Abstract
Signaling through the programmed death 1 (PD-1) inhibitory receptor upon binding its ligand, PD-L1, suppresses immune responses against autoantigens and tumors and plays an important role in the maintenance of peripheral immune tolerance. Release from PD-1 inhibitory signaling revives “exhausted” virus-specific T cells in chronic viral infections. Here we present the crystal structure of murine PD-1 in complex with human PD-L1. PD-1 and PD-L1 interact through the conserved front and side of their Ig variable (IgV) domains, as do the IgV domains of antibodies and T cell receptors. This places the loops at the ends of the IgV domains on the same side of the PD-1/PD-L1 complex, forming a surface that is similar to the antigen-binding surface of antibodies and T cell receptors. Mapping conserved residues allowed the identification of residues that are important in forming the PD-1/PD-L1 interface. Based on the structure, we show that some reported loss-of-binding mutations involve the PD-1/PD-L1 interaction but that others compromise protein folding. The PD-1/PD-L1 interaction described here may be blocked by antibodies or by designed small-molecule drugs to lower inhibitory signaling that results in a stronger immune response. The immune receptor-like loops offer a new surface for further study and potentially the design of molecules that would affect PD-1/PD-L1 complex formation and thereby modulate the immune response.
Keywords: coreceptor, costimulation, inhibitory receptor
Programmed death 1 (PD-1; also CD279) is expressed on activated B cells, T cells, and monocytes and was first cloned from a T cell hybridoma (1). In mice chronically infected with lymphocytic choriomeningitis virus, PD-1 is highly expressed at a time when virus-specific T cells are functionally impaired (2). In these mice, antibodies against either PD-1 or PD-L1 restore the ability of impaired T cells to proliferate, secrete cytokines, and kill infected cells (2). Disruption of the PD-1 gene results in autoimmune diseases in mice such as a lupus-like disorder (3) and autoimmune dilated cardiomyopathy (4). Signaling through PD-1 results in the recruitment of the src homology 2 domain-containing tyrosine phosphatase 2 to tyrosine-based motifs of the intracellular portion of PD-1 (5). PD-L1 (CD274, B7-H1) (6, 7) is expressed widely on both lymphoid and nonlymphoid tissues (8). Disruption of the PD-L1 gene leads to up-regulated T cell responses and the generation of self-reactive T cells (9). Antibody blockade of either PD-1 or PD-L1 leads to increased antitumor immunity (10). In the placenta, antibodies against PD-L1 lead to a breakdown in maternal tolerance to the fetus in a mouse model system (11). A second ligand for PD-1 is PD-L2 (CD273, B7-DC), which has a more restricted expression than PD-L1, being expressed on activated macrophages and dendritic cells (12, 13).
PD-1 is a member of the CD28/CTLA-4 (cytotoxic T lymphocyte antigen) family of coreceptors, which share 25% sequence identity and are type I membrane proteins having a single extracellular Ig variable (V) domain (for reviews, see refs. 14–16). Ligands of the CD28/CTLA-4 family include the B7-1 (CD80), B7-2 (CD86), PD-L1, and PD-L2 molecules and have two extracellular Ig domains, an N-terminal V domain and a C-terminal constant (C) domain. Both CTLA-4 (CD152) and CD28 form disulfide-bonded homodimers. Their ligands B7-1 and B7-2 are also found to be homodimers. Crystal structures have shown that CTLA-4 dimers can bind to B7-1 and to B7-2 dimers in an alternating fashion (17, 18). In contrast, several lines of evidence demonstrate that PD-1 is a monomer, and indeed it lacks the analogous C-terminal cysteine that forms an interchain disulfide in CTLA-4 (19). Mutational results suggest that PD-1 may bind PD-L1 differently from the way that CTLA-4 binds B7-1 and B7-2 (19, 20).
The protein sequence identity between murine and human PD-1 is 64% and between murine and human PD-L1 is 77%. Murine PD-1 binds in vitro to both murine and human PD-L1, and human PD-1 binds to the PD-L1 of each species. The cross-species binding affinities of murine PD-1 to human PD-L1 and of human PD-1 to murine PD-L1 are similar to the affinities of same-species binding (7, 12, 19). Structurally, CTLA-4 uses a conserved hydrophobic sequence, Met-Tyr-Pro-Pro-Pro-Tyr, in the loop connecting the F and G β-strands [FG loop, complementarity-determining region 3 (CDR3)] of its V domain to bind its B7-1 and B7-2 ligands (17, 18, 21). PD-1 does not have such a conserved sequence. The predicted monomeric nature of PD-1 and the reports of the importance of PD-1/PD-L1 in T cell exhaustion led us to choose the PD-1/PD-L1 complex as the target of our structural investigations.
Results and Discussion
Overall Structure of PD-1/PD-L1.
