Abstract
Nearly 1.7 billion people are infected with Mycobacterium tuberculosis. Its ability to survive intracellularly is thought to be central to its success as a pathogen, but how it does this is poorly understood. Using a Drosophila model of infection, we identify three host cell activities, Rab7, CG8743, and the ESCRT machinery, that modulate the mycobacterial phagosome. In the absence of these factors the cell no longer restricts growth of the non-pathogen Mycobacterium smegmatis. Hence, we identify factors that represent unique vulnerabilities of the host cell, because manipulation of any one of them alone is sufficient to allow a nonpathogenic mycobacterial species to proliferate. Furthermore, we demonstrate that, in mammalian cells, the ESCRT machinery plays a conserved role in restricting bacterial growth.
Keywords: Mycobacterium, phagosome, tuberculosis, ubiquitin
One-third of the human population is infected with M. tuberculosis, and each year it causes 2–3 million deaths. The success of M. tuberculosis as a pathogen is due to its ability to survive within macrophages, which normally eradicate intracellular bacteria, a property that has been attributed to resistance to reactive nitrogens, as well as the capacity to alter IFN-γ signaling and phagosome maturation (see reviews in refs. 1 and 2). Virulent mycobacteria are able to alter phagosome maturation, residing in a compartment that resembles an early endosome. The bacteria inhabit a replicative niche that retains early endosomal markers, such as Rab5, but fails to fully acidify or recruit late endosomal markers, such as Rab7, or mature lysosomal hydrolases. M. tuberculosis, as well as a number of species that are used as models, including Mycobacterium avium and the vaccine strain, Mycobacterium bovis bacillus Calmette–Guérin, share the ability to alter phagosome maturation and grow in a variety of evolutionarily divergent phagocytic cells (3–5), suggesting that mycobacteria target evolutionary conserved molecules to survive within macrophages. In contrast, Mycobacterium smegmatis, a useful laboratory tool because of its rapid growth and genetic tractability, does not grow within macrophages and is unable to cause infection in humans. Although there are a number of molecular differences between the phagosomes containing virulent mycobacteria compared with those containing M. smegmatis (or latex beads or heat-killed mycobacteria), the functional importance of these differences in terms of restricting bacterial growth remains unclear.
Previously we performed a functional genomic screen to identify host factors that influence the uptake and growth of mycobacteria (6). We developed a model of infection using Mycobacterium fortuitum and Drosophila S2 cells, a macrophage-like cell line that is amenable to RNAi. Such an approach, using RNAi in combination with a Drosophila tissue culture model of infection, has proved a valuable strategy for dissecting the host contribution to infection for a number of pathogens (7–12). We used M. fortuitum as a model mycobacterial pathogen because, like M. tuberculosis, it restricts phagosome fusion with lysosomes (13), suggesting common virulence properties with other mycobacteria. In addition, it affords the practical advantage of growing relatively rapidly at 25°C and of robustly infecting S2 cells, eventually destroying the monolayer of fly cells. In contrast, S2 cells restrict growth of M. smegmatis (6). This virulence difference is recapitulated in vivo in Drosophila because M. fortuitum kills flies whereas M. smegmatis does not (data not shown). Finally, M. fortuitum is able to induce the expression of macrophage-activated promoters (maps). maps are bacterial genes that are specifically induced when the bacteria grow intracellularly (14). Thus, by visualizing GFP produced under control of the map24 promoter, it is possible to monitor the bacterial response to the intracellular environment. Previously we identified dsRNAs from a genome-wide collection that diminished bacterial GFP expression (6). Here we further characterize three host cell activities identified in this screen, Rab7, CG8743, and the ESCRT machinery. We demonstrate that, when these factors are knocked down by RNAi, the phagosome environment is altered. These factors represent unique vulnerabilities of the host cell; manipulation of any one of them alone is sufficient to allow a nonpathogenic species to proliferate. Furthermore, we demonstrate that the ESCRT machinery plays a conserved function in modulating bacterial phagosomes in mammalian cells.
