucidate host–pathogen points of interaction with more elusive viruses such as HIV. Second, we have confirmed that conserved WNV epitopes are endogenously loaded and presented to CTL by the class I of infected cells. CTL-eliciting vaccines that use endogenous WNV epitopes can be tested, and the presentation of confirmed WNV epitopes by various HLA-A2 supertype members can be ascertained. In summary, the direct discovery and subsequent confirmation of viral CTL epitopes described here provide a solid foundation for viral antigen presentation studies and CTL vaccine design. These data provide an immune system “success story” whereby the nature of viral epitopes that are endogenously sampled has been determined and the immune response to these viral epitopes has been confirmed.
Materials and Methods
Virus and Cell Culture.
WNV strain WNV NY99 was propagated on Vero E6 cell monolayers [American Type Culture Collection (ATCC) CRL-1586]. Infected cell supernatant was cleared of cell debris by centrifugation at 3,000 × g for 15 min and stored at −80°C. HeLa (ATCC CCL-2) and Vero cells were subcultured according to ATCC instructions in DMEM (Caisson Laboratories, Inc.), 10% FBS (Serum Source International), and 1% penicillin/streptomycin (Invitrogen).
sHLA-Secreting Transfectant Cell Line.
HLA-A*0201 cDNA was amplified by using a reverse oligonucleotide primer that truncated the 3′ end of exon 4, deleting the transmembrane and cytoplasmic domains. Furthermore, the 3′ reverse oligonucleotide primer included a VLDLr purification epitope tag (SVVSTDDDLA) that is recognized by the anti-VLDLr mAb (ATCC CRL-2197) (18). The resulting PCR product was cloned into pcDNA 3.1 (Invitrogen) and transfected by FuGENE 6 reagent (Roche) into HeLa cells. Quantification of sHLA in supernatant was performed by using a sandwich ELISA, where an antibody against the VLDLr epitope was the capture antibody, the primary detector antibody was directed against β2-microglobulin (Dako Cytomation).
Viral Detection.
WNV plaque assays were performed on both Vero and HeLa monolayers as described in ref. 19. Rapid detection of WNV RNA in supernatant was completed by using a TaqMan-based reverse-transcriptase RT- PCR. The primers and probes used have been reported to detect the E region of WNV genome (20). RT-PCR was performed by using the Applied Biosystems 7500 RT-PCR system under universal cycling conditions. Approximate plaque-forming units (pfu) were determined from the Ct value using a standard curve [(y = −1.1145 Ln(x) + 29.630); R2 = 0.9899, x range = 1.2 × 101 to 1.2 × 106 pfu/ml].
Class I HLA from WNV-infected cells was required to detect epitopes unique to infected cells. To confirm that HeLa cells were virus-infected, a FISH assay and flow cytometry were performed (21). The probes used in this work were 18S rRNA (5′-[Cy5]TCTAGCGGCGCAATACGAAT-3′) as a positive control, 18S rRNA reverse compliment (5′-[Cy5]ATTCGTATTGCGCCGCTA-3′) as a negative control, and WNVE (5′-[Cy5]GCCCGACCATGGGAGAAGCTC-3′). Hybridized cells were analyzed with a FACSCalibur flow cytometer (BD Biosciences).
Production and Isolation of HLA-A*0201 Peptides.
sHLA-A*0201 peptide complexes were produced in a bioreactor format as described in refs. 22–24. To obtain sHLA from WNV-infected cells, bioreactors containing HeLa cells were infected with 1.5 × 108 pfu (as determined by HeLa plaque assay) in a total volume of 525 ml and recirculated in the bioreactor for 2 h before the harvest of sHLA-containing supernatant was initiated. Supernatant from days 0.5 to 5.5 after infection were pooled for purification because we needed 12 h to flush uninfected supernatant from the system. sHLA complexes were purified by affinity chromatography with the anti-VLDLr epitope antibody. Acid-eluted peptides were lyophilized and then fractionated by RP-HPLC with a Jupiter Proteo 90 Å, 4 μm, 150 × 2 column (Phenomenex) (14, 25).