We produced recombinant extracellular domains of the murine form and the human form of both PD-1 and PD-L1 and demonstrated that they bound to cells transfected with murine or human PD-L1 and PD-1 [supporting information (SI) Fig. 5]. Successful crystallization experiments used murine PD-1 and human PD-L1. Other pairs did not form crystals. Binding experiments of the murine PD-1/human PD-L1 complex using surface plasmon resonance (SPR) yielded a Kd of ≈8 μM, consistent with reported values (19) and our observations that the murine PD-1/human PD-L1 complex partially dissociated at the concentrations achieved during preparative size exclusion chromatography (unpublished results).
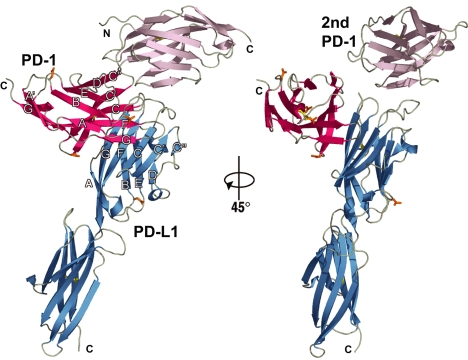
We first determined the structure of uncomplexed human PD-L1 using multiwavelength x-ray data (Methods) and refined the structure to an Rfree of ≈41%. Using the separate Ig domains of the partially refined human PD-L1 model and the available uncomplexed murine PD-1 model [Protein Data Bank (PDB) ID code 1NPU (19)], we determined the structure of the murine PD-1/human PD-L1 complex by molecular replacement to a resolution of 2.65 Å (SI Table 2). Each asymmetric unit of the complex crystal contains two PD-1 molecules and one PD-L1 molecule (Fig. 1). The first PD-1 molecule interacts extensively with PD-L1. The second PD-1 molecule makes limited contacts to the first PD-1 and to PD-L1 that do not appear to be physiologically significant. From the limited surface area buried by the second PD-1 molecule with the first PD-1 (360 Å2 total) and with PD-L1 (620 Å2 total), we conclude that the second PD-1 molecule participated in forming crystal contacts for this arrangement of molecules, but is not part of the biological complex. Therefore, the PD-1:PD-L1 complex is a 1:1 complex of monomeric PD-1 and PD-L1 in the crystal. This contrasts with the 1:1 stoichiometry of homodimers observed in the crystals of the CTLA-4/B7-1 and CTLA-4/B7-2 complexes (17, 18).
Fig. 1.
Two views of the PD-1/PD-L1 complex. The two single-domain PD-1 molecules in the asymmetric unit of the crystal are shown in red (PD-1) and violet (second PD-1). The two-domain PD-L1 molecule is shown in blue. The strands of the two β-sheets of PD-1 are labeled ABED and A′GFCC′C″. The strands of the two β-sheets of the PD-L1 V domain are labeled AGFCC′C″ and BED. Note the tenuous contacts that the second PD-1 makes to PD-1 and PD-L1, seen best on the right side of the figure. N-linked glycosylation sites (gold) are at PD-1 residues 49, 58, 74, and 116 and at PD-L1 residue 35. Carbohydrate at any of the five potential sites is predicted not to interfere with the formation of the complex. The view at right is after a rotation of 45° around the vertical axis. The N and C termini are identified.
PD-1/PD-L1 Interaction Surface.
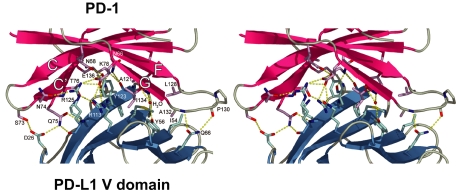
PD-1 and PD-L1 interact using their V-domain A′GFCC′ β-sheets to form a pair of V domains in a Fv-like structure that is similar to the pairing of V domains or Fv of antibodies, T cell receptors (TCRs), and CD8. A total of 87 interatomic contacts (using a 4.0-Å distance cutoff) including 18 hydrogen bonds are formed between 18 murine PD-1 residues and 14 human PD-L1 residues (Fig. 2, Table 1, and SI Table 3). Fifteen of the 18 murine PD-1 residues in the interface with human PD-L1 are identical in the human PD-1 sequence (SI Table 4). The three residues that differ are Met-64 (human Val), Asn-68 (human Tyr), and Val-90 (human Gly). In the complex structure, Met-64 and Val-90 make van der Waals contacts and Asn-68 is involved in one hydrogen bond with PD-L1 residues (Table 1 and SI Tables 3 and 4). It appears from the structure that Tyr-68 in human PD-1 could be accommodated with slight changes in the interface between murine PD-1 and human PD-L1. The Met-64 to Val and Val-90 to Gly differences between the murine and human PD-1 sequences result in a smaller or absent side chain, which would minimally affect the interface. Hydrophobic interactions contribute to the interface with six PD-1 residues forming a surface that packs against a hydrophobic surface formed by five PD-L1 residues (Fig. 2 and SI Fig. 6). PD-1/PD-L1 complex formation buries 1,870 Å2 of solvent-accessible surface area, which is comparable to the buried surface area between the V–V interfaces of antibodies and TCRs (in Fig. 3, 1,660 and 1,820 Å2, respectively).