Results
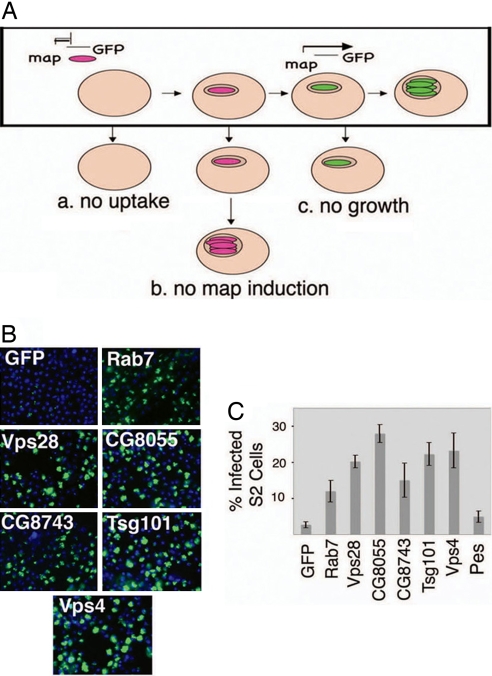
Previously we identified 86 dsRNAs from a genome-wide collection that altered expression from the map24 promoter in M. fortuitum (6). Because GFP expression reflects both the bacterial response to the phagosomal environment and bacterial growth, these dsRNAs could affect the infection by altering bacterial uptake, intracellular bacterial growth, or induction from the map24 promoter (see Fig. 1A). For example, the map24 promoter is responsive to low pH (15), and the screen identified three components of the vacuolar ATPase (V-ATPase), consistent with the hypothesis that depletion of the V-ATPase results in an increase in the phagosomal pH and, hence, diminished map24 induction. We anticipated that there might be additional factors that result in altered map24 promoter activity without affecting intracellular growth (possibilities b versus c in Fig. 1A). It is difficult to measure the intracellular growth of M. fortuitum, because we cannot easily distinguish between intracellular and extracellular bacteria during infection of S2 cells; therefore, we used M. smegmatis to try to discriminate the two possibilities. We reasoned that if the dsRNA treatment resulted in less intracellular bacterial growth of M. fortuitum (Fig. 1A, c), then it should have little effect on infection with M. smegmatis, because these fail to grow in S2 cells. In contrast, if the dsRNA altered the phagosome environment resulting in diminished map induction (Fig. 1A, b), then it is possible that the phagosome might be more permissive for bacterial growth, in which case M. smegmatis might grow.
Fig. 1.
Rab7, CG8743, and ESCRT factors are required to restrict mycobacterial growth in Drosophila S2 cells. (A) Schematic of the M. fortuitum map24::GFP screen. S2 cells were treated with dsRNA to deplete specific host factors, followed by infection with M. fortuitum map24::GFP. The boxed diagram shows infection in control cells. Bacteria are ingested, and intracellular cues, such as low pH and other unknown signals, induce the expression of a macrophage-activated promoter. Bacterial growth occurs over 2 d. Thus, diminished GFP signal can occur from a failure of bacterial uptake (a), map promoter induction (b), or bacterial growth (c). (B) S2 cells were treated with control dsRNA (GFP) or dsRNAs targeting Rab7, CG8743, or ESCRT factors Vps28, CG8055, Tsg101, and Vps4 followed by infection with M. smegmatis hsp60::GFP for 2 d. S2 cell nuclei are stained with Hoechst (shown in blue), and GFP is indicated in green. Images were acquired on the Autoscope (×20). Similar results were seen if the bacteria express GFP under control of the msp12 promoter (data not shown). The percentage of heavily infected cells was quantified (C). Data represent the average from six images from three replicates ± SD.
