Abstract
The adult heart responds to biomechanical stress and neurohormonal signaling by hypertrophic growth, accompanied by fibrosis, diminished pump function, and activation of a fetal gene program. Class II histone deacetylases (HDACs) suppress stress-dependent remodeling of the heart via their association with the MEF2 transcription factor, an activator of heart disease. Protein kinase D (PKD) is a stress-responsive kinase that phosphorylates class II HDACs, resulting in their dissociation from MEF2 with consequent activation of MEF2 target genes. To test whether PKD1 is required for pathological cardiac remodeling in vivo, we generated mice with a conditional PKD1-null allele. Mice with cardiac-specific deletion of PKD1 were viable and showed diminished hypertrophy, fibrosis, and fetal gene activation as well as improved cardiac function in response to pressure overload or chronic adrenergic and angiotensin II signaling. We conclude that PKD1 functions as a key transducer of stress stimuli involved in pathological cardiac remodeling in vivo.
Keywords: cardiac hypertrophy, histone deacetylase, stress-responsive kinase
The adult heart undergoes left ventricular hypertrophy and myocardial remodeling when subjected to pathological stresses, such as increased cardiac afterload due to hypertension, aortic valve stenosis, or acute myocardial infarction. Cardiomyocytes respond to such stress stimuli by increasing cell size, reorganizing sarcomeres to enhance contractility and activating a fetal cardiac gene program (1–3). Although these responses may initially normalize wall stress, prolonged hypertrophy increases the risk for chamber dilation, heart failure and sudden death (4, 5). A complex set of signaling pathways and downstream transcription factors underlie these responses of the heart to acute and chronic injury (6).
Class II histone deacetylases (HDACs) function as negative regulators of pathological cardiac remodeling through association with the myocyte enhancer factor-2 (MEF2) transcription factor and other prohypertrophic transcriptional regulators (7–9). Mice lacking either HDAC5 or HDAC9 are sensitized to cardiac stress (7, 9), whereas mice lacking MEF2D display an impaired response to stress signals that normally lead to hypertrophy, fibrosis, and fetal gene activation (10). Manipulation of the HDAC-MEF2 axis may therefore impact the prognosis and outcome of heart disease.
The actions of class II HDACs are controlled by signal-dependent phosphorylation (11–14). When unphosphorylated, class II HDACs localize to the nucleus where they associate with MEF2 and silence MEF2 target genes. Extracellular stimuli transmitted through G protein-coupled receptors activate protein kinase D (PKD) and Ca2+/calmodulin-dependent kinases (CaMKs), which phosphorylate class II HDACs, triggering their nuclear export, relieving MEF2 repression, and promoting pathological cardiac remodeling (13–15).
Cardiac PKD is activated in response to hypertension, pressure overload, and chronic neurohormonal signaling (15–19). Knockdown of PKD1 expression with siRNA blunts agonist-dependent hypertrophy, whereas in vivo cardiac-specific expression of constitutively active PKD1 causes a brief phase of cardiac hypertrophy, followed by chamber dilation and impaired systolic function and death (16).
To further define the functions of PKD1 in the heart, we generated mice with a cardiac-specific mutation of the Prkcm gene, which encodes PKD1. Mice lacking cardiac PKD1 display an impaired response to stress signals that normally lead to cardiac hypertrophy, fibrosis and fetal gene activation. These findings demonstrate that PKD1 activity plays a key role in mediating stress-dependent remodeling and reprogramming of gene expression in the adult heart.
Results
Cardiac-Specific Deletion of PKD1.
