Abstract
The mechanism by which the Carma1–Bcl10–MALT1 (CBM) complex couples T cell antigen receptor (TCR) signaling to IκB kinase (IKK) and NF-κB activation is not known. Here, we show that Bcl10 undergoes K63-linked polyubiquitination in response to T cell activation and subsequently binds NEMO, the regulatory subunit of IKK. This interaction requires the ubiquitin-binding activity of NEMO. The sites of Bcl10 ubiquitination were mapped to K31 and K63. Mutation of these residues did not affect TCR signaling-induced CBM complex assembly but prevented Bcl10 ubiquitination, NEMO binding, and NF-κB activation. Therefore, the regulated ubiquitination of Bcl10 and its recognition by NEMO are a critical link between the CBM complex, IKK recruitment, and NF-κB activation.
Keywords: IκB kinase, T cell signaling, ubiquitination
The transcription factor NF-κB plays a vital role in many physiological and pathological processes, primarily by controlling the transcription of cytokines, antimicrobial effectors, and genes that regulate cell differentiation, survival, and proliferation (1). Like the large majority of stimuli that activate NF-κB, signaling via the T cell antigen receptor (TCR) utilizes the classical pathway in which IκB kinase (IKK) is activated and phosphorylates the cytosolic inhibitor of NF-κB, IκB (1). IκB is subsequently ubiquitinated and undergoes proteasomal degradation. IKK comprises two catalytic subunits, IKKα and IKKβ, and a regulatory subunit, NEMO. Although possessing no enzymatic activity, NEMO is required for IKK and thus NF-κB activation in most circumstances, including TCR engagement (2).
How TCR-proximal signaling couples to NEMO-dependent IKK activation has been intensively investigated. TCR ligation initiates a cascade of Src (Lck and Fyn) and Syk (ZAP70) family of protein tyrosine kinases, which in turn phosphorylate adaptor proteins such as LAT and SLP76 (3). The phosphorylated adaptor proteins recruit and activate phospholipase Cγ1 (PLCγ1), which catalyzes the production of diacylglycerol and the activation of protein kinase C (PKC) θ, which induces the formation of a complex containing three proteins: Carma1, Bcl10, and MALT1 (CBM) (1, 3–5). Genetic evidence shows that the CBM complex is essential for TCR- and B cell receptor-induced IKK/NF-κB activation (6, 7). Studies also suggest that the CBM complex couples PKCθ to IKK activation and is probably the intermediate in the TCR signaling pathway that is immediately upstream of IKK (4, 8, 9).
Posttranslational modifications of Carma1 and Bcl10 are thought to be mechanisms for up- or down-regulation of IKK activity (10). Stimulation via the TCR induces phosphorylation of the linker region of Carma1 by PKCθ, and a resulting conformational change allows binding of Bcl10 and the assembly of the CBM complex (11). Bcl10 is also phosphorylated in response to TCR activation, which has been reported to regulate IKK/NF-κB activation both positively and negatively (10, 12). Several recent studies have reported that Bcl10 also undergoes ubiquitination in response to the TCR ligation or PKC activation, which has been thought to target Bcl10 for degradation and thus to down-regulate NF-κB activation (13, 14).
We and others have reported that NEMO binds to K63-linked polyubiquitin chains, an activity that is required for binding of IKK to the TNF receptor 1 (TNFR1) signaling intermediate RIP and TNF-induced NF-κB activation (15, 16). Here, we asked whether this paradigm might apply to TCR-induced NF-κB activation as well. We find that after TCR stimulation, Bcl10 acquires K63-linked polyubiquitin chains, which is required for binding to NEMO and activation of IKK/NF-κB. Binding of ubiquitinated Bcl10 to NEMO constitutes an important link between CBM formation, posttranslation modification, and IKK activation.
Results
The Ubiquitin (Ub)-Binding Activity of NEMO Is Required for TCR-Mediated NF-κB Activation.
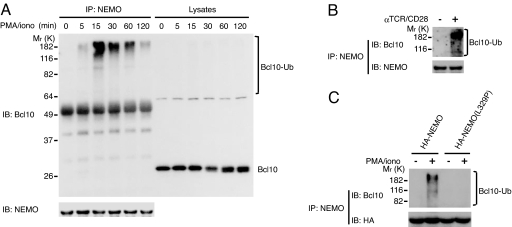
NEMO is a receptor for K63-linked polyubiquitin, and this interaction is essential for TNF-α-induced IKK/NF-κB activation (15, 16). Given that NEMO is also required for T cell activation-induced IKK/NF-κB activation (2, 17), we wanted to determine whether IKK/NF-κB activation in this instance also depends on NEMO binding to K63-linked polyubiquitin. To assess this possibility, NEMO-deficient Jurkat cells were stably reconstituted with either wild-type NEMO or a ubiquitin-binding-defective point mutant [NEMO(L329P) (15)], and multiple independent clones were stimulated with phorbol 12-myristate 13-acetate (PMA) and ionomycin (PMA/iono), which mimics TCR signaling and activates IKK/NF-κB. Consistent with previous reports (2, 17), PMA/iono did not cause IκB degradation in the absence of NEMO (Fig. 1A). Cells in which NEMO was reintroduced, however, responded with robust IκB degradation. Notably, there was little if any IκB degradation in cells reconstituted with NEMO(L329P). The functional significance of NEMO ubiquitin-binding activity was further addressed by transient cotransfection of NEMO-deficient Jurkat cells with cDNA encoding an NF-κB-driven luciferase reporter gene and a retrovirus vector that contains cDNA encoding NEMO or NEMO(L329P). Whereas PMA/iono did not activate NF-κB transcriptional activity in the vector-only transfected cells, NEMO reconstitution dramatically enhanced stimulus-dependent NF-κB transcriptional activity (Fig. 1B). Again, the NEMO(L329P) mutant was unable to restore PMA/iono-induced NF-κB luciferase activity. These results indicate that the ubiquitin-binding activity of NEMO is required for T cell activation-induced IKK/NF-κB activation.