We examined 86 dsRNAs from the original screen for their effect on infection with the nonpathogenic species M. smegmatis. Cells were treated with dsRNAs to deplete the targeted host factor, followed by infection with an M. smegmatis strain that constitutively expresses GFP. Two days after infection cells were examined by microscopy. Four dsRNAs, targeting Rab7, dVps28, CG8055, and CG8743, strongly increased the percentage of heavily infected S2 cells (Fig. 1 B and C). To confirm that the bacterial growth seen by microscopy was occurring intracellularly, we blocked bacterial uptake using dsRNA targeting Pes, a scavenger receptor required for mycobacteria entry in S2 cells (6). Blocking bacterial uptake did not confer increased bacterial growth, demonstrating that extracellular growth does not occur under the conditions of the assay, so growth must be occurring intracellularly (Fig. 1C). Thus, we conclude that Rab7, dVps28, CG8055, and CG8743 play a role in restricting the intracellular growth of M. smegmatis in S2 cells.
M. bovis bacillus Calmette–Guérin blocks phagosome maturation before the recruitment of Rab7, a small GTP-binding protein and marker of late endosomes (16). Because Rab proteins are involved in vesicle maturation (17), one might predict that depletion of Rab7 would block phagosome maturation before lysosome delivery. Indeed, our results confirm that acquisition of Rab7 is required to restrict mycobacterial growth. In contrast, the role of dVps28, CG8055, and CG8743 in bacterial infection is less clear. CG8743 is homologous to a family of predicted ion channels in mammals, encoded by MCOLN1, MCOLN2, and MCOLN3, whereas both dVps28 and CG8055 are members of the ESCRT machinery, which is composed of three protein complexes, ESCRT I, ESCRT II, and ESCRT III (18). dVps28 is part of the ESCRT I complex in Drosophila (19), whereas CG8055 is a predicted homologue of the ESCRT III component SNF7/CHMP4B. The ESCRT machinery is important for receptor trafficking but does not have a well described role in trafficking large cargo such as bacteria. Rather, the ESCRT machinery acts on the membrane of the endosome, functioning in the delivery of ubiquitinated receptors into intraluminal vesicles of multivesicular bodies that are ultimately delivered to the lysosome for degradation. In the absence of ESCRT function, an enlarged endosomal compartment develops, which in yeast has been termed a class E compartment. Because ubiquitinated receptors are not efficiently degraded in the absence of ESCRT function, ubiquitin accumulates on the surface of this aberrant endosomal compartment in Drosophila as in yeast (20–22).
We directly tested the effect of dsRNAs targeting additional ESCRT components (dTsg101, Vps25, CG10711/Vps36, CG9779/Vps24, and CG4071/Vps20) and CG8642/Vps4, the Drosophila homolog of the AAA-type ATPase important in recycling of the ESCRT machinery (23), for their effects on infection. All of the dsRNAs that robustly disrupted ESCRT function, as evidenced by their effects on ubiquitin trafficking (those targeting dTsg101, Vps28, CG8055, and Vps4) [supporting information (SI) Fig. 6], resulted in increased growth of M. smegmatis (Fig. 1 B and C). They also all altered GFP expression in M. fortuitum when expressed from map promoters (SI Fig. 7). Additionally, CG14542, a predicted ESCRT III component, was a weak hit in our initial screen (6), and inhibition of it produced subtle changes in both ubiquitin trafficking and effects on mycobacterial infection (data not shown). For the other dsRNAs tested we were unable to demonstrate disruption of ESCRT function (data not shown), perhaps because of inefficient protein depletion or functional redundancy. However, for all of the dsRNAs that robustly disrupted ESCRT function as evidenced by their effects on ubiquitin trafficking (those targeting dTsg101, Vps28, CG8055, and Vps4), we found that they also altered the phagosome environment, resulting in decreased map24 and map49 induction by M. fortuitum and increased growth of M. smegmatis. These results demonstrate that ESCRT activity modulates the mycobacterial phagosome in S2 cells.