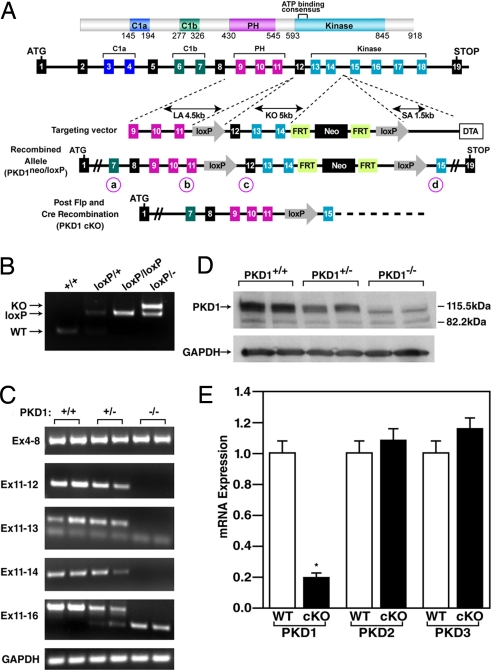
Because PKD1 is expressed in numerous tissues (20), we used the Cre-loxP recombination system to generate a conditional PKD1 (Prkcm) allele that could be deleted specifically in cardiomyocytes. LoxP sites were inserted into the Prkcm locus to flank exons 12 through 14, which encode part of the catalytic domain of PKD1, including the ATP binding motif that is essential for kinase function (Fig. 1A). Expression of Cre recombinase results in deletion of the region between the loxP sites, eliminating the function of PKD1 as a kinase. To determine the outcome of a complete loss of function of PKD1, we deleted the gene using a CAG-Cre transgene, which expresses Cre recombinase in the embryo at the zygote stage (21). The homozygous PKD1 mutant allele caused embryonic lethality with incomplete penetrance, so we generated a cardiomyocyte-specific deletion of PKD1 using α-MHC-Cre transgenic mice (22) that express Cre recombinase specifically in cardiomyocytes.
Fig. 1.
Generation of mice with a conditional PKD1 mutation. (A) Mouse PKD1 locus and targeting strategy. LoxP sites were inserted in the introns flanking exons 12 and 14. Exons 13 and 14 encode the N-terminal region of the kinase domain including the ATP-binding motif. The neomycin resistance cassette (neo) was removed in the mouse germ line by breeding heterozygous mice to hACTB::FLPe transgenic mice, and deletion of exons 12, 13, and 14 was achieved by breeding PKD1loxP/loxP mice to either CAG-Cre or α-MHC-Cre transgenic mice. Positions of PCR primers used for genotyping are labeled a–d and circled. (B) PCR genotyping to distinguish PKD1 alleles. PCR products corresponding to WT (151 bp), PKD1loxP (loxP) (255 bp), and PKD1 KO (359 bp) are shown. The positions of the primers that produce these PCR products are labeled b and c for WT and PKD1loxP and a and d for PKD1 KO and are circled in A. (C) RT-PCR to detect WT and mutant PKD1 transcripts. The PKD1 mutant allele lacks exons 12, 13, and 14. GAPDH was detected as a loading control. Locations of primers used for RT-PCR are shown on the left. (D) Western blot analysis of PKD1 in cardiac extracts from WT and PKD1 mutant mice. GAPDH protein was used as a loading control. (E) Expression of PKD1 transcripts detected by quantitative PCR. Total RNA isolated from ventricles of 8-week-old male mice was used for cDNA synthesis and subsequent quantitative PCR (n = 6 for each genotype). P < 0.01. Error bars indicate ±SEM.
Mice with cardiac-specific deletion of PKD1 (referred to as PKD1 cKO) were indistinguishable from their WT littermates. Deletion of the genomic region between the loxP sites was confirmed by PCR of genomic DNA (Fig. 1B). RT-PCR of RNA from adult heart showed that exon 11 was spliced to exon 15 in the mutant allele, resulting in a frame-shift within the coding region of the PKD1 mRNA (Fig. 1C). Western blot analysis of cardiac extracts of PKD1 cKO mice did not detect PKD1 protein, indicating that the truncated mRNA was unstable or the resulting protein product was rapidly degraded (Fig. 1D). Quantification of PKD1 mRNA by real-time RT-PCR revealed a 5-fold reduction of PKD1 mRNA in PKD1 cKO hearts (Fig. 1E). The observed residual expression of PKD1 mRNA most likely reflects PKD1 expression in fibroblasts, endothelial, smooth muscle, and immune cells within the heart. Quantification of Prkcm2 (encoding PKD2) and Prkcn (encoding PKD3) mRNA expression in PKD1 cKO hearts showed no compensatory up-regulation of these genes (Fig. 1E).
PKD1 cKO Mice Are Resistant to Hypertrophy and Fibrosis in Response to Pressure Overload.