Fig. 1.
NEMO binding to K63-linked polyubiquitin chains is required for TCR-mediated IKK/NF-κB activation. (A) NEMO-deficient Jurkat cells and NEMO-deficient Jurkat cell clones stably transfected with HA-NEMO or HA-NEMO(L329P) were treated with PMA/iono for the indicated times, and IκB levels were determined by immunoblotting. (B) NEMO-deficient Jurkat cells were transiently transfected with NF-κB-driven luciferase reporter and β-gal plasmids in combination with pMSCVpuro or pMSCVpuro encoding HA-NEMO or HA-NEMO(L329P). After 36 h, the cells were treated with or without PMA/iono for 5 h. Cell lysate luciferase activities were determined and normalized to β-gal activity.
Bcl10 Undergoes K63-Linked Polyubiquitination and Binds NEMO in Activated T Cells.
Considering the possibility that NEMO might be a bridge between molecules in the TCR-proximal signaling pathway and IKK in a manner analogous to its role in TNF signaling, we asked whether NEMO has a ubiquitinated binding partner downstream of the TCR and upstream of IKK. Several studies have demonstrated that Bcl10 is ubiquitinated in response to TCR signaling in a PKCθ-dependent manner (13) and suggested that this ubiquitination down-regulates TCR signaling by targeting Bcl10 for degradation (13, 14). However, the presumed ubiquitination-induced Bcl10 degradation was not blocked by proteasome inhibition. We found that Bcl10 ubiquitination was readily apparent in PMA/iono-stimulated Jurkat cells (Fig. 2A). Interestingly, ubiquitination was not observed in Jurkat cells lacking Carmal, which is upstream of Bcl10 in transducing TCR signals to NF-κB activation (11, 18). To assess whether the other essential member of the CBM complex, MALT1, is also required for Bcl10 ubiquitination, MALT1 shRNA was introduced into Jurkat cells. Cell lines stably expressing the shRNA expressed very little MALT1 but normal levels of Bcl10 (Fig. 2B Upper). Notably, stimulation-induced Bcl10 ubiquitination was markedly reduced in the MALT1-deficient Jurkat cells (Fig. 2B Lower). Therefore, the assembly of the complete CBM complex appears to be required for both Bcl10 ubiquitination (Fig. 2) and IKK/NF-κB activation (1, 5, 7).
Fig. 2.
Bcl10 in activated T cells undergoes K63-linked polyubiquitination and is bound by recombinant NEMO. (A) Jurkat cells or Carma1-deficient Jurkat cells were stimulated with or without PMA/iono for 20 min. Bcl10 was immunoprecipitated (IP) from denatured cell lysates and immunoblotted (IB) with anti-ubiquitin. WT, wild type. (B) Jurkat cells stably expressing MALT1 shRNA were immunoblotted with anti-MALT1 (Upper). Jurkat cells or the MALT1 shRNA-expressing Jurkat cells were treated with or without PMA/iono for 15 min and analyzed for Bcl10 ubiquitination as in A (Lower). (C) Jurkat cells transfected with HA-tagged K63-only or K48-only ubiquitin were stimulated or not with PMA/iono for the indicated times, harvested, and lysed. The lysates were treated as in A to disrupt protein–protein interactions, and Bcl10 was immunoprecipitated. The amount of K63-linked or K48-linked polyubiquitinated Bcl10 was determined by immunoblotting with anti-HA (Left). Expression of both HA-Ub mutants was similar, as judged by immunoblotting unstimulated cell lysates with anti-HA (data not shown). (D) Jurkat cells were stimulated with or without PMA/iono or anti-TCR C305/anti-CD28 for 20 min, harvested, and lysed. Cell lysates containing equal amounts of protein were incubated with GST-NEMO-coated beads. The bound material (Upper) and cell lysates (Lower) were immunoblotted with anti-Bcl10. (E) Cell lysates from Jurkat cells treated with or without PMA/iono for 20 min were incubated with GS-beads bound to GST-NEMO or GST-NEMO(L329P). The bound material was immunoblotted with anti-Bcl10. Ponceau S staining showed equal loading of the GST fusion proteins (data not shown).