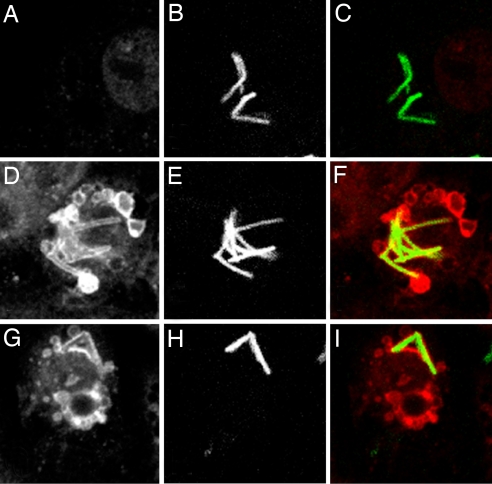
To determine whether the ESCRT machinery acts directly on the mycobacterial phagosome, we examined the localization of bacteria in ESCRT-depleted cells. In Drosophila a lack of ESCRT activity causes the accumulation of enlarged endosomes that are characterized by the accumulation of ubiquitin (20, 22). Therefore, cells that had been depleted of dTsg101 or Vps28 were infected with M. smegmatis, followed by immunofluorescence against ubiquitinated proteins to label this compartment. As early as 3 h postinfection (p.i.) bacteria are found within the heavily ubiquitinated vesicular compartment, demonstrating that the bacteria reside within the compartment on which the ESCRT machinery acts (Fig. 2). In cells that had not been depleted for ESCRT factors there was little vesicular ubiquitin staining and only rare colocalization of bacteria and ubiquitin. In ESCRT-depleted cells some bacteria persist within this compartment, as they continued to colocalize with ubiquitin for up to 24 h p.i. (data not shown).
Fig. 2.
M. smegmatis colocalizes with ubiquitin in ESCRT-depleted cells. S2 cells were treated with control dsRNA (GFP; A–C) or dsRNA targeting Tsg101 (D–F) or Vps28 (G–I) followed by infection with M. smegmatis hsp60::GFP. Three hours p.i., cells were processed for immunofluorescence to detect ubiquitin. Ubiquitin staining is shown in A, D, and G; GFP is shown in B, E, and H; and composite images are shown in C, F, and I. Images were acquired on the Leica confocal microscope.
These results suggest that the ESCRT machinery directly acts on the bacterial phagosome. An alternative possibility is that the bacteria escape the phagosomal system into the cytosol. For example, Salmonella that escape into the cytosol accumulate ubiquitinated proteins on their surface (24). To rule out this possibility, we performed electron microscopy on Tsg101-depleted cells. In both untreated and Tsg101-depleted cells, >90% of the bacteria were confined within a vacuole at these time points (SI Fig. 8). Therefore, the majority of the bacteria in Tsg101-depleted cells reside in a vacuolar location, but, unlike control cells, the compartment is heavily ubiquitinated.
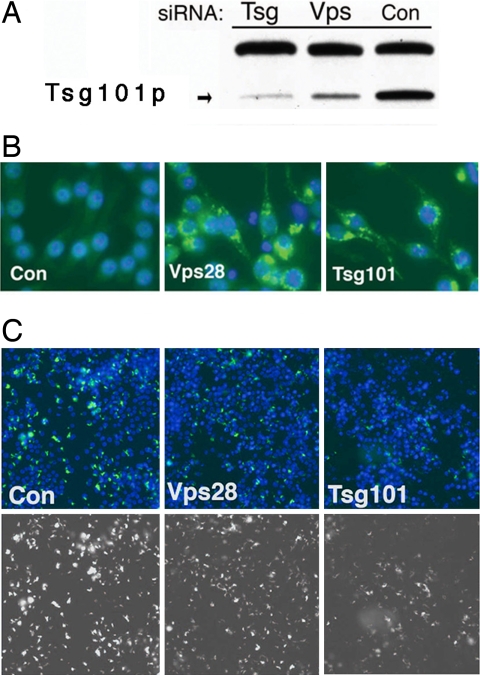
To test whether the ESCRT machinery affects mycobacterial infection in mammalian macrophages, we depleted the ESCRT I components Tsg101 and Vps28 from murine macrophage RAW264.7 cells using siRNA. We found that siRNA treatment against Tsg101 (by Western blot) and Vps28 (by its effect on Tsg101 abundance) (25) (Fig. 3A) lead to efficient depletion. Furthermore, depletion of Vps28 or Tsg101 disrupted ESCRT function as demonstrated by the accumulation of ubiquitinated vesicles (Fig. 3B). As we saw in S2 cells, in Vps28- and Tsg101-depleted cells, there was less GFP detected after the cells were infected with M. fortuitum map24::GFP compared with control cells (Fig. 3C). To determine whether the phagosome in ESCRT-depleted cells was also less restrictive for mycobacterial