Abstract
Epidemiological studies have associated certain human disease outcomes with particular killer cell Ig-like receptor (KIR) and HLA genotypes. However, the functional explanation for these associations is poorly understood, because the KIRs were initially described as natural killer (NK) cell inhibitory receptors with specificity for HLA molecules on their cellular targets. Yet resolution of infections is often associated with genotypic pairing of inhibitory KIRs with their cognate HLA ligands. Recent studies in mice indicate a second role for MHC-specific inhibitory receptors, i.e., self-MHC recognition confers functional competence on the NK cell to be triggered through their activation receptors, a process termed licensing. As a result, licensed NK cells with self-MHC-specific receptors are more readily activated as compared with unlicensed NK cells without self-MHC-specific receptors. Such results predict that human NK cells may undergo a similar process. Here, we examined the human NK cell subset expressing KIR3DL1, the only known KIR specific for HLA-Bw4 alleles. The KIR3DL1+ subset in normal donors with two HLA-B-Bw4 genes displayed increased responsiveness to tumor stimulation compared with the KIR3DL1+ subset from individuals with only one or no Bw4 genes. By contrast, NK cells lacking KIR3DL1 showed no differences. Therefore, these data indicate that particular KIR and HLA alleles are associated with more responsive NK cells, strongly suggesting that human NK cells are also subjected to NK cell licensing, and providing a potential functional explanation for the influence of KIR and HLA genes in disease as well as interindividual differences in NK cell potency.
Keywords: innate immunity, killer cell Ig-like receptor (KIR), NK cells, licensing
Natural killer (NK) cells are a subset of lymphocytes with the defining feature of being able to kill target cells and produce cytokines without the need for prior sensitization. Pathologic targets include virus-infected cells and cells undergoing malignant transformation. In the effector response, the ability of NK cells to discriminate normal from pathologic self-tissues is largely explained by the inhibitory function of NK cell surface receptors that recognize and bind specific MHC class I molecules on potential targets (1, 2). In many pathologic conditions, MHC class I molecules are down-modulated, releasing NK cells from the inhibitory receptor effect and permitting the action of activation receptors that recognize target cells. In humans, the killer Ig-like receptors (KIRs) are such inhibitory receptors (3). Their genes are clustered on chromosome 19q; display extensive polymorphism, at both the haplotype and individual gene allele levels; and are typically expressed in a stochastic manner (4, 5). Moreover, an individual NK cell may simultaneously express multiple different receptors, each of which may have different ligand specificities. The KIR ligands are specific HLA molecules encoded by genes on chromosome 6 and are even more genetically diverse. Therefore, the genes for KIR and their cognate HLA ligands display extensive polymorphism and segregate independently, and NK cells display a diversity of receptors.
There is significant heterogeneity between individuals with respect to the NK cell response to pathogens and tumor targets (6, 7). The response of individual NK cells likely contribute to overall NK cell potency that in turn may contribute to individual variation in host immune response to infections and malignancy. Although potentially a large number of determinants may be responsible for the spectrum of NK cell potency, none has been clearly identified.
Recently, epidemiological studies have associated specific KIR and HLA genotypes with certain disease processes, such as clearance of infections (8). Counterintuitive to the known inhibitory role of KIR at the effector level, the best associations surprisingly indicate improved outcomes from infections in individuals with the pairing of genes for specific inhibitory KIR and their cognate HLA ligands. For example, coinheritance of inhibitory KIR2DL3 and its cognate ligand HLA-CAsn-80 is associated with increased clearance of hepatitis C virus (9). Protection from progression of cervical carcinoma in situ to invasive carcinoma, a disease caused by human papilloma virus, is also correlated with the genotype for KIR3DL1 in combination with that of its ligand HLA-Bw4 or the genotype of KIR2DL1 with its HLA-CLys-80 ligand (10). Furthermore, the combined genotype of KIR3DL1 and its ligand HLA-Bw4 was found to have a strong association with slower progression to AIDS in HIV infection (11, 12). However, epidemiological studies had previously implicated the related activating isoforms of KIRs, such as KIR2DS1, with HLA-B alleles in protection from HIV (13). Although the known interaction between KIR and HLA molecules has thus implicated a role for NK cells in these diseases, it has been challenging to provide a mechanistic explanation for these correlations. Moreover, the presence of both the genes for an inhibitory receptor and its ligand is ironically associated with protection in many studies (14).