The nature of the polyubiquitin chain linkage was determined by transfecting the cells with hemagglutinin (HA)-tagged ubiquitin mutants in which all of the lysines except either K63 or K48 were replaced with arginine. The transfected cells were stimulated with PMA/iono, detergent lysates were boiled in the presence of SDS to disrupt noncovalent protein–protein interactions, and Bcl10 was immunoprecipitated and immunoblotted with anti-HA. K48-linked polyubiquitinated Bcl10 was detected 20 min after stimulation and K63-linked polyubiquitinated Bcl10 somewhat earlier, being evident as early as 5 min after stimulation (Fig. 2C). Given the affinity of NEMO for K63-linked polyubiquitin chains, we assessed whether the ubiquitinated Bcl10 is bound by NEMO. To do so, GST-NEMO-coated beads were used to pull down Bcl10 from lysates of cells stimulated with nothing, PMA/iono, or agonistic anti-TCR (C305) and anti-CD28 antibodies. GST-NEMO did not bind Bcl10 from untreated cells but pulled down polyubiquitinated Bcl10 from lysates of cells stimulated with PMA/iono or anti-TCR/CD28 (Fig. 2D Upper). Notably, although easily detectable in the cell lysates (Fig. 2D Lower), unmodified Bcl10 was not pulled down by GST-NEMO. Binding required the contribution from polyubiquitination because the ubiquitin-binding-defective NEMO(L329P) was unable to pull down ubiquitinated Bcl10 from lysates of PMA/iono-stimulated cells (Fig. 2E).
To determine whether NEMO binds Bcl10 in vivo, Jurkat cells were stimulated with PMA/iono and lysed, and NEMO was immunoprecipitated. Whereas no endogenous Bcl10 was coimmunoprecipitated with endogenous NEMO in quiescent cells, anti-Bcl10 detected high-molecular-weight bands as early as 5 min after stimulation, peaking at 15 min, and declining thereafter (Fig. 3A). Again, despite the vast predominance of unmodified Bcl10 in these cells (see immunoblots of total lysates), nonubiquitinated Bcl10 was not detected in the material coimmunoprecipitated with NEMO (Fig. 3A). Ubiquitinated Bcl10 was also coimmunoprecipitated with NEMO from lysates of Jurkat cells stimulated via the TCR (Fig. 3B). The importance of ubiquitin binding was addressed in NEMO-deficient Jurkat cells that were stably reconstituted with wild-type NEMO or NEMO(L329P) and stimulated with PMA/iono, lysed, and immunoprecipitated with anti-NEMO. Polyubiquitinated Bcl10 was coimmunoprecipitated with wild-type NEMO but not with NEMO(L329P) (Fig. 3C). These results strongly argue that NEMO interacts only with ubiquitin-modified Bcl10 in activated T cells.
Fig. 3.
NEMO binds to activation-induced polyubiquitinated Bcl10 in vivo. (A) Lysates of Jurkat cells that were stimulated with PMA/iono for the indicated times and lysates immunoprecipitated (IP) with anti-NEMO, resolved on SDS/PAGE, and immunoblotted (IB) with anti-Bcl10 or anti-NEMO. (B) Jurkat cells were treated with or without C305/anti-CD28 for 15 min, and lysates were immunoprecipitated and analyzed for Bcl10 and NEMO as in A. (C) NEMO-deficient Jurkat cells stably reconstituted with HA-NEMO (C1, see Fig. 1A) or HA-NEMO(L329P) (C1, see Fig. 1A) were stimulated with PMA/iono for 15 min, immunoprecipitated, and analyzed for Bcl10 and HA as in A.
Polyubiquitination of Human Bcl10 Occurs on K31 and K63.
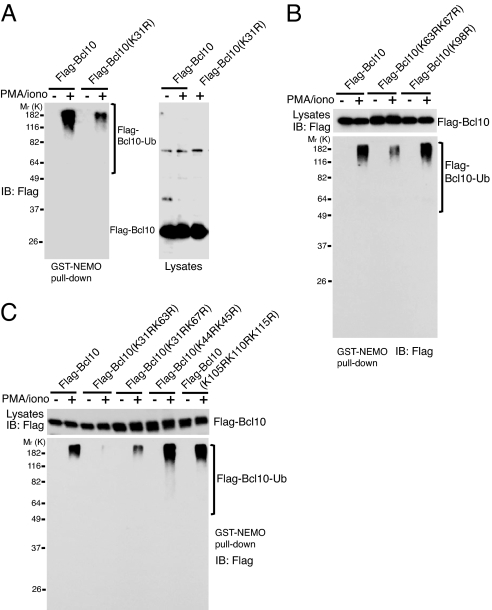
Given the results above, one would predict that disruption of Bcl10 polyubiquitination would negatively affect TCR-mediated IKK activation. We therefore attempted to map the Bcl10 lysine residues that are ubiquitin-modified after TCR stimulation. Human Bcl10 consists of 233 aa, 15 of which are lysines. A large percentage of the lysine residues are conserved among species, almost all of them in the N-terminal caspase recruitment domain (CARD; amino acids 1–103) and in C-terminal CARD-adjacent residues [supporting information (SI) Fig. 7]. These regions are important for Bcl10 function, and amino acids 1–119 constitute the minimal region that is essential and sufficient for Bcl10 overexpression to activate NF-κB (19, 20). We mutated nine conserved lysine residues to arginine in five different constructs and inserted them into a retroviral expression vector, pMSCVpuro. This retroviral vector uses a weak promoter (LTR) to drive protein expression, and we have found that their transient introduction by electroporation results in protein levels similar to those of the endogenous protein. Therefore, the wild-type and mutant Bcl10 retroviral plasmids were transiently transfected into Jurkat cells by electroporation. SI Fig. 8 shows a typical example of transfection efficiency, indicated by the percentage of GFP-positive cells, and Bcl10 protein expression was analyzed by immunoblotting. Transient transfection yielded Bcl10 levels similar to that of endogenous Bcl10. The lysates of transfected cells that had been stimulated with medium alone or PMA/iono were incubated with GST-NEMO-coated beads. As with endogenous Bcl10 (see Fig. 2C), only transiently expressed ubiquitinated Bcl10 from activated cells was pulled down by GST-NEMO (Fig. 4A). This strategy was therefore used to locate the lysine residues required for the Bcl10-Ub–NEMO interaction. Whereas Bcl10(K98R) retained the ability to bind to NEMO, the K31R mutation and the dual K63R and K67R mutation impaired Bcl10-Ub–NEMO interactions (Fig. 4 A and B). Similarly, with K31R/K67R there was a substantial reduction but not complete loss of Bcl10-Ub–NEMO binding (Fig. 4C). The combination of K31R and K63R, however, completely disrupted NEMO binding (Fig. 4C). In contrast, another double lysine mutation, K44R/K45R, and a triple substitution, K105R/K110R/K115R, did not affect the Bcl10-Ub–NEMO binding at all (Fig. 4C). The K31R/K63R double mutation also disrupted the Bcl10-Ub–NEMO interaction when anti-TCR/CD28 was used to stimulate Jurkat cells (SI Fig. 9).