growth, RAW264.7 cells were infected with M. smegmatis hsp60::GFP. Bacterial growth was assessed by microscopy and by plating for CFU. In control cells, 24 h p.i., there was little bacterial growth as assessed by the GFP signal. In contrast, there was substantial growth in Tsg101- and Vps28-depleted cells (Fig. 4A). When analyzed by CFU there was no difference in bacterial uptake in ESCRT-depleted cells compared with controls, but bacterial growth was enhanced (Fig. 4B). To rule out an off-target effect of one of the siRNAs contained within the pools, we individually tested the four siRNAs comprising the Tsg101 pool. All of the siRNAs had equivalent effects on Tsg101 protein levels and behaved similarly in their ability to promote growth of M. smegmatis (SI Fig. 9). We conclude that the ESCRT machinery is required to restrict the growth of M. smegmatis in mammalian macrophages, suggesting an evolutionarily conserved function of the ESCRT machinery in mycobacterial phagosome maturation.
Fig. 3.
Depletion of Tsg101 or Vps28 alters mycobacterial growth in mammalian macrophages. RAW264.7 cells were transfected with control siRNA or siRNAs targeting Tsg101 or Vps28. (A) Western blot analysis shows depletion of Tsg101 protein. (B) Ubiquitinated vesicles, shown in green, accumulate in cells depleted for Tsg101 or Vps28. Autoscope images are ×40. Two days after RNAi treatment, cells were infected with M. fortuitum map24::GFP (C). (Upper) Composite images of RAW cell nuclei (stained with Hoechst and shown in blue) and GFP (shown in green). (Lower) The GFP channel alone. Images were acquired on the Autoscope (×20).
Fig. 4.
ESCRT factors are required to restrict the growth of M. smegmatis in RAW cells. RAW cells were treated with control siRNA or siRNA to deplete Tsg101, Vps28, EEA1, or Hrs. Cells were infected with M. smegmatis and analyzed 1 d later by microscopy (A) or plating for CFU (B). Cells were stained with Hoechst (in blue) and TRITC-phalloidin (red), and GFP is in green in composite panels (A Left). A Right shows GFP alone. Autoscope images are ×40. (B) CFU immediately after infection (blue) and 22 h later (red). Values are normalized to control wells at the respective time points. Data are the average of two experiments performed in triplicate ± SD. Cells were treated with cytochalasin (indicated by +) or solvent control (−) before bacterial uptake. At 22 h p.i., only cells treated with cytochalasin or siRNA targeting Tsg101 or Vps28 showed a statistically significant difference from control cells (P = 1.1 × 10−4, 6.9 × 10−7, and 1.2 × 10−4, respectively). (C) siRNA targeting EEA1 depletes EEA1 protein. (D) When 10 nM siRNA targeting Hrs was used, there was considerable residual protein. siRNA at 25 nM leads to more efficient depletion, which could not be enhanced at 100 nM. EEA1 and Hrs proteins are indicated by arrows in C and D, respectively. Nonspecific background bands serve as loading controls. (E) Hrs siRNA (25 nM) treatment caused punctate accumulation of ubiquitinated proteins.