In addition to their inhibitory function at the effector level, another critical role for these MHC-specific inhibitory receptors was recently described (15), a process that we have termed licensing. We showed that the ability to activate murine NK cells depends on the interaction between these receptor(s) and their cognate MHC class I ligand(s), expressed as a self-MHC molecule. The licensing effect is manifested with both target cell-mediated and target cell-free stimuli when the self-MHC-specific inhibitory receptor is not engaged. A recent study of selected donors showed a relatively enhanced activity of KIR2DL1+ and KIR2DL2/3+ human NK cells in the presence of their respective HLA-C ligands, suggesting that the licensing process may also be relevant for human NK cells (16). Both studies suggest that there are two distinct classes of NK cells within a given mouse or human subject: a functionally competent (licensed) class that expresses receptors recognizing self-MHC class I molecules and another functionally incompetent (unlicensed) class that does not (17, 18). We also previously observed that subsets of NK cells, expressing the same MHC-recognizing receptors, show clear differences in responsiveness when compared between mice with different MHC class I but otherwise identical genetic backgrounds (15). However, interindividual variation between different human donors has not been previously addressed but is necessary because of the profound genetic variation between humans. Thus, it is possible that licensing effects may contribute to the heterogeneity of overall NK cell potency between different individuals, even in humans, which may affect disease susceptibility and outcome.
Herein, we addressed whether HLA and KIR genotypes, as a single set of genetic variables, can influence differences in NK cell functional responses among normal unrelated individuals. For the examined combination of a KIR and its HLA ligand, we find enhanced ability of NK cells to be triggered, consistent with human NK cell licensing. We propose that these findings may be extrapolated to the broader universe of HLA-specific receptors and their cognate ligands, thus providing a basis for differences in overall NK cell potency in individuals and a potential explanation for the epidemiological association of KIR–HLA combinations with resolution from viral infections.
Results
We chose to focus our study on the KIR3DL1 subset of NK cells because KIR3DL1 has been widely studied and has important associations with disease in many correlative clinical studies (8). Moreover, there is a specific antibody for the KIR3DL1 receptor that can distinguish the inhibitory KIR3DL1 isoform from the activating KIR3DS1 isoform (19, 20). Furthermore, there is a well established specificity of this receptor for HLA-B molecules belonging to the Bw4 group, whereas those of the other epitope group, Bw6, are poorly recognized by this receptor (21, 22). These two public epitopes comprise all HLA-B alleles and are mutually exclusive. Finally, KIR3DL1 is the only known KIR with HLA-B specificity (8). Thus, our studies specifically addressed the effects of HLA-B allele differences on the functional capacity of KIR3DL1+ NK cells among 39 healthy, unrelated donors.
Phenotypic and genotypic analyses showed that 36 donors expressed detectable KIR3DL1 and 2 donors (nos. 44–06 and 46–05) did not have the gene for KIR3DL1 [Table 1 and supporting information (SI) Table 2]. The remaining donor (no. 66–10) had the gene, but the receptor was not detected on the cell surface, suggesting that it was for a subtype reported to be retained intracellularly (23).
Table 1.
HLA-B and KIR genotypes of study subjects
| Donor ID no. | HLA-B | HLA-B | 3DL1 | 3DS1 |
|---|---|---|---|---|
| 44–01 | 3701 | 4403 | + | − |
| 44–02 | 1302 | 4403 | + | − |
| 44–03 | 4402 | 5101 | + | + |
| 44–04 | 3701 | 4403 | + | − |
| 44–05 | 4402 | 5801 | + | − |
| 44–06 | 2705 | 5701 | − | + |
| 44–07 | 4901 | 5301 | + | − |
| 44–08 | 1302 | 2705 | + | − |
| 44–09 | 4402 | 5801 | + | − |
| 44–10 | 4402 | 5201 | + | + |
| 44–11 | 2705 | 5801 | + | − |
| 44–12 | 4403 | 5101 | + | − |
| 66–01 | 1402 | 4001 | + | − |
| 66–02 | 0801 | 1801 | + | − |
| 66–03 | 0801 | 3501 | + | − |
| 66–04 | 0702 | 1501 | + | + |
| 66–05 | 0801 | 4001 | + | − |
| 66–06 | 0702 | 0801 | + | − |
| 66–07 | 0801 | 3502 | + | + |
| 66–08 | 1501 | 3901 | + | − |
| 66–09 | 0702 | 4001 | + | + |
| 66–10 | 0702 | 1801 | + | − |
| 66–11 | 0702 | 3503 | + | − |
| 66–12 | 0702 | 1501 | + | − |
| 66–13 | 1501 | 4801 | + | + |
| 66–14 | 0726 | 1801 | + | − |
| 46–01 | 0801 | 2705 | + | − |
| 46–02 | 1801 | 5701 | + | + |
| 46–03 | 0801 | 4701 | + | − |
| 46–04 | 1301 | 3501 | + | + |
| 46–05 | 4001 | 4402 | − | − |
| 46–06 | 2705 | 3906 | + | + |
| 46–07 | 3802 | 4002 | + | + |
| 46–08 | 1501 | 5201 | + | − |
| 46–09 | 0702 | 5701 | + | − |
| 46–10 | 3501 | 4403 | + | − |
| 46–11 | 1501 | 4402 | + | − |
| 46–12 | 4006 | 4403 | + | + |
| 46–13 | 1401 | 3801 | + | − |
HLA-B alleles with Bw4 epitopes are shown in boldface type. + and − signs denote the presence or the absence of the indicated KIR genes.