Fig. 4.
Bcl10 K31 and K63 are required for activation-induced Bcl10 ubiquitination and binding to NEMO. Jurkat cells transiently expressing (A) FLAG-Bcl10 or FLAG-Bcl10(K31R), or (B) FLAG-Bcl10, FLAG-Bcl10(K63R/K67R), or FLAG-Bcl10(K98R), or (C) FLAG-Bcl10, FLAG-Bcl10(K31R/K63R), FLAG-Bcl10(K31R/K67R), FLAG-Bcl10(K44R/K45R), or FLAG-Bcl10(K105R/K110R/K115R) for 36 h were treated with PMA/iono for 15 min. Cell lysates were incubated with bead-bound GST-NEMO, and the proteins pulled down were immunoblotted (IB) with anti-FLAG. Cell lysates used for GST pulldown were also immunoblotted with anti-FLAG to determine the expression of the transfected FLAG-Bcl10 constructs. Equal amounts of GST-NEMO were used in each pulldown as confirmed by Ponceau S staining or immunoblotting with anti-GST (data not shown).
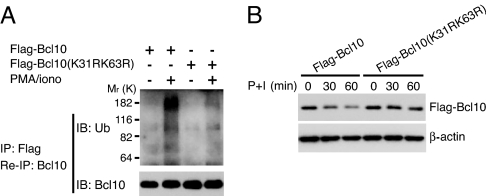
To confirm that K31 and K63 are indeed the ubiquitinated Bcl10 residues in activated cells, lysates from Jurkat cells that were transfected with FLAG-Bcl10 or FLAG-Bcl10(K31R/K63R) were immunoprecipitated with anti-FLAG. The immunoprecipitates were eluted by heating in the presence of SDS, reimmunoprecipitated with anti-Bcl10, and immunoblotted with anti-ubiquitin. Although wild-type ubiquitinated FLAG-Bcl10 was easily detectable, very little ubiquitinated FLAG-Bcl10(K31R/K63R) was found (Fig. 5A). Moreover, the individual K31R and K63R mutations diminished the in vivo Bcl10-Ub–NEMO interaction detected by anti-FLAG immunoblotting (for FLAG-Bcl10) of anti-NEMO immunoprecipitates of lysates from activated cells (data not shown). We conclude that K31 and K63 are the major sites of Bcl10 K63-linked polyubiquitin addition after stimulation. To determine whether K31 and K63 are required for TCR signaling-induced Bcl10 degradation, Jurkat cells were transfected with FLAG-Bcl10 or FLAG-Bcl10(K31R/K63R), stimulated with PMA/iono, and analyzed for Bcl10 levels by immunoblotting (Fig. 5B). As observed by others (13, 14), wild-type Bcl10 protein levels decreased after PMA/iono stimulation. However, there was little if any reduction in Bcl10(K31R/K63R) (Fig. 5B).
Fig. 5.
Bcl10 K31 and K63 are required for activation-induced Bcl10 ubiquitination and Bcl10 degradation. (A) Jurkat cells were transfected with FLAG-Bcl10 or FLAG-Bcl10(K31R/K63R) for 36 h and stimulated with PMA/iono for 15 min. Cell lysates were immunoprecipitated (IP) with anti-FLAG. The immunoprecipitates were eluted in lysis buffer containing SDS by heating, diluted with lysis buffer, reimmunoprecipitated with anti-Bcl10, and immunoblotted with anti-Ub or anti-Bcl10. (B) Jurkat cells were transfected with FLAG-Bcl10 and FLAG-Bcl10(K31R/K63R) for 36 h, then stimulated with PMA/iono (P+I) for the indicated times. Cell lysates were immunoblotted with anti-Bcl10.
Bcl10 Ubiquitination Is Required for TCR-Induced NF-κB Activation.