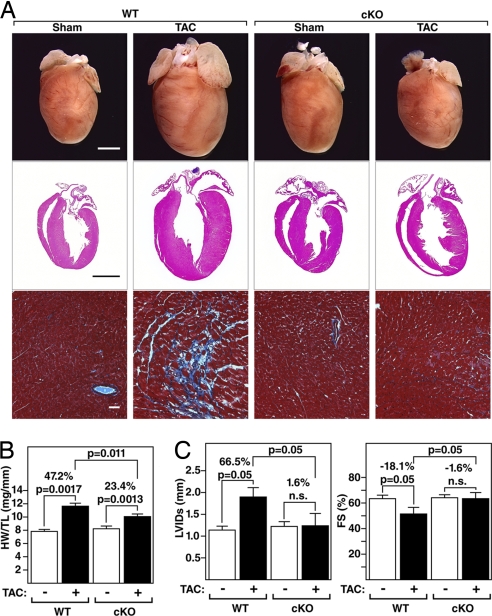
The hearts of WT and PKD1 cKO mice were comparable in size in the absence of stress (Fig. 2 A and B). Twenty-one days after thoracic aortic constriction (TAC), WT mice showed a 47% increase in heart weight/tibia length (HW/TL) with pronounced thickening of the left ventricular free wall and interventricular septum (Fig. 2 A and B). In contrast, PKD1 cKO mice showed only a 23% increase in HW/TL with a minimal increase in wall thickness. Pressure overload hypertrophy in WT mice is accompanied by extensive fibrosis of the ventricular wall, as detected by Masson's trichrome staining (Fig. 2A). There was a dramatic reduction in Masson's trichrome staining of cKO hearts compared with WT littermates (Fig. 2A).
Fig. 2.
Diminished hypertrophy of PKD1 cKO mice after TAC. (A) Hearts from WT and PKD1 mutant mice subjected to either a sham operation (WT and PKD1 cKO, n = 6) or TAC (Top; WT, n = 12; PKD1 cKO, n = 11). Histological sections stained with H&E (Middle) or Masson's trichrome to detect fibrosis (Bottom). (Scale bars: Top and Middle, 2 mm; Bottom, 40 μm.) (B) Heart weight/tibia length (HW/TL) ratios (±SEM) of WT and PKD1 cKO mice were determined 21 days after TAC. (C) PKD1 cKO mice display less left ventricular dilation during systole (LVIDs) and a less pronounced decrease in fractional shortening (FS) in response to TAC than WT mice.
Cardiac Function in PKD1 cKO Mice.
At baseline, no significant differences were seen in the left ventricular end-diastolic diameter (LVIDd), left ventricular end-systolic diameter (LVIDs), heart rate, or fractional shortening (FS) between WT and PKD1 cKO mice, as measured by echocardiography (Fig. 2C and Table 1). Three weeks after TAC, WT mice showed a dramatic increase in LVIDs accompanied by a pronounced reduction in cardiac contractility, as indicated by decreased FS. In contrast, PKD1 cKO animals were remarkably resistant to left ventricular dilation and its concomitant decrease in contractility (Fig. 2C and Table 1). Additionally, WT animals experienced a reduction in heart rate, indicative of cardiac demise, which was not observed in PKD1 cKO mice. These data demonstrate that PKD1 is required for a maximal cardiac remodeling response and functional deterioration of the heart in response to pressure overload.
Table 1.
Echocardiographic analysis of PKD1 mutant mice after TAC
| Animal | LVIDd, mm | LVIDs, mm | FS, % | HR, min−1 | PWd, mm |
|---|---|---|---|---|---|
| WT SHAM | 3.10 ± 0.1 | 1.15 ± 0.11 | 63.2 ± 2.5 | 660 ± 27 | 0.73 ± 0.05 |
| WT TAC | 3.92 ± 0.15 | 1.92 ± 0.29 | 51.7 ± 5 | 635 ± 35 | 1.05 ± 0.1 |
| cKO SHAM | 3.48 ± 0.1 | 1.24 ± 0.09 | 64.1 ± 2.3 | 660 ± 17 | 0.83 ± 0.03 |
| cKO TAC | 3.31 ± 0.28 | 1.26 ± 0.26 | 63.1 ± 4.6 | 720 ± 15 | 0.99 ± 0.05 |
Data are given ± SEM (n = 5–7 per group). LVIDd, left ventricular end-diastolic diameter; LVIDs, left ventricular end-systolic diameter; FS, fractional shortening; HR, heart rate; PWd, posterior wall thickness in diastole.