Fig. 2.
The PD-1/PD-L1 interface. Shown is a stereoview of the PD-1/PD-L1 interface showing side chains of residues on β-strands (CC′FG) of PD-1 (red) and on β-strands (GFCC′, left to right) of PD-L1 (blue) that make contacts. Interacting PD-1 side chains (pink) and PD-L1 (light blue) are shown; for clarity a few side chains are not shown. Dotted lines (yellow) indicate hydrogen bonds formed in the interface and with a water molecule.
Table 1.
Contacts between PD-1 and PD-L1 (distances < 4.0 Å)
| PD-1 contact residue | PD-1 residue location | PD-L1 contact residue | PD-L1 residue location |
|---|---|---|---|
| M 64 | C strand | A121 | G strand |
| N 66 | C strand | A121, D122 | G strand |
| N 68 | C strand | Y123 | G strand |
| S 73 | CC′ loop | D26 | A strand |
| N 74 | CC′ loop | R125 | G strand |
| Q 75 | CC′ loop | D26, K124, R125 | A strand, G strand |
| T 76 | C″ strand | Y123, K124, R125 | G strand |
| K 78 | C′ strand | F19, A121, D122 | N terminus, G strand |
| V 90 | C″D loop | T20 | N terminus |
| L 122 | F strand | R125 | G strand |
| G 124 | F strand | Y123 | G strand |
| I 126 | F strand | Y123 | G strand |
| L 128 | FG (CDR3) loop | I54, M115, S117 | C strand, F strand |
| P 130 | FG (CDR3) loop | Q66 | C′ strand |
| K 131 | FG (CDR3) loop | Q66 | C′ strand |
| A 132 | FG (CDR3) loop | Y56, Q66 | C strand, C′ strand |
| I 134 | G strand | Y56, M115 | C strand, F strand |
| E 136 | G strand | R113, Y123, R125 | F strand, G strand |
Fig. 3.
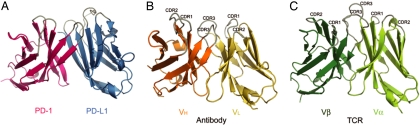
The IgV domains of PD1/PD-L1 are similar to antigen receptors. (A) The loops at the ends of the PD-1 domain and the first domain of PD-L1 are located on the same side of the complex. (B) For comparison, the Fv or VHVL portion of a representative antibody [PDB ID code 1G7J (31)] found in the search of the database was superimposed on the Fv-like portion of the PD-1/PD-L1 complex to yield an rms difference of 2.6 Å over 180 pairs of α-carbon pairs. (C) The VαVβ portion of a representative TCR [PDB ID code 2BNU (32)] was superimposed on the Fv-like part of the PD-1/PD-L1 complex with an rms difference of 3.1 Å over 191 Cα pairs. In B and C, after superimposing on to PD-1/PD-L1, the models were translated apart for viewing. The loops that are antigen receptor CDRs are labeled (CDR1, CDR2, and CDR3). The search of the database was performed with the PD-1/PD-L1 Fv-like domain as query and all of the targets considered as two-domain rigid bodies.
Similarities with Antigen Receptors.
We searched the protein structure database with the Fv-like portion of the PD-1/PD-L1 complex after removing the C domain of PD-L1 (22). The Fv portion was considered in the search as a single domain. This search yielded >600 antibody and 25 TCR Fv molecules, with rms differences between 2.6 and 3.8 Å calculated on a total of 150–210 pairs of α-carbon positions of query and target Fv-like domains. The only non-antigen receptor found in the search was the CD8 homodimer. It appears that only rarely do non-antigen receptor variable domains interact in an Fv-like manner. The V domains of antibody heavy chains and light chains overlay onto PD-1 and PD-L1, respectively. The BC (“CDR1”), C′C″ (“CDR2”), and FG (“CDR3”) loops of PD-1/PD-L1 are in positions similar to those of the antigen-binding loops of the representative antigen receptors (Fig. 3). Like antibodies and TCRs, the FG loops of PD-1 and PD-L1 are centrally located and the other loops are positioned outwards from the center. In antibody and TCR molecules, these loops bind antigens, and, in the case of CD8, they bind class I major histocompatibility molecules. This raises the possibility that the loops of the PD-1/PD-L1 complex may bind another molecule.
Comparison with CTLA-4/B7 Structures.