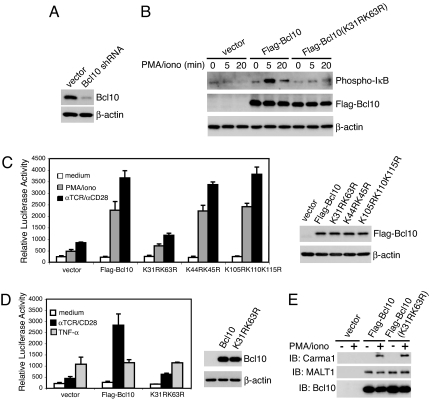
Because we are unaware of any Bcl10-deficient T cell lines, we tried to stably knock down Bcl10 with shRNA. Jurkat cells were infected with retrovirus transcribing a Bcl10 shRNA that has been shown to reduce Bcl10 levels in human B cells (21) and selected expressing cells with puromycin. The initial shRNA-transfected population had a considerable reduction in Bcl10 expression (data not shown), and after subcloning a cell with markedly reduced Bcl10 levels (Bcl10KD) was chosen for further analysis (Fig. 6A). Bcl10KD cells were transiently transfected with wild-type Bcl10 or Bcl10(K31R/K63R) and analyzed for PMA/iono-stimulated IκB phosphorylation (Fig. 6B). Whereas Bcl10KD cells transfected with vector had little response, the cells reconstituted with wild-type Bcl10 had a large increase in IκB phosphorylation after stimulation, confirming that the loss of IκB phosphorylation in the Bcl10KD cells was caused by Bcl10 deficiency. However, the restoration of IκB phosphorylation by reconstitution with Bcl10(K31R/K63R) was markedly impaired. The functional significance of Bcl10 K63-linked polyubiquitination was further addressed by cotransfecting Bcl10KD cells with cDNA encoding an NF-κB-driven luciferase reporter and shRNA-resistant wild-type Bcl10 or Bcl10 mutants in which lysines were replaced with arginine (Fig. 6C). Bcl10KD cells transfected with vector alone exhibited only slight increases in luciferase activity in response to PMA/iono or anti-TCR/CD28. When wild-type Bcl10 expression was restored, however, Bcl10KD cells responded robustly, confirming that the loss of the NF-κB response to T cell signaling was caused by Bcl10 deficiency. The NF-κB activity of Bcl10(K44R/K45R) or Bcl10(L105R/K110R/K115R)-expressing cells increased to the same extent in response to these stimuli as that of wild-type Bcl10-transfected cells (Fig. 6 C and D). In contrast, the K31R/K63R Bcl10 mutant did not restore activation-inducible NF-κB activity (Fig. 6 C and D). As expected, Bcl10 reconstitution did not change the NF-κB response of Bcl10KD cells to TNF-α, a TCR-independent activation pathway (Fig. 6D).
Fig. 6.
Bcl10 K31 and K63 are required for activation-induced IKK/NF-κB activation but not CBM assembly. (A) Jurkat cells were infected with virus containing pRSMX-Bcl10 shRNA. A stable clone with very low levels of Bcl10 (Bcl10KD) as detected by immunoblotting was selected for further analysis. (B) Bcl10 shRNA-expressing Bcl10KD cells were transfected with a plasmid encoding EGFP plus pMSCVpuro or pMSCVpuro encoding Bcl10 shRNA-resistant FLAG-Bcl10 or FLAG-Bcl10(K31R/K63R) for 24 h, and EGFP-positive cells were isolated by cell sorting. Twenty-four h later, the EGFP-positive cells were treated with PMA/iono for the indicated times, and cell lysates were immunoblotted with anti-phospho-IκB or anti-FLAG. (C and D) Bcl10 shRNA-expressing Bcl10KD cells were transiently transfected with an NF-κB luciferase reporter and a β-gal plasmid, in combination with vector, Bcl10 shRNA-resistant FLAG-Bcl10, or the indicated FLAG-Bcl10 mutants for 36 h. The transfected cells were treated with PMA/iono or C305/anti-CD28 (C), or C305/anti-CD28 or TNF-α (D), for 6 h, and NF-κB luciferase activity was measured and normalized to β-gal activity. The experiment was performed in triplicate, and error bars represent the SEM. FLAG-Bcl10 expression from one set of transfected cells is shown. (E) Jurkat cells were transfected with the indicated plasmids and stimulated with PMA/iono for 15 min. Cell lysates were subjected to immunoprecipitation with anti-FLAG and immunoblotting with anti-Carma1, anti-MALT1, and anti-Bcl10.
It is possible that loss-of-function lysine mutations in Bcl10 could conceivably affect NF-κB activation by interfering with the assembly of the CBM complex. To test this possibility, coimmunoprecipitation studies were used to examine the association of FLAG-Bcl10 with endogenous Carma1 and MALT1 (Fig. 6E). No Carmal or MALT1 was detected in the anti-FLAG immunoprecipitate from vector-transfected cells stimulated with nothing or PMA/iono. As others have observed (12), we easily detected constitutive association of MALT1 with Bcl10, and PMA/iono-inducible recruitment of Carma1 to the complex. Importantly, Bcl10(K31R/K63R) associated with both MALT1 and, in PMA/iono-simulated cells, Carma1. These results indicate that activation-induced Bcl10 polyubiquitination and activation-induced Bcl10-Ub–NEMO interaction are not required for CBM complex formation but are essential for coupling it to NF-κB activation.