MS Analysis and Peptide-Binding Assay.
MS ion maps were generated for HPLC fractions containing peptides. Fractions were ionized by using nanoelectrospray capillaries (Proxeon) into a QSTAR elite (Applied Biosystems) electrospray quadrupole time-of-flight mass spectrometer. Peptide sequences were determined manually by using the MASCOT software package (Matrix Science). Peptide binding to HLA-A*0201 was determined by using the florescence polarization method (Pure Protein L.L.C.) (16). WNV peptide epitope sequence alignments were made by using CLC Free Workbench 3 (CLC Bio).
WNV Patient Samples.
For the early sample group, peripheral blood was collected from consenting donors (four men, four women) acutely infected with WNV in heparinized tubes in accordance with a protocol approved by Research Ethics Board at McMaster University. The median patient age was 62.5 years, with a range of 23–81 years (mean = 57.9 ± 7.1 years). The median time between onset of disease and the blood draw was 28.5 days, with a range of 12–46 days (mean = 26.9 ± 4.0 days). PBMCs were isolated and cryopreserved in RPMI containing 12.5% human serum albumin (Sigma) and 10% DMSO (Sigma) (26). HLA-A*02 expression was determined by flow cytometry by using phycoerythrin-conjugated HLA-A*02 antibody (clone BB7.2; Becton Dickinson).
For the late sample group, peripheral blood was collected from 32 WNV antibody-positive subjects, recruited from high-incidence areas in the central and southwest health districts of the State of Idaho. All persons had been diagnosed with WNV in the summer of 2006, with confirmatory testing conducted by the Idaho State Health Department. Subjects were recruited between March and June 2007, and all gave informed consent following approved Pittsburgh University IRB protocols. The median age of the Idaho cohort was 50 years, and the range was 31–88 years. After collection, the PBMC fraction was isolated on Ficoll–Hypaque density gradients and cryopreserved in 10% DMSO. Negative controls were anonymous donors obtained from Pittsburgh Central Blood Bank. All subjects were prescreened for HLA-A*02 by staining with antibody (clone BB7.2; Becton Dickinson) and confirmed by HLA DNA-based typing.
IFN-γ ELISPOT.
For the early sample group, ELISPOT assays were carried out by using the human IFN-γ ELISPOT set (BD Biosciences) according to the manufacturer's instructions with 2 μg/ml peptide. As a positive control, PBMC samples were restimulated with peptide pools containing class I restricted viral peptides specific for Epstein–Bar virus (EBV; 91 peptides), cytomegalovirus CMV; 40 peptides), or influenza (47 peptides), respectively. The flu peptides were selected from the Immune Epitope database (27). The EBV and CMV peptides were described by Bihl et al. (28). For the late sample group, human IFN-γ ELISPOT assays were carried out as described in ref. 29. Briefly, PBMC were thawed, allowed to recover overnight at 37°C, and then added to nitrocellulose-coated filter plates (Millipore) at 1 × 105 cells per well (triplicate wells per treatment). After -overnight incubation with WNV peptides (10 μg/ml) or control pool of 4 CMV, 15 EBV, and 12 influenza virus (CEF) peptides (1 μg/ml) representing a cross-section of MHC class I dominant epitopes (30) or medium control, the plates were developed, and spots were counted with an automated ELISPOT reader (AID GmbH). All data are presented as SFUs per 105 PBMCs.
Statistical Analysis.
For Fig. 3, significance was determined by Kruskal–Wallis one-way ANOVA followed by Dunn's method. A P value of <0.05 was considered significant. Statistical analysis was performed by using SigmaStat 3.0 software (SPSS, Inc.).
ACKNOWLEDGMENTS.