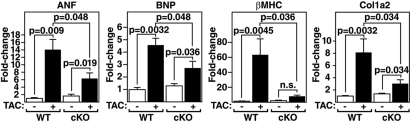
Fetal Gene Activation Is Blunted in PKD1 cKO Hearts in Response to TAC.
PKD1 was also essential for maximal fetal gene activation in response to TAC. Up-regulation of the hypertrophic gene markers, atrial natriuretic factor (ANF), brain natriuretic peptide (BNP) and myosin, heavy polypeptide 7, cardiac muscle, beta (β-MHC) was severely blunted in mutant mice (Fig. 3). Induction of procollagen, type I, alpha 2 (Col1a2), which is up-regulated during cardiac fibrosis, was also compromised in PKD1 cKO mice. Baseline expression of fetal cardiac genes was unaltered in PKD1 cKO mice, suggesting that deletion of PKD1 does not itself impose a stress on the heart.
Fig. 3.
Diminished fetal gene activation in PKD1 cKO mice after TAC. Transcripts for markers of hypertrophy in hearts from WT and PKD cKO mice were detected by quantitative PCR 21 days after TAC (n = 3–9 per group). Values indicate relative expression level to a WT sham-operated group (±SEM). ANF, atrial natriuretic factor; BNP, brain natriuretic peptide; β-MHC, β-myosin heavy chain; Col1a2, procollagen, type I, α2.
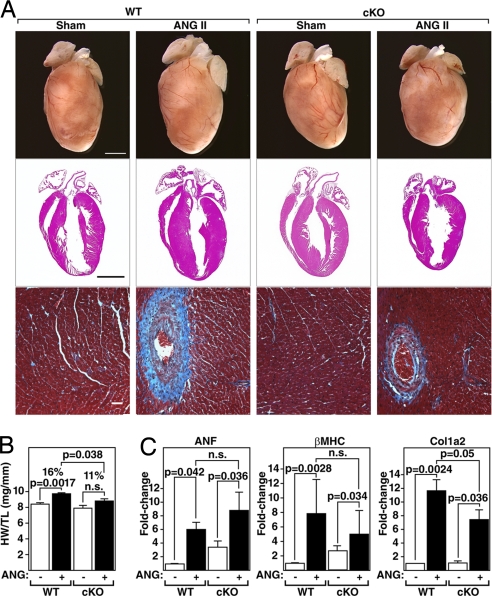
PKD1 cKO Mice Are Resistant to Angiotensin II-Dependent Hypertrophy.
PKD1 is activated by angiotensin II (AngII) in vitro (23). We therefore examined whether PKD1 was necessary for cardiac hypertrophy in response to chronic AngII stimulation by administering AngII to mice over 14 days using osmotic minipumps (3.0 mg/kg/day). Compared with WT mice, which showed a 16% increase in HW/TL, PKD1 cKO mice showed only an 11% increase in HW/TL (Fig. 4 A and B). There was also a reduction in fibrosis as seen by Masson's trichrome staining of cKO hearts compared with WT littermates (Fig. 4A). Up-regulation of ANF, β-MHC, and Col1α2 expression was also compromised in PKD1 cKO mice (Fig. 4C). We conclude that PKD1 acts as a mediator of the hypertrophic effects of AngII on the heart. However, because the dose of AngII used in these experiments has been shown to induce hypertension (24, 25), we cannot distinguish whether the loss of PKD1 in the heart prevents left ventricular hypertrophy by specifically antagonizing AngII signaling in cardiomyocytes or by preventing hypertrophy in response to hypertension.
Fig. 4.
Diminished hypertrophic response of PKD1 cKO mice to AngII infusion. (A) Hearts of WT and PKD1 cKO mice treated with either saline vehicle (WT and PKD1 cKO, n = 6) or AngII (3.0 mg/kg per day) for 14 days (Top; WT, n = 9; PKD1 cKO, n = 8), histological sections stained with H&E (Middle), or Masson's trichrome to detect fibrosis (Bottom). (Scale bars: Top and Middle, 2 mm; Bottom, 40 μm.) (B) Heart weight/tibia length (HW/TL) ratios (±SEM) are shown as bar graphs (n = 7–9). (C) Transcripts for markers of hypertrophy in hearts from WT and PKD cKO mice treated with either saline vehicle or AngII. Values indicate relative expression level to a WT sham-operated group (±SEM). ANF, atrial natriuretic factor; β-MHC, β-myosin heavy chain; Col1α2, procollagen, type I, α2.