Discussion
Given the recent finding that NEMO is a sensor of K63-linked polyubiquitin and that this function is important in detecting RIP ubiquitination downstream of TNF receptor signaling (15, 16), we asked whether an analogous process might occur in the case of the TCR. Notably, we found that NEMO binds to ubiquitin-modified Bcl10 in the CBM complex and that disruption of Bcl10 ubiquitination markedly inhibits IKK activation. It has been suggested that K63-linked protein ubiquitination, specifically of NEMO itself, has a role in TCR signaling-induced NF-κB activation (22). It is an attractive possibility that NEMO may bind to ubiquitinated NEMO, enhancing NEMO oligomerization, which has been suggested to favor lipopolysaccharide (LPS)-induced IKK activation (23). However, the NEMO(K399R) mutation, which abolished NEMO ubiquitination in response to TCR signaling, had only modest effects on TCR-mediated NF-κB activation (22). In contrast, mutation of the key lysines for Bcl10 ubiquitination profoundly disrupted TCR signaling-induced NF-κB activation. Therefore, although it is possible that ubiquitination of several intermediary proteins in the TCR signaling cascade may contribute to IKK activation, the interaction between ubiquitinated Bcl10 and NEMO would appear to be of particular importance.
We found that Bcl10 undergoes both K63- and K48-linked polyubiquitination in response to TCR signaling. Although these in vivo results do not allow one to determine whether one particular ubiquitin linkage is required for signaling, we favor the notion that K63-linked polyubiquitination is required. First, although both were detected, K63-linked Bcl10 was observed earlier than K48-linked ubiquitinated Bcl10. More importantly, NEMO has a marked binding preference for K63-linked compared with K48-linked polyubiquitin chains (15). Several studies have suggested that Bcl10 ubiquitination targets it for degradation, resulting in down-regulation of TCR-induced NF-κB activation (13, 14). Therefore, it is possible that, like RIP in the TNF receptor signaling pathway, ubiquitination of Bcl10 may have two discrete roles: induction of IKK activation through K63-linked ubiquitination and then degradation of Bcl10 through K48-linked ubiquitination to limit further signaling. However, unlike RIP, it appears that Bcl10 is not degraded by proteasomes (13). Instead, Bcl10 was found to translocate to lysosomal vesicles, and it was suggested that ubiquitinated Bcl10 was degraded in that compartment. Our results demonstrate that polyubiquitin chains acquired on K31 and K63 are required for Bcl10 binding to NEMO. The finding that these mutants at these sites were also less sensitive to Bcl10 down-regulation after stimulation suggests that either they are the sites of both K63- and K48-linked ubiquitination or that as for MHC class I and IFN-α receptors, K63-linked polyubiquitination is required for lysosomal degradation (24, 25).
In addition to involvement in NF-κB activation, the Carmal–Bcl10 complex selectively mediates c-Jun N-terminal kinase (JNK)2 activation in the TCR signaling pathway (26). Interestingly, the regulation of JNK2 activation by Carma1–Bcl10 is also dependent on Bcl10 ubiquitination (SI Fig. 10). It has been reported that K63-linked protein polyubiquitination is required for TNF-α-induced JNK activation (27, 28). Although the molecular mechanism remains unclear, it may involve recognition of polyubiquitin chains by ubiquitin-binding proteins such as TAB2/TAB3 and/or other unidentified mediators in the JNK pathway (29).
The ubiquitin protein ligase(s) responsible for K63-linked polyubiquitination of Bcl10 remains to be identified and may be one of those implicated in Bcl10 degradation. MALT1 and MALT1-associated TRAF6 or TRAF2 have the capacity to catalyze K63-linked polyubiquitination (22, 30). Interestingly, we detected a large reduction in PMA/iono-induced Bcl10 ubiquitination in cells where MALT1 levels were reduced with shRNA (Fig. 2B). Whether MALT1 is an E3 ligase for Bcl10 ubiquitination or acts to couple other E3 ligases to Bcl10 remains to be determined. In any case, the finding that MALT1 is required for Bcl10 ubiquitination may provide a mechanism for its role in TCR signaling.
The findings in this work establish a connection between stimulation via the TCR, formation of the CBM complex, and IKK activation. Together with previous observations with regard to TNF signaling (15), they suggest that the ubiquitin-binding activity of NEMO is a conserved mechanism for linking receptor-proximal signaling to IKK/NF-κB activation. Both Bcl10 and NEMO are also required for B cell receptor-mediated NF-κB activation (7). Like the TCR, B cell receptor-proximal signaling utilizes the CBM complex to mediate IKK/NF-κB activation (18). It will be of interest to determine whether a Bcl10-Ub–NEMO interaction occurs in other signaling pathways and, if so, whether it is required as well for NF-κB activation.
Materials and Methods
Cell Lines, Plasmids, and Reagents.