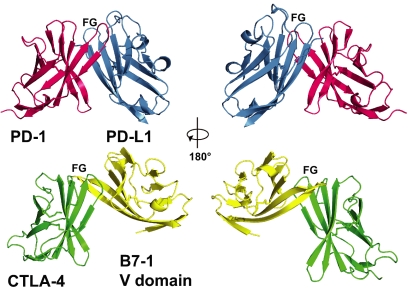
In the PD-1/PD-L1 complex, the FG and other loops are mainly free and are not involved in the binding between domains (Table 1). This contrasts with the CTLA-4/B7 complexes where the FG loop of CTLA-4 makes many contacts to B7 through its conserved proline-rich sequence and where the elongated FG loop of B7 binds to CTLA-4 (Fig. 4). Compared with other members of the CD28/B7 costimulatory family, the PD-1/PD-L1 complex has a significantly larger buried surface area (1,870 Å2) than the ≈1,200-Å2 buried surface areas of the CTLA-4/B7-1 [PDB ID code 1I8L (17)] and CTLA-4/B7-2 [PDB ID code 1I85 (18)] complexes. An analogous search of the database of protein structures using the CTLA-4/B7-1 complexed V domains did not return antibody or TCR Fv domains. The pairings of the V domains of PD-1/PD-L1 and CTLA-4/B7 are different: B7-1 and CTLA-4 β-strands closest to the interface are nearly orthogonal to each other, whereas the analogous β-strands of PD-1/PD-L1 are at an acute angle to each other (Fig. 4). However, the V domains are similar when compared one domain at a time. The rms difference between the V domain of PD-1 and that of CTLA-4 is 2.2 Å over 104 α-carbon pairs. The rms difference between the V domain of PD-L1 and that of B7.1 is 1.7 Å over 93 α-carbon pairs.
Fig. 4.
A comparison of the PD-1/PD-L1 and CTLA-4/B7-1 variable-like (V) domain interactions. (Left) the two complexes were overlaid by superimposing PD-1 (red) on to CTLA-4 (green), then translated apart vertically for viewing. (Right) The two complexes were rotated by 180°. Note the marked difference in the location of the B7-1 V domain (yellow) in comparison to the V domain of PD-L1 (blue).
PD-L1 is composed of one N-terminal V domain and one C domain joined by a short linker. A comparison of the PD-L1 molecule in the complex with the PD-L1 molecule that was crystallized alone revealed that the hinge angles between the V and C domains of complexed PD-L1 and of free PD-L1 differ by 38°, indicating that there is conformational flexibility between the two domains (SI Fig. 7). This flexibility would allow PD-L1 to accommodate to the orientation of PD-1 during binding. There is a recent report that PD-L1 can bind directly to B7-1 (23). The flexibility of the PD-L1 domains may assist in achieving B7-1 binding. Considered alone, the V domain of PD-L1 did not undergo significant conformational changes upon binding to PD-1 because the V domains of complexed and uncomplexed PD-L1 exhibit a rms difference of 0.50 Å over 111 α-carbon pairs (SI Fig. 7). In the crystal of PD-L1 alone, two PD-L1 molecules are present in the asymmetric unit but do not appear to form a plausible dimer. In solution, no dimer or higher oligomer of PD-L1 was detected in analytical ultracentrifugation sedimentation equilibrium experiments or during size exclusion chromatography (data not shown). PD-L1 appears to be a monomer in solution.
The two PD-1 molecules found in the asymmetric unit of the crystal and the uncomplexed PD-1 (19) are almost identical to each other with rms differences between 0.7 and 0.9 Å over 114 Cα pairs. In the three crystallographically independent molecules, the CC′ loop at the base of the domains and the FG loops at the top are the only regions that differ, because they make contacts to PD-L1 in the complex. Residues (Ser-71 to Gln-75) of the complexed PD-1 CC′ loop located at the bottom of the domain move an average of 2.0 Å relative to their positions in the second PD-1 and in the uncomplexed PD-1 (SI Fig. 7). The PD-1 FG loop residues (Leu-128 to Ala-132) are displaced an average of 1.3 Å relative to the second PD-1 and 2.3 Å relative to uncomplexed PD-1. No other conformational differences in PD-1 and PD-L1 are observed in the complex when compared with the free molecules.
Interpretation of Mutations in Light of the Complex Structure.