To examine the potential effects of HLA-B polymorphisms on NK cell activation, the 36 KIR3DL1-expressing donors were divided into three groups based on their HLA genotypes: HLA-Bw4/4 homozygotes (n = 11); HLA-Bw4/6 heterozygotes (n = 12); and HLA-Bw6/6 homozygotes (n = 13). Peripheral blood mononuclear cells (PBMC) were freshly isolated, incubated with MHC-deficient 721.221 tumor cells, and analyzed for IFN-γ production as a measure of NK cell activation. In all Bw4/4 donors, the KIR3DL1+ subset produced IFN-γ to a greater degree than the KIR3DL1− subset did (Fig. 1a and data not shown), consistent with licensing through the interaction of KIR3DL1 and its ligand. In contrast, this difference in IFN-γ production between KIR3DL1+ and KIR3DL1− subsets was not apparent in the Bw6/6 group. These measurable differences were also seen in killing assays (Fig. 1b), again demonstrating that the KIR3DL1+ subset was more responsive than the KIR3DL1− subset when compared within a given individual homozygous for the Bw4 genotype but not the Bw6 genotype.
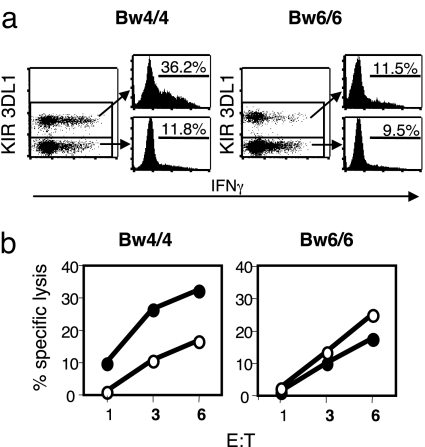
Fig. 1.
Enhanced potency of KIR3DL1+ NK cells in individuals with HLA-Bw4. (a) Freshly isolated PBMC from the indicated donors were incubated with MHC-deficient 721.221 tumor cells and analyzed for intracellular IFN-γ. Gated NK (CD3− CD56+) cells are shown, and the numbers represent the percentages of IFN-γ+ cells among the KIR3DL1+ or KIR3DL1− NK cell populations. In the dot plots, note that the KIR3DL1− subset is much larger than the KIR3DL1+ subset, giving the illusion of more response by the KIR3DL1− cells. The histograms more accurately show the responses by the different subsets. (b) KIR3DL1+ (solid circles) and KIR3DL1− (open circles) NK cells were sorted from freshly isolated PBMC of the indicated donors and used in standard 4-h killing assays against 721.221 target cells. Effector-to-target (E:T) ratios are indicated on the x axis. This panel is representative of two Bw4/Bw4 and five Bw6/Bw6 donors. One donor from each group was retested, and the killing patterns were essentially identical in these two independent experiments. For the donors tested for both assays, IFN-γ production generally correlated with killing activity (data not shown).