Jurkat cells were obtained from American Tissue Culture Collection. NEMO-deficient Jurkat cells reconstituted with wild-type NEMO or NEMO(L329P) have been described (15). Carma1-deficient Jurkat cells were kindly provided by Xin Lin (M. D. Anderson Cancer Center, Houston, TX). Phoenix Ampho cells were obtained from Gary Nolan (Stanford University, Palo Alto, CA). Human Bcl10 cDNA was obtained from Srinivasa Srinivasula (National Cancer Institute, National Institutes of Health, Bethesda, MD). pMSCVpuro-FLAG-Bcl10 was generated by cloning a sequence-verified PCR fragment of FLAG-NEMO into the pMSCVpuro vector (BD Clontech) digested with XhoI and EcoRI. The Bcl10 cDNA in pMSCVpuro-FLAG-Bcl10 contained five “wobble” mutations that maintained amino acid fidelity but made the transcript resistant to the effect of pRSMX-Bcl10shRNA (see below). Bcl10 point mutations were generated with a QuikChange kit (Stratagene). pGEX-4T1-NEMO, pGEX-4T1-NEMO(L329P), pMSCV-HA-NEMO, and pMSCV-HA-NEMO(L329P) have been described (15). The retroviral plasmids for Bcl10 shRNA and MALT1 shRNA (pRSMX-Bcl10shRNA and pRSMX-MALT1shRNA) have been described (21). pSUPER.retro.puro was obtained from OligoEngine. Anti-Bcl10, anti-MALT1, anti-ubiquitin, anti-IκBα, and polyclonal anti-NEMO antibodies were purchased from Santa Cruz Biotechnology. Anti-CD28 and monoclonal anti-NEMO antibodies were from BD PharMingen. Anti-Carma1 was from Cell Signaling, anti-β-actin from Sigma, and anti-HA from Roche Applied Science. Anti-Jurkat TCR C305 was obtained by collecting the culture supernatant of hybridoma cells.
Transient Transfections and NF-κB-Driven Luciferase Reporter Assay.
Jurkat cells were transiently transfected by electroporation with a BTX electroporator (Harvard Apparatus, Inc.) at 320 V for 10 s. For the NF-κB luciferase assay, NEMO-deficient Jurkat cells were transfected with the indicated vectors plus pNF-κB luc and pcDNA3.1/His/lacZ for 36 h. The cells were stimulated with 20 ng/ml PMA and 160 ng/ml ionomycin, 20% (vol/vol) C305 supernatant plus 1 μg/ml anti-CD28, or 40 ng/ml TNF-α for 6 h, and luciferase activity was determined (31). The relative light units were normalized to β-galactosidase activity (β-Gal kit; Invitrogen).
Retroviral Transduction and Derivation of Stable shRNA-Transfected Cells.
Jurkat cells were infected for 48 h with viral supernatants from Phoenix Ampho cells transfected with pRSMX containing Bcl10 shRNA or MALT1 shRNA. The infected cells were selected with puromycin (1 μg/ml) for 3–7 days. Surviving cells analyzed for protein expression by immunoblotting. Jurkat cells stably expressing MALT1 shRNA were established as a line. Bcl10 shRNA-transfected cells were cloned by limiting dilution.
Immunoprecipitation, Coimmunoprecipitation, and Immunoblotting.
After the indicated treatments, cells were lysed as described and normalized to protein concentration (15). For analyzing Bcl10 ubiquitination, SDS was added to the cell lysates to a concentration of 1%, and the lysates were heated at 95°C for 5 min, diluted with lysis buffer to a final concentration of 0.1% SDS, and incubated with antibodies for 16 h at 4°C followed by incubation with protein A-conjugated Sepharose-4B. For analyzing in vivo protein interactions, coimmunoprecipitation was performed as described in ref. 15. The immunoprecipitates or cell lysates were resolved by SDS/PAGE and transferred onto nitrocellulose membranes that were incubated with the indicated antibodies. Immunoblots were visualized by using horseradish peroxidase-conjugated secondary antibodies (Amersham) and enhanced chemiluminescence (Pierce).
In Vitro Protein-Binding Assays.
GST proteins were expressed in Escherichia coli and purified by binding to GSH beads. Purified GST proteins were incubated with Jurkat cell lysates for 3 h at 4°C. The bead-bound complexes were eluted with sample buffer, resolved by SDS/PAGE, and immunoblotted with the indicated antibodies.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Bei Dong for expert help with mutagenesis, Barbara Taylor (National Cancer Institute FACS Core Laboratory) for help with cell sorting, Srinivasa Srinivasula and Dietrich Conze for critical reading of the manuscript and helpful discussion, Shao-Cong Sun (M. D. Anderson Cancer Center, Houston, TX) for NEMO-deficient Jurkat cells, Xin Lin for Carma1-deficient Jurkat cells, and Louis Staudt and Vu Ngo (National Cancer Institute, National Institutes of Health) for Bcl10 and MALT1 shRNA. This work was supported by the Intramural Research Program of the National Institutes of Health, Center for Cancer Research, National Cancer Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712313105/DC1.