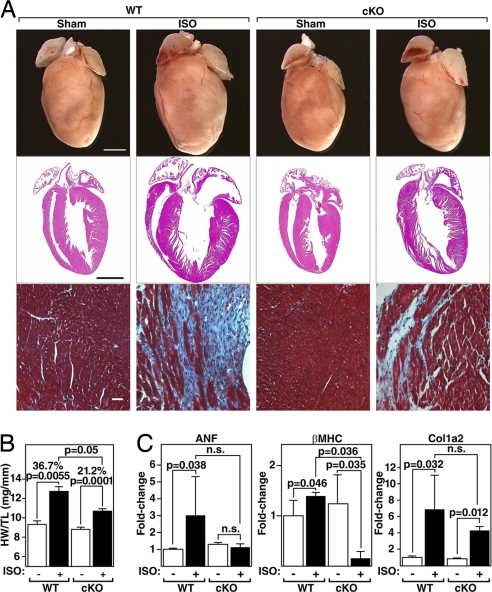
PKD1 cKO Mice Are Resistant to Isoproterenol-Dependent Hypertrophy.
In contrast to Ang II, isoproterenol (ISO), a β-adrenergic agonist, does not activate PKD1 in vitro (16). To examine whether PKD1 is necessary for cardiac hypertrophy in response to β-adrenergic stimulation, ISO was administered to mice over 7 days by using osmotic minipumps (8.7 mg/kg/day). Unexpectedly, we observed that the hearts of PKD1 cKO mice showed only a 21% increase in HW/TL compared with the 37% increase in WT mice (Fig. 5 A and B) and diminished expression of stress-response genes (Fig. 5C). Thus, contrary to in vitro findings, these results suggest that PKD1 is necessary for cardiac hypertrophy in response to chronic β-adrenergic stimulation in vivo. It is possible that chronic long-term administration of ISO indirectly activates other pathways such as the α-adrenergic receptor pathway.
Fig. 5.
Diminished hypertrophic response of PKD1 cKO mice to ISO infusion. (A) Hearts of WT and PKD1 cKO mice chronically infused with either saline vehicle (WT and PKD1 cKO; n = 6) or isoproterenol (8.7 mg/kg per day) for 7 days (Top; n = 11; PKD1 cKO, n = 16), histological sections stained with H&E (Middle) or Masson's trichrome to detect fibrosis (Bottom). (Scale bars: Top and Middle, 2 mm; Bottom, 40 μm.) (B) Heart weight/tibia length (HW/TL) ratios (±SEM) are shown as bar graphs (n = 7–9). (C) Transcripts for markers of hypertrophy in hearts from WT and PKD cKO mice infused with either saline vehicle or isoproterenol. Values indicate relative expression level to a WT sham-operated group (±SEM). ANF, atrial natriuretic factor; β-MHC, β-myosin heavy chain; Col1α2, procollagen, type I, α2.
Discussion
The results of this study demonstrate that PKD1, the major PKD isoform in the heart (19), is a critical component of the signaling pathways through which pressure overload, AngII, and adrenergic signaling drive pathological cardiac remodeling. PKD is a potent kinase for class II HDACs (15, 16), which function as signal-responsive repressors of cardiac hypertrophy, at least in part through their repressive influence on MEF2 (7–9). These findings provide the first evidence that deletion of a class II HDAC kinase in vivo diminishes stress-induced hypertrophy. The blunted hypertrophic response of PKD1 cKO mice is similar to that of mice lacking MEF2 (10), further substantiating the hypertrophic signaling pathway from PKD1 to MEF2 in vivo.
Signal-Dependent Control of PKD.
The canonical pathway for activation of PKD involves PKC-mediated phosphorylation of two serine residues within an activation loop, which relieves PKD from repression by the amino-terminal pleckstrin homology (PH) domain (26). Ca2+-independent, nonconventional PKCs, the dominant regulators of PKD activity, have been linked to cardiomyocyte hypertrophy and death (17, 18, 27, 28).