Residues of the PD-L1 V domain that bind to PD-1 are conserved across species. The residues Ala-121, Asp-122, Tyr-123, and Lys-124 in the G strand of PD-L1 make intimate contacts with PD-1 (SI Fig. 6) and are conserved in all available PD-L1 sequences from mammals and birds. Published mutational results confirm the importance of PD-L1 residues 121–124. In murine PD-L1, a mutation of Lys-124 to Ser abolishes binding to murine PD-1 as determined by SPR, ELISA, and flow cytometry (SI Table 5) (20). Mutations of Phe-67 and Ile-126 also showed no binding to PD-1, but Phe-67 and Ile-126 have buried side chains and are not in the interface (SI Table 5). It is likely that the Phe-67 and Ile-126 mutations prevented binding indirectly by disrupting the folding of PD-L1. In the PD-1/PD-L1 complex, atoms from nine residues of PD-1 contact PD-L1 residues 121–124. Five of these nine PD-1 residues have been studied by mutation in the binding of murine PD-1 to murine PD-L1 (SI Table 4) (19). When three (Lys-78, Ile-126, and Glu-136) of the five PD-1 residues were in turn mutated to alanine, PD-1 binding to PD-L1 was lost as observed in a SPR binding assay (SI Table 4) (19).
A second ligand for PD-1 is PD-L2, with a sequence identity of 38% to PD-L1. Through comparison with the sequence of PD-L1, PD-L2 appears to lack the V domain C″ strand (SI Fig. 8), but the C″ strand of PD-L1 does not contact PD-1 in the complex crystal. The residues in PD-L2 that correspond to PD-L1 residues 121–124 are identical; thus, PD-L2 would be expected to bind in a manner similar to that of PD-1. In support of this interpretation, when murine PD-L2 Asp-111 and Lys-113, residues that are analogous to Asp-122 and Lys-124 of PD-L1, were mutated to serine no binding of PD-L2 to PD-1 was detected by SPR, ELISA, and flow cytometry (20).
The V domains of the PD-1/PD-L1 complex display an array of loops that resemble those of an antigen receptor. This raises the possibility that when a PD-1 receptor on an immune cell binds to a PD-L1 ligand, a binding site for a third molecule may be formed. Generating a receptor in this manner at the surface of a cell or between the surfaces of interacting cells would have novel implications for immune cell signaling and its regulation.
Methods
Preparation of Recombinant Murine PD-1 and Human PD-L1.
Expression constructs encoding the extracellular domains of murine (m) PD-1 (from Leu-25 to Ser-157 with the unpaired Cys-83 mutated to Ser and a Met-Ala added to the N terminus) and of human (h) PD-L1 (from Ala-18 to Thr-239 with a Met added to the N terminus) were designed. PCRs were carried out with 0.05 units/ml ExTaq polymerase (Takara Shuzo) in 0.25 ml with 0.5 mM primers through 35 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 2 min. The PCR products were purified from a 1% (wt/vol) agarose gel by the use of QIAquick Gel Extraction Kit (Qiagen), cloned into pLM1 (24), and transformed into the Escherichia coli Rosetta 2 (DE3) pLysS strain (Novagen). Colonies were inoculated into TB broth (Difco Laboratories) supplemented with 50 mg/liter ampicillin (Meiji Seika Kaisha), 34 mg/liter chloramphenicol, and 50 mg/liter carbenicillin. After 5 h, isopropyl-β-d-thiogalactopyranoside (1 mM; Wako Pure Chemical) was added to induce protein expression. The cells were further cultured for 6 h and harvested at 17,000 × g for 10 min at 4°C.
The pellets were washed once with 10 mM Tris·HCl buffer (pH 8.0) (Nacalai Tesque), resuspended in 50 mM Tris·HCl buffer (pH 8.0), 25% (wt/vol) sucrose, 1 mM ethylenediamine-N,N,N,N′-tetraacetic acid disodium salt dihydrate (NaEDTA; Wako Pure Chemical), 0.1% (wt/vol) sodium azide, and 10 mM DTT (Wako Pure Chemical), and then treated with lysozyme (Sigma–Aldrich) at room temperature for 1 h. Lysis buffer consisting of 50 mM Tris·HCl, 1% (vol/vol) Triton X-100 (Nacalai Tesque), 1% (wt/vol) sodium deoxycholate (Wako Pure Chemical), 100 mM NaCl, 0.1% sodium azide, 10 mM DTT, and 1 mM NaEDTA (pH 8.0) was added to the suspended cells and incubated at room temperature for 1 h with vigorous stirring, then stored at −80°C. The frozen samples were thawed in warm water, and 1 mg of DNase I (Sigma–Aldrich) and MgCl2 (6 mM) was added to the solutions. The samples were allowed to stand at room temperature for 8 h with stirring and centrifuged at 27,000 × g for 15 min at 4°C. The inclusion bodies were washed with buffer consisting of 50 mM Tris·HCl, 0.5% (vol/vol) Triton X-100, 100 mM NaCl, 1 mM NaEDTA (pH 8.0), 0.1% (wt/vol) sodium azide, and 1 mM DTT, then homogenized by using a motor-driven pestle. After washing two times, the inclusion body pellets were washed once with detergent-free buffer consisting of 50 mM Tris·HCl, 1 mM NaEDTA, 0.1% (wt/vol) Na azide, and 1 mM DTT (pH 8.0). The washed inclusion body pellets were dissolved in 25 mM Mes-NaOH (pH 6.0), 10 mM NaEDTA, 1 M urea (Nacalai Tesque), 6 M guanidine-HCl (Wako Pure Chemical), and 1 mM DTT and ultracentrifuged at 180,000 × g for 30 min at room temperature. The resulting clear supernatants were supplemented with 50 mM DTT and allowed to stand at 37°C for 1 h.