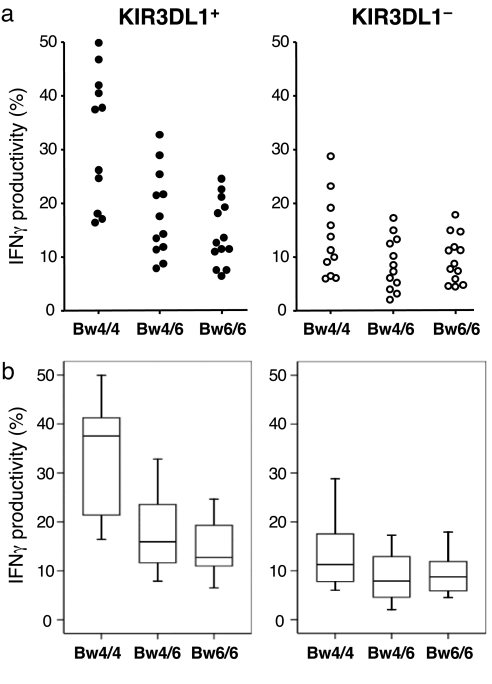
Comparison of the entire population of donors revealed a strong correlation between the HLA-B genotype and NK cell responses. Upon tumor stimulation, the responsiveness of KIR3DL1+ NK cells was significantly different among the Bw4/4, Bw4/6, and Bw6/6 groups (H = 12.91, P = 0.002; Fig. 2). In the Bw4/4 group, there was robust production of IFN-γ by the KIR3DL1+ subset that was strikingly and significantly greater than that of the Bw4/6 group (U = 22.0, P = 0.007) and the Bw6/6 group (U = 15.0, P = 0.001). There appeared to be a slightly higher degree of responsiveness by the KIR3DL1+ NK cells from the Bw4/6 group than by the same NK cell subset from the Bw6/6 group, but this was not statistically significant (U = 56.0, P = 0.247). Nevertheless, there did appear to be a gene dosage effect by Bw4 because there was a statistically significant trend when comparing the IFN-γ by the KIR3DL1+ NK cells across all three groups (τ = −0.471, P < 0.0001). In contrast to the KIR3DL1+ subset, there was no significant difference in the responsiveness of the KIR3DL1− subset among the three groups (H = 3.24, P = 0.198; Fig. 2). Taken together, these data indicate that the functional capacity of KIR3DL1+ NK cells is primarily determined by HLA-B polymorphisms.
Fig. 2.
Interindividual heterogeneity in the potency of KIR3DL1+ NK cells is determined by HLA-B alleles. Freshly isolated PBMC were incubated with 721.221 tumor cells and analyzed for intracellular IFN-γ. On the basis of HLA-B genotypes, donors were divided into three groups (Bw4/4 homozygotes, n = 11; Bw4/6 heterozygotes, n = 12; Bw6/6 homozygotes, n = 13) as indicated on the x axis. The y axis reflects the percentage of cells expressing IFN-γ among the KIR3DL1+ (Left) or KIR3DL1− (Right) NK cell populations. (a) Individual results from each donor are shown. (b) Statistical representation of results. Individual results from a are shown as boxes representing the interquartile (25th to 75th percentile) group. The horizontal bar within the box represents the median value, and whiskers represent the range of values. For the KIR3DL1+ subset (Left), statistically significant differences were seen between the Bw4/4 group and the Bw4/6 group (U = 22.0, P = 0.007) and between the Bw4/4 group and the Bw6/6 group (U = 15.0, P = 0.001). A statistically significant trend was observed across the three groups (τ = −0.471, P < 0.0001).
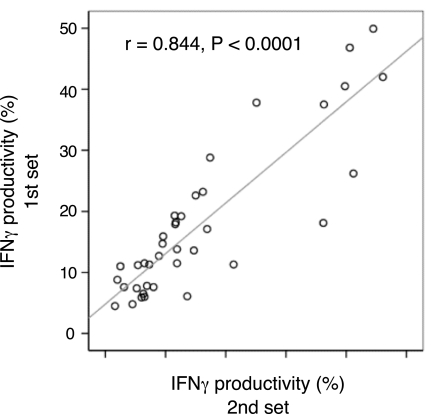
Because these results were collected from an aggregate of seven independent experiments, we retested 19 available subjects from the Bw4/4 and Bw6/6 groups to ensure that interexperimental variation was not responsible for the differences. This subsequent retesting of the 19 subjects was performed at least 6 months after the initial testing, and the results demonstrated that measurements of IFN-γ productivity were unaffected by experimental variation (r = 0.844, P < 0.0001; Fig. 3 and SI Fig. 5). Again, the KIR3DL1+ subset from the Bw4/4 group produced significantly more IFN-γ than that of the Bw6/6 group (U = 0.00, P < 0.0001; SI Fig. 5). These data confirm that the robust difference in the responsiveness of KIR3DL1+ NK cells toward 721.221 targets is strongly associated with the presence of HLA-Bw4 alleles.
Fig. 3.
Consistency of KIR3DL1+ NK cell potency in HLA-Bw4 individuals. Nineteen of the original donors were retested at least 6 months after the initial set of assays for IFN-γ production after tumor target stimulation. These donors were either Bw4/4 homozygotes (n = 9) or Bw6/6 homozygotes (n = 10). Both KIR3DL1+ and KIR3DL1− subsets are represented on the graph. Spearman's rank correlation coefficient was calculated to assess similarity between the first and second set of data. Raw data are available in SI Fig. 5.