In cultured cardiomyocytes, PKD1 is activated by a subset of stress stimuli, such as AngII and phenylephrine but not by isoproterenol (16, 23). Surprisingly, however, both AngII- and isoproterenol-induced cardiac hypertrophy was blunted in mice harboring the PKD1 deletion. The apparent requirement of PKD1 for cardiac hypertrophy in response to isoproterenol in vivo might reflect secondary signals leading to PKD1 activation, which do no exist after short-term exposure of cultured cardiomyocytes to isoproterenol.
Potential Redundancy of PKD Isoforms.
PKD1 is one of three PKD family members that share homology in two amino-terminal cysteine-rich domains that mediate binding to diacylglycerol, an internal PH domain and carboxyl-terminal catalytic domains (29). Each PKD isoform is capable of phosphorylating the class II HDACs 4, 5, 7, and 9 on the serines that mediate nuclear export via 14–3-3, suggesting the potential for redundant control of class II HDACs by PKD family members. Indeed, siRNA knockdown of PKD1 expression in cultured cardiomyocytes blunts but does not eliminate HDAC5 nuclear export (15). In addition, disruption of both PKD1 and PKD3 is necessary to block HDAC5 phosphorylation in response to antigen receptor signaling in chicken B lymphocytes (30). The blunted hypertrophic response of PKD1 cKO mice indicates that PKD2 and PKD3 cannot fully compensate for the loss of PKD1. The residual hypertrophy and fetal gene activation in PKD1 cKO animals likely reflects redundant functions of PKD2 and 3 as well as other stress-responsive kinases such as CaMK. In this regard, deletion of calcineurin and its target transcription factor NFAT also blunts, but does not abolish hypertrophy (31, 32) further suggesting redundancy in hypertrophic signaling pathways.
Additional Cardiac Functions of PKD.
PKD has also been implicated in the phosphorylation of cardiac troponin I (33, 34) and numerous other substrates (19). Interestingly, PKD phosphorylates the same sites in troponin I as PKA, resulting in reduced myofilament Ca2+ sensitivity (34), whereas other PKA sites in phospholamban and cardiac myosin-binding protein C are not targeted by PKD. However, for the majority of PKA targets that play a role in excitation–contraction coupling, such as the ryanodine receptor and the L-type calcium channel, it is unknown whether PKD is capable of phosphorylating their PKA sites and thereby regulating their activity.
Circumstantial evidence also suggests a role for PKD in the control of cardiac fibrosis. The profibrotic mineralocorticoid, aldosterone, can activate PKD (35), and PKD signaling was recently shown to stimulate aldosterone production in adrenal cells through up-regulation of aldosterone synthase (36). These findings suggest that PKD contributes to a positive-feedback loop that promotes cardiac fibrosis.
The activation of PKD by diverse stimuli that lead to pathological cardiac remodeling, its activation in failing human hearts, ability to drive hypertrophy and heart failure in transgenic mice (17, 18), and its apparent requirement for a maximal hypertrophic remodeling response in mice, as shown in the present study, point to PKD as a promising therapeutic target for cardiac hypertrophy and heart failure. PKD has also been implicated in myocardial responses to ischemia, angiogenesis, and platelet activation (19), pointing to the potential of PKD inhibitors as therapeutics for diverse cardiovascular disorders.
Materials and Methods
Generation of PKD1 Knockout Mice and Animal Experiments.
Details of mouse breeding schemes and generation of mutant mice are described in supporting information (SI) Materials and Methods.
Surgical Manipulations and Echocardiography.
Methods for TAC and implantation of AngII and Iso pumps are described in SI Materials and Methods.
Histology.
Methods for histology are described in SI Materials and Methods.
RNA analysis.
Methods for RNA analysis are described in SI Materials and Methods.
Statistical Methods.
Values are presented as mean ± SEM. Gene expression was normalized to GAPDH mRNA and calculated as fold change over the respective sham-treated group. Differences in morphologic and biochemical parameters between groups were analyzed by Mann–Whitney U test or two-sided Student's t test. Statistics were calculated with Excel and SPSS software. A P value <0.05 was considered to be statistically significant.
Supplementary Material
ACKNOWLEDGMENTS.
We thank M. Avkiran and T. McKinsey for comments on the manuscript, M. D. Schneider (Baylor College of Medicine, Houston, TX) for the α-MHC-Cre mouse line, and K. Song for helpful discussions. E.N.O. was supported by grants from the National Institutes of Health, the Donald W. Reynolds Center for Clinical Cardiovascular Research, and the Robert A. Welch Foundation. J.F. was supported by a fellowship from the Muscular Dystrophy Association and the Pfizer Fellowship of the German Society of Cardiology.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712265105/DC1.