The solubilized mPD-1 protein was refolded by dilution in buffer consisting of 1 M arginine-HCl, 100 mM Tris·HCl (pH 8.0), 0.5 mM (p-amidinophenyl) methane sulfonylfluoride-HCl, 2 mM NaEDTA, 0.5 mM reduced glutathione, and 0.05 mM oxidized glutathione at 4°C overnight. hPD-L1 was refolded in the same Tris-based refolding buffer except that 0.25 mM reduced glutathione and 0.25 mM oxidized glutathione were present. The proteins were dialyzed three times against 10 volumes of 10 mM Tris·HCl (pH 8.0) at 4°C for 48–96 h and subjected to DE52 anion-exchange column chromatography (Whatman).
The refolded proteins were purified by using consecutive column chromatographies. Murine PD-1 was eluted from a Q Sepharose HP column (0.8 cm × 97 cm) at a conductivity of 14.6 mS and migrated on Superdex 200 (0.8 cm × 60 cm) at 0.80 of the column volume, and, similarly, hPD-L1 was eluted at 20.0 mS and migrated at 0.68 of the column volume. The elution positions of the proteins during size exclusion chromatography corresponded to sizes of 15 kDa and 25 kDa, respectively, nearly equal to the predicted sizes of 1,5061 Da for mPD-1 and 25,337 Da for hPD-L1. N-terminal amino acid sequencing, mass spectrometry, native PAGE, and SDS/PAGE confirmed the identity and homogeneity of the mPD-1 and hPD-L1 recombinant proteins.
Crystallization.
Purified mPD-1 and hPD-L1 proteins were buffer-exchanged into 10 mM Tris (pH 8.0) and concentrated to 8 mg/ml and 4 mg/ml, respectively. Crystallization screens at room temperature were carried out by sitting-drop vapor diffusion by mixing 1 μl of each crystallization condition and 1 μl of hPD-L1 alone or the mPD-1/hPD-L1 complex. Crystals of hPD-L1 alone formed in 20–25% PEG3350, 0.1 M NaCacodylate, and 0.2 M ammonium formate or ammonium fluoride (pH 6.0–6.8) at room temperature.
To form the complex crystals, mPD-1 and hPD-L1 were mixed together in a molar ratio of 2:1 mPD-1:hPD-L1. Diffraction-quality crystals were grown by sitting-drop vapor diffusion in a solution composed of 20% (wt/vol) PEG3350 and 200 mM NH4H2PO4. Crystals were cryoprotected in 20% (wt/vol) PEG3350, 200 mM NH4H2PO4, and 20% (vol/vol) glycerol for 10 s before plunging into liquid nitrogen.
Structure Determination of hPD-L1 Alone.
The hPD-L1 structure was determined by using multiwavelength x-ray data measured from native crystals and crystals incorporating selenomethionine and platinum atoms. Heavy atom sites were refined and phases were calculated in SHARP (25) (Global Phasing).
Structure Determination of the PD-1/PD-L1 Complex.
X-ray data, 335 frames of 0.5° each, were acquired at a wavelength of 0.97943 Å at the 19-BM (Structural Biology Center–Collaborative Access Team) beamline at the Advanced Photon Source at Argonne National Laboratory (Chicago, IL). The 2.65-Å-resolution data were integrated and scaled with HKL2000 (26), resulting in the statistics presented in SI Table 2. The complex structure was solved by molecular replacement with PHASER (27), as implemented in the CCP4 software suite (28), using the 2.0-Å crystal structure of mPD-1 [PDB ID code 1NPU (19)] and our partially refined hPD-L1 structure (Rfree = 41%; unpublished results) as search models. The model was refined by using the CNS (29) and Refmac (28) software packages, resulting in Rwork/Rfree values of 21.4% and 26.8%. Electron density was continuous from Arg-33 to Ile-148 for the first PD-1, from Gly-30 to Arg-147 for the second PD-1, and from Ala-18 to Leu-229 for PD-L1, numbered from the initiating methionines. Figures were produced with PyMOL software (30). The unpaired cysteine residue (Cys-83) of mPD-1 that was mutated to a serine for protein expression purposes is located at the C terminus of the C′ β-strand and does not interact with PD-L1. Structure comparisons used the protein structure comparison service, secondary structure matching (SSM), at the European Bioinformatics Institute (22).