We thank Dr. Ken Jackson of the University of Oklahoma Health Sciences Left Molecular Biology Proteomics Facility for technical assistance. Also, we thank the Idaho Central District Health Department, the Idaho Southwest District Health Department, and the participants with a history of WNV infection. We thank Dr. Matt Fogle, Oriana Hawkins, and Dr. Daryl Cox for their review of the manuscript. This work was supported by National Institutes of Health Contract HHSN266200400027C (to W.H.H.) and National Institutes of Health Contract N01-AI-40066 (to J.B. and M.L.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Hayes EB, et al. Epidemiology and transmission dynamics of West Nile virus disease. Emerging Infect Dis. 2005;11:1167–1173. doi: 10.3201/eid1108.050289a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bunning ML, et al. Experimental infection of horses with West Nile virus and their potential to infect mosquitoes and serve as amplifying hosts. Ann NY Acad Sci. 2001;951:338–339. doi: 10.1111/j.1749-6632.2001.tb02712.x. [DOI] [PubMed] [Google Scholar]
- 3.Sejvar JJ, et al. Neurologic manifestations and outcome of West Nile virus infection. J Am Med Assoc. 2003;290:511–515. doi: 10.1001/jama.290.4.511. [DOI] [PubMed] [Google Scholar]
- 4.Brinton MA. The molecular biology of West Nile virus: A new invader of the Western hemisphere. Annu Rev Microbiol. 2002;56:371–402. doi: 10.1146/annurev.micro.56.012302.160654. [DOI] [PubMed] [Google Scholar]
- 5.Jerzak G, Bernard KA, Kramer LD, Ebel GD. Genetic variation in West Nile virus from naturally infected mosquitoes and birds suggests quasispecies structure and strong purifying selection. J Gen Virol. 2005;86:2175–2183. doi: 10.1099/vir.0.81015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samuel MA, Diamond MS. Pathogenesis of West Nile virus infection: A balance between virulence, innate and adaptive immunity, and viral evasion. J Virol. 2006;80:9349–9360. doi: 10.1128/JVI.01122-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Nathan D, et al. Prophylactic and therapeutic efficacy of human intravenous immunoglobulin in treating West Nile virus infection in mice. J Infect Dis. 2003;188:5–12. doi: 10.1086/376870. [DOI] [PubMed] [Google Scholar]
- 8.Diamond MS, Shrestha B, Marri A, Mahan D, Engle M. B cells and antibody play critical roles in the immediate defense of disseminated infection by West Nile encephalitis virus. J Virol. 2003;77:2578–2586. doi: 10.1128/JVI.77.4.2578-2586.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliphant T, et al. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat Med. 2005;11:522–530. doi: 10.1038/nm1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kesson AM, Blanden RV, Mullbacher A. The primary in vivo murine cytotoxic T cell response to the flavivirus, West Nile. J Gen Virol. 1987;68:2001–2006. doi: 10.1099/0022-1317-68-7-2001. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Lobigs M, Lee E, Koskinen A, Mullbacher A. CD8(+) T cell-mediated immune responses in West Nile virus (Sarafend strain) encephalitis are independent of γ-interferon. J Gen Virol. 2006;87:3599–3609. doi: 10.1099/vir.0.81306-0. [DOI] [PubMed] [Google Scholar]
- 12.Shrestha B, Diamond MS. Role of CD8+ T cells in control of West Nile virus infection. J Virol. 2004;78:8312–8321. doi: 10.1128/JVI.78.15.8312-8321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shrestha B, Samuel MA, Diamond MS. CD8+ T cells require perforin to clear West Nile virus from infected neurons. J Virol. 2006;80:119–129. doi: 10.1128/JVI.80.1.119-129.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wahl A, Weidanz J, Hildebrand W. Direct class I HLA antigen discovery to distinguish virus-infected and cancerous cells. Expert Rev Proteomics. 2006;3:641–652. doi: 10.1586/14789450.3.6.641. [DOI] [PubMed] [Google Scholar]