References
- 1.Chien KR. Stress pathways and heart failure. Cell. 1999;98:555–558. doi: 10.1016/s0092-8674(00)80043-4. [DOI] [PubMed] [Google Scholar]
- 2.Marks AR. A guide for the perplexed: Towards an understanding of the molecular basis of heart failure. Circulation. 2003;107:1456–1459. doi: 10.1161/01.cir.0000059745.95643.83. [DOI] [PubMed] [Google Scholar]
- 3.Olson EN, Schneider MD. Sizing up the heart: Development redux in disease. Genes Dev. 2003;17:1937–1956. doi: 10.1101/gad.1110103. [DOI] [PubMed] [Google Scholar]
- 4.Gardin JM, Lauer MS. Left ventricular hypertrophy: The next treatable, silent killer? J Am Med Assoc. 2004;292:2396–2398. doi: 10.1001/jama.292.19.2396. [DOI] [PubMed] [Google Scholar]
- 5.Vakili BA, Okin PM, Devereux RB. Prognostic implications of left ventricular hypertrophy. Am Heart J. 2001;141:334–341. doi: 10.1067/mhj.2001.113218. [DOI] [PubMed] [Google Scholar]
- 6.McKinsey TA, Olson EN. Toward transcriptional therapies for the failing heart: Chemical screens to modulate genes. J Clin Invest. 2005;115:538–546. doi: 10.1172/JCI24144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang S, et al. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol Cell Biol. 2004;24:8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song K, et al. The transcriptional coactivator camta2 stimulates cardiac growth by opposing class II histone deacetylases. Cell. 2006;125:453–466. doi: 10.1016/j.cell.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 9.Zhang CL, et al. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Y, et al. The mef2d transcription factor mediates stress-dependent cardiac remodeling. J Clin Invest. 2008;118:124–132. doi: 10.1172/JCI33255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Backs J, Olson EN. Control of cardiac growth by histone acetylation/deacetylation. Circ Res. 2006;98:15–24. doi: 10.1161/01.RES.0000197782.21444.8f. [DOI] [PubMed] [Google Scholar]
- 12.Verdin E, Dequiedt F, Kasler HG. Class II histone deacetylases: Versatile regulators. Trends Genet. 2003;19:286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- 13.McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKinsey TA, Zhang CL, Olson EN. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14–3-3 to histone deacetylase 5. Proc Natl Acad Sci USA. 2000;97:14400–14405. doi: 10.1073/pnas.260501497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vega RB, et al. Protein kinases c and d mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol Cell Biol. 2004;24:8374–8385. doi: 10.1128/MCB.24.19.8374-8385.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison BC, et al. Regulation of cardiac stress signaling by protein kinase d1. Mol Cell Biol. 2006;26:3875–3888. doi: 10.1128/MCB.26.10.3875-3888.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haworth RS, Goss MW, Rozengurt E, Avkiran M. Expression and activity of protein kinase d/protein kinase c mu in myocardium: Evidence for alpha1-adrenergic receptor- and protein kinase c-mediated regulation. J Mol Cell Cardiol. 2000;32:1013–1023. doi: 10.1006/jmcc.2000.1143. [DOI] [PubMed] [Google Scholar]
- 18.Haworth RS, Roberts NA, Cuello F, Avkiran M. Regulation of protein kinase d activity in adult myocardium: Novel counter-regulatory roles for protein kinase cepsilon and protein kinase a. J Mol Cell Cardiol. 2007;43:686–695. doi: 10.1016/j.yjmcc.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Avkiran M, Rowland AJ, Cuello F, Haworth RS. Protein kinase d in the cardiovascular system: Emerging roles in health and disease. Circ Res. 2008 doi: 10.1161/CIRCRESAHA.107.168211. in press. [DOI] [PubMed] [Google Scholar]
- 20.Oster H, Abraham D, Leitges M. Expression of the protein kinase d (pkd) family during mouse embryogenesis. Gene Expr Patterns. 2006;6:400–408. doi: 10.1016/j.modgep.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Sakai K, Miyazaki J. A transgenic mouse line that retains cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem Biophys Res Commun. 1997;237:318–324. doi: 10.1006/bbrc.1997.7111. [DOI] [PubMed] [Google Scholar]