The 721.221 target cell is a well characterized MHC-deficient EBV-transformed human B cell line that is readily recognized and lysed by human NK cells. Although stimulation of primary human NK cells by this target occurs mainly through the activating receptor NKp46 and the coactivating receptors 2B4 and NTB-A (24, 25), no consistent difference in these receptor expression levels was seen between the different HLA-B groups (data not shown). Furthermore, when examined within individuals, the KIR3DL1− subset displayed a generally higher level of NKp46 expression compared with the KIR3DL1+ NK cell subset, even though the former subset had a poorer response to 721.221 stimulation (data not shown). Thus, the enhanced responsiveness of the KIR3DL1+ subset in the Bw4/4 group is not due to expression of these activating or coactivating receptors, suggesting that they are due to intrinsic differences downstream of these receptors.
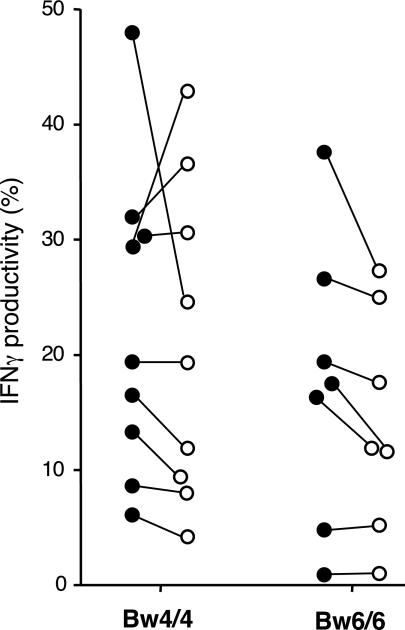
Interestingly, the apparent licensing through KIR3DL1 did not universally affect NK cell responsiveness through all stimuli. With some donors, we were also able to examine NK cells stimulated by CD16 cross-linking, which is sufficient to stimulate freshly isolated NK cells to produce IFN-γ (16, 26). Unexpectedly, there was no significant difference in IFN-γ production by the KIR3DL1+ subset between Bw4/4 and Bw6/6 groups, unlike what was seen with tumor cell stimulation (U = 24.5, P = 0.470; Fig. 4). Furthermore, there were no consistent differences between the KIR3DL1+ and KIR3DL1− subsets even when compared within a given Bw4/4 individual (P = 0.477; Fig. 4). These results contrast those from a recent study in which CD16 cross-linking was able to stimulate preferentially the KIR2DL1+ and KIR2DL2/3+ subsets in a small number of individuals having the cognate HLA-C ligands (16). Whether this discrepancy is due to differences in receptors examined (KIR2DL1/2/3 vs. KIR3DL1) or experimental conditions (e.g., incubation time) may need to be explored. Although previous murine studies indicate that licensing affects NK cell responsiveness to various target cell-free stimuli, such as antiactivation receptor cross-linking (15, 27), the results presented here suggest that there is a higher level of complexity in humans that needs to be explored further.
Fig. 4.
Responsiveness of KIR3DL1+ NK cells to CD16 cross-linking is not affected by host HLA-B genes. Freshly isolated PBMC from the indicated donors (Bw4/4 homozygotes, n = 9; Bw6/6 homozygotes, n = 7) were stimulated with plate-bound anti-CD16 and analyzed for intracellular IFN-γ production by KIR3DL1+ (solid circles) or by KIR3DL1− (open circles) NK cell population. Each dot represents the percentage of IFN-γ+ cells among the KIR3DL1+ or KIR3DL1− NK cell population from each individual; the percentages from the same individual are connected by a line. The IFN-γ production by KIR3DL1+ subsets between HLA-B groups was not significantly different (Bw4/4 vs. Bw6/6, U = 24.5, P = 0.470); the IFN-γ production by KIR3DL1− subsets between HLA-B groups was also not significantly different (Bw4/4 vs. Bw6/6, U = 23.5, P = 0.408). Comparison of KIR3DL1+ vs. KIR3DL1− cells in Bw4/4 homozygotes showed no significant difference in IFN-γ production (P = 0.477). Comparison of KIR3DL1+ vs. KIR3DL1− cells in Bw6/6 homozygotes also showed no significant difference in IFN-γ production (P = 0.063).