- 15.Buchli R, et al. Development and validation of a fluorescence polarization-based competitive peptide-binding assay for HLA-A*0201: A new tool for epitope discovery. Biochemistry. 2005;44:12491–12507. doi: 10.1021/bi050255v. [DOI] [PubMed] [Google Scholar]
- 16.Brien JD, Uhrlaub JL, Nikolich-Zugich J. Protective capacity and epitope specificity of CD8(+) T cells responding to lethal West Nile virus infection. Eur J Immunol. 2007;37:1855–1863. doi: 10.1002/eji.200737196. [DOI] [PubMed] [Google Scholar]
- 17.Purtha WE, et al. Antigen-specific cytotoxic T lymphocytes protect against lethal West Nile virus encephalitis. Eur J Immunol. 2007;37:1845–1854. doi: 10.1002/eji.200737192. [DOI] [PubMed] [Google Scholar]
- 18.Hickman-Miller HD, et al. Rhesus macaque MHC class I molecules present HLA-B-like peptides. J Immunol. 2005;175:367–375. doi: 10.4049/jimmunol.175.1.367. [DOI] [PubMed] [Google Scholar]
- 19.Papin JF, Vahrson W, Dittmer DP. SYBR green-based real-time quantitative PCR assay for detection of West Nile virus circumvents false-negative results due to strain variability. J Clin Microbiol. 2004;42:1511–1518. doi: 10.1128/JCM.42.4.1511-1518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanciotti RS, et al. Rapid detection of West Nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J Clin Microbiol. 2000;38:4066–4071. doi: 10.1128/jcm.38.11.4066-4071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borzi RM, et al. A fluorescent in situ hybridization method in flow cytometry to detect HIV-1-specific RNA. J Immunol Methods. 1996;193:167–176. doi: 10.1016/0022-1759(96)00070-1. [DOI] [PubMed] [Google Scholar]
- 22.Hickman HD, et al. Toward a definition of self: Proteomic evaluation of the class I peptide repertoire. J Immunol. 2004;172:2944–2952. doi: 10.4049/jimmunol.172.5.2944. [DOI] [PubMed] [Google Scholar]
- 23.Prilliman K, et al. Large-scale production of class I bound peptides: Assigning a signature to HLA-B*1501. Immunogenetics. 1997;45:379–385. doi: 10.1007/s002510050219. [DOI] [PubMed] [Google Scholar]
- 24.Prilliman K, et al. HLA-B15 peptide ligands are preferentially anchored at their C termini. J Immunol. 1999;162:7277–7284. [PubMed] [Google Scholar]
- 25.Hickman HD, et al. C-terminal epitope tagging facilitates comparative ligand mapping from MHC class I-positive cells. Hum Immunol. 2000;61:1339–1346. doi: 10.1016/s0198-8859(00)00216-0. [DOI] [PubMed] [Google Scholar]
- 26.Disis ML, et al. Maximizing the retention of antigen-specific lymphocyte function after cryopreservation. J Immunol Methods. 2006;308:13–18. doi: 10.1016/j.jim.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Peters B, et al. The immune epitope database and analysis resource: From vision to blueprint. PLoS Biol. 2005;3:e91. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bihl FK, et al. Simultaneous assessment of cytotoxic T lymphocyte responses against multiple viral infections by combined usage of optimal epitope matrices, anti- CD3 mAb T cell expansion, and “RecycleSpot.”. J Transl Med. 2005;3:20–39. doi: 10.1186/1479-5876-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang XL, et al. Processing and presentation of exogenous HLA class I peptides by dendritic cells from human immunodeficiency virus type 1-infected persons. J Virol. 2005;79:3052–3062. doi: 10.1128/JVI.79.5.3052-3062.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Currier JR, et al. A panel of MHC class I restricted viral peptides for use as a quality control for vaccine trial ELISPOT assays. J Immunol Methods. 2002;260:157–172. doi: 10.1016/s0022-1759(01)00535-x. [DOI] [PubMed] [Google Scholar]