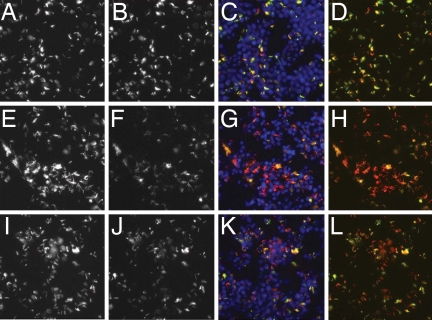
We suggest that the ESCRT machinery plays an evolutionarily conserved role in modulating the mycobacterial phagosome, creating an environment where map promoters are efficiently induced and growth of M. smegmatis is restricted. To validate that the diminished GFP signal seen in cells infected with M. fortuitum map24::GFP was in fact due to diminished expression, as opposed to decreased bacterial growth, we used an M. fortuitum strain that expresses both red fluorescent protein (dsRed2) and GFP under control of two distinct promoters. These bacteria constitutively express dsRed2 under control of the msp12 promoter, whereas GFP is expressed under control of the map49 promoter (26). map49 is induced during intracellular growth, but, unlike map24, the stimulus does not appear to be low pH (15). When ESCRT I-depleted cells were infected with the dual reporter bacteria, we found diminished GFP expression. This difference was detectable as early as 3 h p.i. At that point, bacteria infecting control cells begin to have detectable GFP expression, which was diminished in bacteria infecting ESCRT-depleted cells (data not shown). By 24 h p.i., bacteria in control cells had strongly induced GFP expression, but there was less intense fluorescence in Tsg101- and Vps28-treated cells. Examination of red fluorescence, however, demonstrated that bacterial uptake was similar to control-treated cells, and bacterial growth appeared similar or slightly increased (Fig. 5). Similar results were seen when Drosophila cells were infected with the dual reporter strain (SI Fig. 10). These results suggest that there is a difference in the phagosomal environment in cells that lack ESCRT function, such that induction from the map24 and map49 promoters is diminished and bacterial growth is not effectively restricted.
Fig. 5.
Depletion of Tsg101 or Vps28 results in diminished induction from the map49 promoter in M. fortuitum. RAW264.7 cells were transfected with control siRNA (A–D) or siRNAs targeting Tsg101 (E–H) or Vps28 (I–L) and infected with M. fortuitum msp12::dsRed2 map49::GFP (25) for 1 d. dsRed2 is shown in A, E, and I; GFP is shown in B, F, and J. Composite images showing RAW cell nuclei (stained with Hoechst and shown in blue), dsRed2, and GFP are in C, G, and K; D, H, and L are composites of both dsRed2 and GFP channels. Images are ×20 from the Autoscope.
Virulent mycobacteria are able to alter phagosome maturation and reside in a compartment that resembles an early endosome. This may be achieved by interfering with the accumulation of phosphatidylinositol 3-phosphate (PI3P) on the maturing phagosome, perhaps by altering the localization and activity of VPS34, a PI3 kinase, as well as by secretion of SapM, a PI3P phosphatase (27–31). Diminished PI3P is thought to interfere with the recruitment of downstream effectors, thereby preventing phagosome maturation. Because two PI3P binding proteins, EEA1 and Hrs, are diminished on mycobacterial phagosomes, it has been postulated that their absence may account in part for the arrest in phagosome maturation and survival of M. tuberculosis intracellularly (30, 32). Hrs is required for sorting receptors to MVBs and for recruiting ESCRT I by directly interacting with Tsg101 (25). However, although we achieved sufficient knock-down of Hrs to alter ubiquitin trafficking (Fig. 4 D and E), we found no difference compared with control cells in terms of their ability to restrict the growth of M. smegmatis (Fig. 4 A and B). Similarly, although we were able to deplete EEA1, we saw no effect on bacterial growth (Fig. 4 A–C). Although Hrs and ESCRT components are thought to act sequentially in the same process, their loss results in distinct phenotypes in mammalian cells in vitro and in Drosophila in vivo (20, 21, 33), and our results demonstrate distinct effects on the growth of intracellular bacteria, although we cannot exclude the possibility that the differences are attributable to the level of protein knock-down.