- 22.Agah R, et al. Gene recombination in postmitotic cells. Targeted expression of cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Invest. 1997;100:169–179. doi: 10.1172/JCI119509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan M, Xu X, Ohba M, Cui MZ. Angiotensin II-induced protein kinase d activation is regulated by protein kinase cdelta and mediated via the angiotensin II type 1 receptor in vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2004;24:2271–2276. doi: 10.1161/01.ATV.0000148449.92035.3a. [DOI] [PubMed] [Google Scholar]
- 24.Collins AR, et al. Osteopontin modulates angiotensin II-induced fibrosis in the intact murine heart. J Am Coll Cardiol. 2004;43:1698–1705. doi: 10.1016/j.jacc.2003.11.058. [DOI] [PubMed] [Google Scholar]
- 25.Wesseling S, et al. Resistance to oxidative stress by chronic infusion of angiotensin II in mouse kidney is not mediated by the at2 receptor. Am J Physiol. 2005;288:F1191–F1200. doi: 10.1152/ajprenal.00322.2004. [DOI] [PubMed] [Google Scholar]
- 26.Waldron RT, Rozengurt E. Protein kinase c phosphorylates protein kinase d activation loop ser744 and ser748 and releases autoinhibition by the pleckstrin homology domain. J Biol Chem. 2003;278:154–163. doi: 10.1074/jbc.M208075200. [DOI] [PubMed] [Google Scholar]
- 27.Asada A, et al. The calcium-independent protein kinase c participates in an early process of cd3/cd28-mediated induction of thymocyte apoptosis. Immunology. 2000;101:309–315. doi: 10.1046/j.1365-2567.2000.00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asada A, Zhao Y, Kondo S, Iwata M. Induction of thymocyte apoptosis by ca2+-independent protein kinase c (npkc) activation and its regulation by calcineurin activation. J Biol Chem. 1998;273:28392–28398. doi: 10.1074/jbc.273.43.28392. [DOI] [PubMed] [Google Scholar]
- 29.Johannes FJ, Prestle J, Eis S, Oberhagemann P, Pfizenmaier K. Pkcu is a novel, atypical member of the protein kinase c family. J Biol Chem. 1994;269:6140–6148. [PubMed] [Google Scholar]
- 30.Matthews SA, et al. Essential role for protein kinase d family kinases in the regulation of class II histone deacetylases in b lymphocytes. Mol Cell Biol. 2006;26:1569–1577. doi: 10.1128/MCB.26.4.1569-1577.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bueno OF, et al. Impaired cardiac hypertrophic response in calcineurin abeta-deficient mice. Proc Natl Acad Sci USA. 2002;99:4586–4591. doi: 10.1073/pnas.072647999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilkins BJ, et al. Targeted disruption of nfatc3, but not nfatc4, reveals an intrinsic defect in calcineurin-mediated cardiac hypertrophic growth. Mol Cell Biol. 2002;22:7603–7613. doi: 10.1128/MCB.22.21.7603-7613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuello F, et al. Protein kinase d selectively targets cardiac troponin i and regulates myofilament ca2+ sensitivity in ventricular myocytes. Circ Res. 2007;100:864–873. doi: 10.1161/01.RES.0000260809.15393.fa. [DOI] [PubMed] [Google Scholar]
- 34.Haworth RS, et al. Protein kinase d is a novel mediator of cardiac troponin i phosphorylation and regulates myofilament function. Circ Res. 2004;95:1091–1099. doi: 10.1161/01.RES.0000149299.34793.3c. [DOI] [PubMed] [Google Scholar]
- 35.McEneaney V, Harvey BJ, Thomas W. Aldosterone rapidly activates protein kinase d via a mineralocorticoid receptor/egfr trans-activation pathway in the m1 kidney ccd cell line. J Steroid Biochem Mol Biol. 2007;107:180–190. doi: 10.1016/j.jsbmb.2007.03.043. [DOI] [PubMed] [Google Scholar]
- 36.Romero DG, et al. Angiotensin II-mediated protein kinase d activation stimulates aldosterone and cortisol secretion in h295r human adrenocortical cells. Endocrinology. 2006;147:6046–6055. doi: 10.1210/en.2006-0794. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.