Discussion
To our knowledge, this is the first functional demonstration of how HLA polymorphisms can determine differences in innate immune cell responses between individuals. We have shown that interindividual differences in the response of KIR3DL1+ NK cells to their targets can be determined by polymorphisms in HLA-B genes. Our data are consistent with a licensing effect of self-HLA on human NK cell functional maturation through inhibitory receptors for a given HLA because the cognate ligand for KIR3DL1 is HLA-Bw4. Surprisingly, this effect is robust despite the diversity of individual KIRs expressed by each NK cell and the overlapping subsets of NK cells based on KIR expression. Moreover, there are likely to be other genes that contribute to NK cell potency. For example, it remains formally possible that other KIR and HLA combinations contribute to the apparent licensing effect because the KIR genes are linked to each other, and the HLA alleles are similarly linked. Indeed, in a limited analysis, we found the KIR2DL2/3+ KIR3DL1+ double positive subset to be somewhat more responsive to tumor stimulation than the KIR2DL2/3− KIR3DL1+ NK cells (data not shown), suggesting that the KIR2DL2/3 receptors contribute to some degree, but minimally, to NK cell potency. Similarly, it is possible that other inhibitory receptors may contribute to these effects, such as CD94/NKG2A, or even receptors without HLA specificity such as NKRP1A. To determine the isolated contribution of individual KIR and HLA allele pairs to NK cell licensing, future studies will require much larger cohorts of donors. Nonetheless, our data support the general conclusion that KIR-HLA ligand combinations can produce a licensing effect, permitting the hypothesis that the combined effect of an individual's KIR and HLA alleles in licensing the total pool of NK cells will contribute to overall NK cell potency in that given individual and thus play a major role in determining the heterogeneity of NK cell function between humans.
Our findings that the KIR3DL1+ subset of NK cells is more responsive in individuals with Bw4, as opposed to Bw6, alleles, is consistent with licensing through KIR3DL1 by HLA-Bw4. These findings markedly extend and add insights to previous studies on the relationship of KIR expression to human NK cell function (16). Recently, one of our groups also described a hierarchy of NK cell responses as determined by inhibitory NK cell receptor repertoire and self-MHC (28). In both cases, selected individuals were studied in greater detail than shown here. Our studies reported here focused on determining the licensing effect of a single receptor–ligand interaction on NK cell responses among a larger panel of donors and between different individuals. There was no apparent difference in IFN-γ production by KIR3DL1+ NK cells between donors who are homo- or heterozygous for KIR3DL1 (data not shown), suggesting that there was no apparent gene dosage effect of KIR3DL1. Perhaps this result was not unexpected because there should be very few NK cells that express both alleles of KIR3DL1 in the KIR3DL1 homozygous donor population, if KIR3DL1 is expressed in a stochastic manner, as expected. However, our current studies indicate that there may be a gene dosage effect relating the number of copies of HLA-Bw4 genes with NK cell potency. In particular, the KIR3DL1+ NK cell subset in the Bw4/4 homozygote group of individuals had a much greater degree of potency compared with that of either the Bw4/6 heterozygote or the Bw6/6 homozygote groups, suggesting that avidity between KIR3DL1 and HLA-Bw4 may be an important determinant of immune potency. Likewise, avidity factors may influence the contribution of each KIR–HLA receptor–ligand pair in terms of licensing effects.
The robust responsiveness of the KIR3DL1+ subset in Bw4/4 homozygotes may be particularly relevant to recent clinical observations demonstrating that Bw4/4 homozygotes have a very strong protection against HIV infection compared with Bw4/6 heterozygotes or Bw6/6 homozygotes (29). Because HIV is known to down-modulate HLA-B through the virally encoded Nef protein (30) (releasing KIR3DL1+ NK cells from inhibitory effects of HLA-B), the more potent KIR3DL1+ NK cells in these Bw4/4 homozygote individuals may be providing stronger protection. In addition to possible avidity effects, there may also be influences by the differences in affinity between the many Bw4 alleles (>100) and the many KIR3DL1 subtypes (at least 10) (31, 32). Consistent with this possibility, recent studies have shown that HIV patients with high-affinity KIR3DL1/HLA-Bw4 pairs had a significantly delayed progression to AIDS (12). To examine these possible avidity and affinity effects on NK cell potency, future studies will, therefore, need to consider the KIR3DL1 subtype, as well as specification of HLA-Bw4 alleles, in functional analyses of a much greater number of samples. Nonetheless, our observations that differences in NK cell potency between individuals may be influenced by KIR and HLA genotypes suggest that licensing of NK cells may be an important factor in the control of HIV, as well as other virus-related diseases (9, 10).
Therefore, further studies to assess relative contributions by other KIRs, as well as other ITIM-containing receptors, will be important to understand more completely how various KIR and HLA alleles affect NK cell potency. It is conceivable that certain KIR and HLA combinations contribute more to overall NK cell potency than other combinations. Our studies thus provide a novel approach to understand the effects of HLA alleles (and KIR alleles) on disease susceptibility and outcome.