References
- 1.Hayden MS, Ghosh S. Signaling to NF-κB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 2.Rudolph D, et al. Severe liver degeneration and lack of NF-κB activation in NEMO/IKKγ-deficient mice. Genes Dev. 2000;14:854–862. [PMC free article] [PubMed] [Google Scholar]
- 3.Clements JL, Boerth NJ, Lee JR, Koretzky GA. Integration of T cell receptor-dependent signaling pathways by adapter proteins. Annu Rev Immunol. 1999;17:89–108. doi: 10.1146/annurev.immunol.17.1.89. [DOI] [PubMed] [Google Scholar]
- 4.Weil R, Israel A. Deciphering the pathway from the TCR to NF-κB. Cell Death Differ. 2006;13:826–833. doi: 10.1038/sj.cdd.4401856. [DOI] [PubMed] [Google Scholar]
- 5.Thome M, Tschopp J. TCR-induced NF-κB activation: A crucial role for Carma1, Bcl10, and MALT1. Trends Immunol. 2003;24:419–424. doi: 10.1016/s1471-4906(03)00177-7. [DOI] [PubMed] [Google Scholar]
- 6.Wang D, et al. A requirement for CARMA1 in TCR-induced NF-κB activation. Nat Immunol. 2002;3:830–835. doi: 10.1038/ni824. [DOI] [PubMed] [Google Scholar]
- 7.Ruland J, et al. Bcl10 is a positive regulator of antigen receptor-induced activation of NF-κB and neural tube closure. Cell. 2001;104:33–42. doi: 10.1016/s0092-8674(01)00189-1. [DOI] [PubMed] [Google Scholar]
- 8.Hayden MS, West AP, Ghosh S. NF-κB and the immune response. Oncogene. 2006;25:6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- 9.Lin X, Wang D. The roles of CARMA1, Bcl10, and MALT1 in antigen receptor signaling. Semin Immunol. 2004;16:429–435. doi: 10.1016/j.smim.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 10.Thome M, Weil R. Post-translational modifications regulate distinct functions of CARMA1 and Bcl10. Trends Immunol. 2007;28:281–288. doi: 10.1016/j.it.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Matsumoto R, et al. Phosphorylation of CARMA1 plays a critical role in T cell receptor-mediated NF-κB activation. Immunity. 2005;23:575–585. doi: 10.1016/j.immuni.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Wegener E, et al. Essential role for IκB kinase β in remodeling Carma1–Bcl10–Malt1 complexes upon T cell activation. Mol Cell. 2006;23:13–23. doi: 10.1016/j.molcel.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 13.Scharschmidt E, Wegener E, Heissmeyer V, Rao A, Krappmann D. Degradation of Bcl10 induced by T cell activation negatively regulates NF-κB signaling. Mol Cell Biol. 2004;24:3860–3873. doi: 10.1128/MCB.24.9.3860-3873.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu S, et al. cIAP2 is a ubiquitin protein ligase for BCL10 and is dysregulated in mucosa-associated lymphoid tissue lymphomas. J Clin Invest. 2006;116:174–181. doi: 10.1172/JCI25641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys-63-linked polyubiquitination by NEMO is a key event in NF-κB activation. Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 16.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNF-α requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt-Supprian M, et al. NEMO/IKKγ-deficient mice model incontinentia pigmenti. Mol Cell. 2000;5:981–992. doi: 10.1016/s1097-2765(00)80263-4. [DOI] [PubMed] [Google Scholar]
- 18.Schulze-Luehrmann J, Ghosh S. Antigen-receptor signaling to nuclear factor κB. Immunity. 2006;25:701–715. doi: 10.1016/j.immuni.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Koseki T, et al. CIPER, a novel NF-κB-activating protein containing a caspase recruitment domain with homology to Herpesvirus-2 protein E10. J Biol Chem. 1999;274:9955–9961. doi: 10.1074/jbc.274.15.9955. [DOI] [PubMed] [Google Scholar]
- 20.Lucas PC, et al. Bcl10 and MALT1, independent targets of chromosomal translocation in malt lymphoma, cooperate in a novel NF-κB signaling pathway. J Biol Chem. 2001;276:19012–19019. doi: 10.1074/jbc.M009984200. [DOI] [PubMed] [Google Scholar]
- 21.Ngo VN, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 22.Zhou H, et al. Bcl10 activates the NF-κB pathway through ubiquitination of NEMO. Nature. 2004;427:167–171. doi: 10.1038/nature02273. [DOI] [PubMed] [Google Scholar]
- 23.Tegethoff S, Behlke J, Scheidereit C. Tetrameric oligomerization of IκB kinase γ (IKKγ) is obligatory for IKK complex activity and NF-κB activation. Mol Cell Biol. 2003;23:2029–2041. doi: 10.1128/MCB.23.6.2029-2041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duncan LM, et al. Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. EMBO J. 2006;25:1635–1645. doi: 10.1038/sj.emboj.7601056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar KG, et al. Site-specific ubiquitination exposes a linear motif to promote interferon-α receptor endocytosis. J Cell Biol. 2007;179:935–950. doi: 10.1083/jcb.200706034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blonska M, et al. The CARMA1–Bcl10 signaling complex selectively regulates JNK2 kinase in the T cell receptor-signaling pathway. Immunity. 2007;26:55–66. doi: 10.1016/j.immuni.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi CS, Kehrl JH. Tumor necrosis factor (TNF)-induced germinal center kinase-related (GCKR) and stress-activated protein kinase (SAPK) activation depends upon the E2/E3 complex Ubc13-Uev1A/TNF receptor-associated factor 2 (TRAF2). J Biol Chem. 2003;278:15429–15434. doi: 10.1074/jbc.M211796200. [DOI] [PubMed] [Google Scholar]
- 28.Habelhah H, et al. Ubiquitination and translocation of TRAF2 is required for activation of JNK but not of p38 or NF-κB. EMBO J. 2004;23:322–332. doi: 10.1038/sj.emboj.7600044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanayama A, et al. TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 30.Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol Cell. 2004;14:289–301. doi: 10.1016/s1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- 31.Petrak D, Memon SA, Birrer MJ, Ashwell JD, Zacharchuk CM. Dominant negative mutant of c-Jun inhibits NF-AT transcriptional activity and prevents IL-2 gene transcription. J Immunol. 1994;153:2046–2051. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.