Discussion
We previously conducted a genome-wide RNAi screen in Drosophila to identify host factors that alter infection with the model mycobacterial pathogen M. fortuitum. This screen identified >80 host factors that altered infection. Interestingly, a small subset of the factors identified in the original screen (Rab7, CG8743, and ESCRT factors) also play a role in restricting growth of a nonpathogenic mycobacteria.
The mechanism by which the ESCRT machinery restricts the growth of bacteria remains to be defined. Although the phagosomal environment is clearly altered in ESCRT-depleted cells (as evidenced by altered map induction, bacterial growth, and ubiquitin colocalization), the functionally important difference is not clear, although it is probably not simply a higher pH. First, ESCRT depletion similarly affected activity of the pH-sensitive map24 and the insensitive map49 promoters. Second, LysoTracker staining did not reveal an obvious difference in the acidified compartment in ESCRT-depleted cells compared with controls. Finally, RNAi treatment targeting the V-ATPase in S2 cells did not similarly allow growth of M. smegmatis as ESCRT depletion (data not shown).
ESCRT factors could play several roles. The ESCRT machinery might be required for delivery of the bacteria to the lysosome, and, in its absence, the bacteria reside in an earlier, more permissive compartment. Alternatively, the ESCRT machinery could play an indirect role, for example, by affecting cellular signaling. Finally, in cells that lack ESCRT activity bacteria might traffic to the lysosome, but lysosomal constituents might not be properly localized so that the environment is more permissive. One interesting possibility is that the bactericidal arsenal of the phagosomal system is diminished because of altered ubiquitin trafficking. Ubiquitin has been shown to be a bactericidal component of the lysosome (34), and, in the absence of ESCRT function, there may be less ubiquitin entering MVBs to be liberated into the lysosome. Furthermore, with ubiquitin trapped around the aberrant endosomal compartment in ESCRT-depleted cells, free ubiquitin pools may be diminished, which could affect any number of cellular processes that influence bacterial infection. A role for ubiquitin in mycobacterial infection is also suggested by the fact that a human susceptibility locus for leprosy maps to an E3 ubiquitin-protein ligase, parkin (35).
While the ESCRT machinery restricts the growth of M. smegmatis, genome-wide screens in Drosophila found that CG8055, Vps28, and Vps4 were required for intracellular growth of Listeria monocytogenes (8, 9), suggesting that diminished ESCRT activity does not make cells generally susceptible to bacterial infection. However, when these cells were infected with a mutant strain of L. monocytogenes that normally gets trapped in the vacuole because of a defect in the cytolysin, which is required for escape from the phagosome (LLO), the bacteria were better able to escape. This shows that diminished ESCRT activity affects other vacuolar bacteria, and it raises the possibility that our results are also due to escape of the bacteria into the cytosol. Arguing against this possibility, phagosomal escape of LLO-minus Listeria requires the bacterial phospholipases. Thus, loss of phagosome integrity in ESCRT-depleted cells still depends on bacterial factors (9). Furthermore, we examined by electron microscopy the intracellular location of M. smegmatis in S2 cells that had been depleted of Tsg101 (SI Fig. 8). These results are consistent with bacterial growth occurring within a vacuolar compartment, although we cannot exclude some degree of membrane disruption.
It has been previously shown that infection with virulent mycobacteria involves a number of host factors whose functions might relate to ESCRT. For example, both Hrs, a protein involved in MVB formation, and EEA1, a factor involved in phagosome maturation, appear to be important for infection with M. bovis bacillus Calmette–Guérin. Although we do not see a phenotype associated with knockdown of these proteins, this by no means suggests that they have no role. Differences in cell types and degree of inhibition might well account for discrepancies. Alternatively, the conditions induced by knockdown of these proteins might have different effects on the growth of pathogenic and nonpathogenic bacteria.