Materials and Methods
Samples.
Peripheral blood from 39 normal, healthy, unrelated volunteers was obtained with informed consent, approved by the Institutional Review Board at Washington University School of Medicine. PBMC were isolated by Ficoll-Hypaque density gradient centrifugation within 24 h of obtaining the blood in the first set of experiments (n = 39) and within 12 h in the second set of experiments (n = 19).
Cytokine Assay.
For stimulation with tumor targets, PBMC (2 × 106 cells) were mixed with MHC-deficient 721.221 target cells (1 × 105 cells) in RPMI1640 complete medium containing 10% FCS. For stimulation through CD16, PBMC (5 × 106 ≈1 × 107 cells) were incubated with in six-well plates directly coated with 5 μg/ml of anti-CD16 (3G8). Cells were incubated for 1 h at 37°C, and they were incubated for an additional 6–7 h in the presence of the protein transport inhibitor brefeldin A. After stimulation, cells were first stained for cell surface markers by using anti-CD56, anti-KIR3DL1 (DX9), and anti-CD3, and then they were stained for intracellular IFN-γ after fixation with 2% formaldehyde. To exclude T cells from the analysis, CD3+ cells were gated out in all experiments, and CD19+ cells were also gated out in the second set of experiments. Because of the technical and logistic limitations of human studies involving freshly isolated samples, the results are the aggregate of seven independent stimulation assay experiments in the first set of experiments. In the second set of experiments, the results are the aggregate of five independent experiments.
Cytotoxicity Assay.
KIR3DL1+ CD56+ CD3− and KIR3DL1− CD56+ CD3− NK cells were sorted from freshly isolated PBMC to a purity of >90% and were directly tested in standard 4 h 51Cr-release assays against 721.221 target cells in 96-well V-bottom plates.
HLA and KIR Genotyping.
HLA typing was performed by low-resolution PCR-sequence-specific oligonucleotide typing for HLA-A, -B, and -C, followed by high-resolution sequence-specific oligonucleotide probe (SSOP) analysis. KIR genotyping was performed as described in ref. 33.
Statistical Analysis.
Nonparametric statistics were used because of the sample size and nonnormal distribution of data. In the tumor stimulation assays, Kruskal–Wallis tests were used to compare the differences in IFN-γ productivity across all three HLA-B groups, and Mann–Whitney U pairwise comparison posttests were used to assess differences between pairs of groups. A Bonferroni correction was applied to adjust the alpha level for multiple comparisons (α = 0.017). Kendall's tau-b statistic was computed to evaluate trend across the three HLA groups. Spearman's rank correlation coefficient was calculated to assess similarity between test (first set of experiments) and retest (second set of experiments) subjects (n = 19) in HLA-Bw4/4 and HLA-Bw6/6 homozygote groups. In the CD16 stimulation assays, differences in IFN-γ productivity between HLA-Bw4/4 and HLA-Bw6/6 homozygote groups were analyzed by using Mann–Whitney tests, and differences in IFN-γ productivity between paired (KIR3DL1+ and KIR3DL1−) NK cells from the same donor were compared by using Wilcoxon signed rank tests. All tests were two-tailed tests with α = 0.05. SPSS 14.0 (SPSS Inc.) was used to conduct statistical tests and generate figures. All statistical analyses were conducted by the Clinical Outcomes Research Office, Department of Otolaryngology, Washington University School of Medicine.
Supplementary Material
ACKNOWLEDGMENTS.
We thank D. Higuchi and J. Gao for technical assistance and P. Klekotka and H. Jonsson for critical review of the manuscript. This study was supported by National Institutes of Health grants to W.M.Y, who is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712229105/DC1.