Establishment of Cell Lines Expressing Murine and Human PD-1 and PD-L1.
cDNAs encoding mPD-1 and hPD-1 and ones encoding mPD-L1 and hPD-L1 were ligated into the plasmids pApuroXS and pEF-BOSneoSE, respectively. The PD-1 expression plasmids were transfected into the IIA1.6 murine B cell line, and the PD-L1 expression plasmids were transfected into the P815 murine mastocytoma cell line by electroporation at 360 V and 500 mF in a cuvette with a 0.4-mm gap (Bio-Rad). The resultant cells were incubated in a 75-cm2 flask containing 30 ml of RPMI medium 1640 with 10% (vol/vol) FCS, 10 mM 2-mercaptoethanol, and penicillin/streptomycin at 37°C with 5% CO2. After 2 days, the cell suspension was centrifuged at 600 × g for 5 min and the supernatant was discarded. The pellet was resuspended in 400 ml of RPMI medium 1640 supplemented with 10% (vol/vol) FCS, 10 mM 2-mercaptoethanol, penicillin/streptomycin, and 3 mg/ml puromycin, plated into 20 round-bottom 96-well tissue culture plates, and incubated at 37°C with 5% CO2 for 2 weeks. Several colonies were transferred into 24-well tissue culture plates and maintained in the RPMI medium 1640/puromycin medium. The PD-1 clones were examined for surface mPD-1 expression by flow cytometry using the biotin-conjugated anti-mPD-1 rat monoclonal antibody 4-7 generated in our laboratory and streptavidin-PE. The PD-L1 clones were examined for surface hPD-L1 by flow cytometry using the biotin-conjugated anti-hPD-L1 rat monoclonal antibody 27A2 generated in our laboratory and streptavidin-PE.
Flow Cytometry.
Cells (5 × 105) transfected with mPD-1 or hPD-L1 were transferred to round-bottom 96-well plates and washed once with Isoflow sheath solution (Beckman Coulter) containing 2% (vol/vol) FCS. After centrifuging at 4°C for 2 min at 600 × g and discarding the supernatants, the pellets were tapped gently and 50 ml of biotin-conjugated mPD-1 or hPD-L1 was added. The plate was incubated on ice in the dark for 20 min and was centrifuged again at 4°C for 2 min at 600 × g. The cells were washed three times with sheath solution containing 2% FCS and stained with 100 ml of streptavidin-PE. After again incubating for 20 min on ice in the dark, the cells were washed two times with sheath solution containing 2% (vol/vol) FCS and two more times with sheath solution without FCS. Finally, the pellets were suspended in 200 ml of sheath solution with 1% (wt/vol) paraformaldehyde, then diluted with an additional 500 ml of sheath solution. The cells were applied to the flow cytometer (FACSCalibur; Becton Dickinson), and 10,000 events were analyzed. Results of cell staining are presented as histograms with cell number on the vertical axis and relative fluorescence on the logarithmic horizontal axis.
SPR.
A cDNA encoding the mPD-1/human Fc fusion protein in the vector pEF-BOSneoSE (kindly provided by Shigekazu Nagata, Osaka University Medical School, Osaka) was transfected into the 293T cell line. Cell culture supernatants were loaded on a Protein G column (Amersham Biosciences). The chimeric protein was eluted with citric acid at pH 4 and neutralized with Tris. The purified mPD-1/Fc protein and mPD-L1/Fc, hPD-1/Fc, and hPD-L1/Fc obtained from R & D Systems were stored in PBS at 4°C. Fusion proteins were coupled to the CM5 sensor chip by the standard amine-coupling protocol and used in a BIAcore 3000 instrument.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Andrea Balbo and Peter Schuck for analysis of human PD-L1 by analytical ultracentrifugation, Takao Yoshida and Shiro Shibayama (Ono Pharmaceutical) for SPR analysis, and Timothy J. Allison for early contributions to this work. X-ray data were collected at the Structural Biology Center–Collaborative Access Team 19-ID/BM and Southeast Regional–Collaborative Access Team 22-ID beamlines at the Advanced Photon Source of Argonne National Laboratory supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract W-31-109-Eng-38. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Culture, Sports, and Technology, Japan, by the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation, by the Inamori Foundation, Japan, and by the Intramural Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 3BIK (PD-1/PD-L1 complex) and 3BIS (PD-L1 alone)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0712278105/DC1.