References
- 1.Karlhofer FM, Ribaudo RK, Yokoyama WM. MHC class I alloantigen specificity of Ly-49+ IL-2-activated natural killer cells. Nature. 1992;358:66–70. doi: 10.1038/358066a0. [DOI] [PubMed] [Google Scholar]
- 2.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 3.Long EO, Rajagopalan S. HLA class I recognition by killer cell Ig-like receptors. Semin Immunol. 2000;12:101–108. doi: 10.1006/smim.2000.0212. [DOI] [PubMed] [Google Scholar]
- 4.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 5.Kelley J, Walter L, Trowsdale J. Comparative genomics of natural killer cell receptor gene clusters. PLoS Genet. 2005;1:129–139. doi: 10.1371/journal.pgen.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korbel DS, et al. Heterogeneous human NK cell responses to Plasmodium falciparum-infected erythrocytes. J Immunol. 2005;175:7466–7473. doi: 10.4049/jimmunol.175.11.7466. [DOI] [PubMed] [Google Scholar]
- 7.Imai K, et al. Natural cytotoxic activity of peripheral-blood lymphocytes and cancer incidence: An 11-year follow-up study of a general population. Lancet. 2000;356:1795–1799. doi: 10.1016/S0140-6736(00)03231-1. [DOI] [PubMed] [Google Scholar]
- 8.Bashirova AA, Martin MP, McVicar DW, Carrington M. The killer immunoglobulin-like receptor gene cluster: Tuning the genome for defense. Annu Rev Genomics Hum Genet. 2006;7:277–300. doi: 10.1146/annurev.genom.7.080505.115726. [DOI] [PubMed] [Google Scholar]
- 9.Khakoo SI, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 10.Carrington M, et al. Hierarchy of resistance to cervical neoplasia mediated by combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci. J Exp Med. 2005;201:1069–1075. doi: 10.1084/jem.20042158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Vazquez A, et al. Interaction between KIR3DL1 and HLA-B*57 supertype alleles influences the progression of HIV-1 infection in a Zambian population. Hum Immunol. 2005;66:285–289. doi: 10.1016/j.humimm.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Martin MP, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin MP, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 14.Rajagopalan S, Long EO. Understanding how combinations of HLA, KIR genes influence disease. J Exp Med. 2005;201:1025–1029. doi: 10.1084/jem.20050499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 16.Anfossi N, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 17.Yokoyama WM, Kim S. How do natural killer cells find self to achieve tolerance? Immunity. 2006;24:249–257. doi: 10.1016/j.immuni.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Raulet DH, Vance RE. Self-tolerance of natural killer cells. Nat Rev Immunol. 2006;6:520–531. doi: 10.1038/nri1863. [DOI] [PubMed] [Google Scholar]
- 19.O'Connor GM, et al. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J Immunol. 2007;178:235–241. doi: 10.4049/jimmunol.178.1.235. [DOI] [PubMed] [Google Scholar]
- 20.Trundley A, et al. Allelic expression patterns of KIR3DS1 and 3DL1 using the Z27 and DX9 antibodies. Eur J Immunol. 2007;37:780–787. doi: 10.1002/eji.200636773. [DOI] [PubMed] [Google Scholar]
- 21.Cella M, et al. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med. 1994;180:1235–1242. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gumperz JE, et al. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995;181:1133–1144. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pando MJ, et al. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J Immunol. 2003;171:6640–6649. doi: 10.4049/jimmunol.171.12.6640. [DOI] [PubMed] [Google Scholar]
- 24.Parolini S, et al. X-linked lymphoproliferative disease: 2B4 molecules displaying inhibitory rather than activating function are responsible for the inability of natural killer cells to kill Epstein–Barr virus-infected cells. J Exp Med. 2000;192:337–346. doi: 10.1084/jem.192.3.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bottino C, et al. NTB-A, a novel SH2D1A-associated surface molecule contributing to the inability of natural killer cells to kill Epstein–Barr virus-infected B cells in X-linked lymphoproliferative disease. J Exp Med. 2001;194:235–246. doi: 10.1084/jem.194.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bryceson YT, March ME, Ljunggren HG, Long EO. Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion. Blood. 2006;107:159–166. doi: 10.1182/blood-2005-04-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandez NC, et al. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood. 2005;105:4416–4423. doi: 10.1182/blood-2004-08-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu J, et al. Hierarchy of the human natural killer cell response is determined by class and quantity of inhibitory receptors for self-HLA-B and HLA-C ligands. J Immunol. 2007;179:5977–5989. doi: 10.4049/jimmunol.179.9.5977. [DOI] [PubMed] [Google Scholar]
- 29.Flores-Villanueva PO, et al. Control of HIV-1 viremia and protection from AIDS are associated with HLA-Bw4 homozygosity. Proc Natl Acad Sci USA. 2001;98:5140–5145. doi: 10.1073/pnas.071548198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohen GB, et al. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 1999;10:661–671. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 31.Thananchai H, et al. Cutting Edge: Allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol. 2007;178:33–37. doi: 10.4049/jimmunol.178.1.33. [DOI] [PubMed] [Google Scholar]
- 32.Carr WH, Pando MJ, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol. 2005;175:5222–5229. doi: 10.4049/jimmunol.175.8.5222. [DOI] [PubMed] [Google Scholar]
- 33.Hsu KC, et al. Improved outcome in HLA-identical sibling hematopoietic stem-cell transplantation for acute myelogenous leukemia predicted by KIR, HLA genotypes. Blood. 2005;105:4878–4884. doi: 10.1182/blood-2004-12-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.