References
- 1.Ishida Y, Agata Y, Shibahara K, Honjo T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992;11:3887–3895. doi: 10.1002/j.1460-2075.1992.tb05481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barber DL, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 3.Nishimura H, Nose M, Hiai H, Minato N, Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 1999;11:141–151. doi: 10.1016/s1074-7613(00)80089-8. [DOI] [PubMed] [Google Scholar]
- 4.Nishimura H, et al. Autoimmune dilated cardiomyopathy in PD-1 receptor-deficient mice. Science. 2001;291:319–322. doi: 10.1126/science.291.5502.319. [DOI] [PubMed] [Google Scholar]
- 5.Okazaki T, Maeda A, Nishimura H, Kurosaki T, Honjo T. PD-1 immunoreceptor inhibits B cell receptor-mediated signaling by recruiting src homology 2-domain-containing tyrosine phosphatase 2 to phosphotyrosine. Proc Natl Acad Sci USA. 2001;98:13866–13871. doi: 10.1073/pnas.231486598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong H, Zhu G, Tamada K, Chen L. B7–H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat Med. 1999;5:1365–1369. doi: 10.1038/70932. [DOI] [PubMed] [Google Scholar]
- 7.Freeman GJ, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okazaki T, Honjo T. The PD-1-PD-L pathway in immunological tolerance. Trends Immunol. 2006;27:195–201. doi: 10.1016/j.it.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Latchman YE, et al. PD-L1-deficient mice show that PD-L1 on T cells, antigen-presenting cells, and host tissues negatively regulates T cells. Proc Natl Acad Sci USA. 2004;101:10691–10696. doi: 10.1073/pnas.0307252101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwai Y, et al. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc Natl Acad Sci USA. 2002;99:12293–12297. doi: 10.1073/pnas.192461099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guleria I, et al. A critical role for the programmed death ligand 1 in fetomaternal tolerance. J Exp Med. 2005;202:231–237. doi: 10.1084/jem.20050019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latchman Y, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol. 2001;2:261–268. doi: 10.1038/85330. [DOI] [PubMed] [Google Scholar]
- 13.Tseng SY, et al. B7-DC, a new dendritic cell molecule with potent costimulatory properties for T cells. J Exp Med. 2001;193:839–846. doi: 10.1084/jem.193.7.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenwald RJ, Freeman GJ, Sharpe AH. The B7 family revisited. Annu Rev Immunol. 2005;23:515–548. doi: 10.1146/annurev.immunol.23.021704.115611. [DOI] [PubMed] [Google Scholar]
- 15.Chen L. Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity. Nat Rev Immunol. 2004;4:336–347. doi: 10.1038/nri1349. [DOI] [PubMed] [Google Scholar]
- 16.Okazaki T, Honjo T. PD-1 and PD-1 ligands: From discovery to clinical application. Int Immunol. 2007;19:813–824. doi: 10.1093/intimm/dxm057. [DOI] [PubMed] [Google Scholar]
- 17.Stamper CC, et al. Crystal structure of the B7-1/CTLA-4 complex that inhibits human immune responses. Nature. 2001;410:608–611. doi: 10.1038/35069118. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz JC, Zhang X, Fedorov AA, Nathenson SG, Almo SC. Structural basis for co-stimulation by the human CTLA-4/B7-2 complex. Nature. 2001;410:604–608. doi: 10.1038/35069112. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, et al. Structural and functional analysis of the costimulatory receptor programmed death-1. Immunity. 2004;20:337–347. doi: 10.1016/s1074-7613(04)00051-2. [DOI] [PubMed] [Google Scholar]
- 20.Wang S, et al. Molecular modeling and functional mapping of B7–H1 and B7-DC uncouple costimulatory function from PD-1 interaction. J Exp Med. 2003;197:1083–1091. doi: 10.1084/jem.20021752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwartz JC, Zhang X, Nathenson SG, Almo SC. Structural mechanisms of costimulation. Nat Immunol. 2002;3:427–434. doi: 10.1038/ni0502-427. [DOI] [PubMed] [Google Scholar]
- 22.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 23.Butte MJ, Keir ME, Phamduy TB, Sharpe AH, Freeman GJ. Programmed death-1 ligand 1 interacts specifically with the B7-1 costimulatory molecule to inhibit T cell responses. Immunity. 2007;27:111–122. doi: 10.1016/j.immuni.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sodeoka M, Larson CJ, Chen L, LeClair KP, Verdine GL. A multifunctional plasmid for protein expression by ECPCR: Overproduction of the p50 subunit of NF-kB. Bioorg Med Chem Lett. 1993;3:1089–1094. [Google Scholar]
- 25.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. In: Methods in Enzymology. Carter CW, Sweet RM, editors. Vol 276. New York: Academic; 1997. pp. 307–326. [Google Scholar]
- 27.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 28.Collaborative Computational Project Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 29.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.DeLano WL. The PyMOL User's Manual. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- 31.Sundberg EJ, et al. Estimation of the hydrophobic effect in an antigen-antibody protein-protein interface. Biochemistry. 2000;39:15375–15387. doi: 10.1021/bi000704l. [DOI] [PubMed] [Google Scholar]
- 32.Chen JL, et al. Structural and kinetic basis for heightened immunogenicity of T cell vaccines. J Exp Med. 2005;201:1243–1255. doi: 10.1084/jem.20042323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.