We found that an alteration in a single host activity is sufficient to allow a nonpathogenic species to grow intracellularly. This identifies a point of host cell vulnerability that could be manipulated by bacterial pathogens to grow, raising the possibility that virulent mycobacteria or other vacuolar pathogens target the ESCRT machinery for their growth. Although no bacteria are known to target ESCRT machinery, numerous enveloped viruses do impinge upon ESCRT function (36). For example, HIV-1 Gag binds Tsg101, thereby hijacking ESCRT activity for viral budding, a process topologically identical to MVB formation. Because expression of Gag can disrupt ESCRT activity (37), it is tempting to speculate that in coinfected cells the effect of HIV on ESCRT may contribute to the enhanced susceptibility that HIV-infected individuals exhibit to M. tuberculosis and opportunistic species such as M. avium. Thus, the intracellular lifestyle of both enveloped viruses and vacuolar pathogens may converge on a common core of cellular machinery.
Materials and Methods
Tissue Culture.
S2 cells (Schneider's line S2 cells/SL2) were grown at 25°C in Schneider's medium (SM; GIBCO) with 10% heat-inactivated (h.i.) FBS (JRH Biosciences). Description of the SL2 cell line is at www.flyrnai.org. RAW264.7 cells were grown in DMEM (GIBCO) with 20 mM Hepes, 2 mM l-glutamine (DMEM complete), and 10% h.i. FBS at 37°C in a humidified 5% CO2 atmosphere. Penicillin/streptomycin (GIBCO) was added for passaging but omitted during infections.
Bacterial Strains and Growth Conditions.
M. smegmatis strain mc2-155 and M. fortuitum strain EJR154 were used (6). Bacteria were grown at 37°C to log phase in Middlebrook 7H9 broth, 0.5% Tween, BBL Middlebrook ADC Enrichment, and 0.2% glycerol. Kanamycin (50 μg/ml) was added when selecting plasmids in EJR154, whereas 5 μg/ml kanamycin was used to select plasmids in mc2-155. Plasmids used were hsp60::GFP (38), map24::GFP, map49::GFP, and pDL4912 (msp12::dsRed2, map49::GFP) (14, 15, 26).
dsRNA and siRNA Reagents.
Tsg101, Vps28, Hrs, and EEA1 siGENOME SMARTpools were used. siCONTROL NonTargeting siRNA pool and siCONTROL NonTargeting siRNA no. 1 were controls. siGLO RISC-Free siRNA was used to visually confirm transfection (Dharmacon). The sequences of the four siRNAs comprising the Tsg101 SMARTpool are in SI Table 1. dsRNA was generated as described (39), and details are in SI Table 2.
S2 Cell dsRNA Treatments.
RNAi treatment was performed in either 384- or 96-well optically clear plates (Costar). For 384-well plates, 5 μl of dsRNA (50 ng/μl) was aliquoted into each well. A total of 2 × 104 cells in 10 μl of SM (serum-free, antibiotic-free) were incubated with the dsRNA for 50 min after which 20 μl of SM with serum was added. In 96-well plates, 8 × 104 cells were incubated with 20 μl of dsRNA (50 ng/μl) for 50 min before the addition of 80 μl of serum-containing SM. Cells were infected or analyzed by immunofluorescence 3 d later.
RAW Cell siRNA Treatments.
For protein analysis, 5 × 105 RAW cells were grown overnight in six-well dishes in DMEM complete with 10% h.i. FBS followed by transfection using HiPerFect (Qiagen). Before addition of transfection mix, cells were washed once in DMEM complete with 10% h.i. FBS, and 500 μl of DMEM complete with 10% h.i. FBS was added to each well. A total of 500 μl of transfection mix (DMEM without serum, 20 nM siRNA, and 30 μl of HiPerFect) was added to each well for siCON, Tsg101, and Vps28 siRNAs. In the case of Hrs, maximal knock-down required 50 nM siRNA. To assess transfection efficiency, siGLO was used at 200 nM. Five hours later, 3 ml of DMEM complete was added. Cells were analyzed by immunofluorescence or Western analysis 2 d after transfection. For infection experiments, 4 × 104 cells per well were plated in 96-well plates (Costar). The following day they were transfected as described above, except that volumes were 1/20th of those used for six-well dishes. Cells were infected 2 d after transfection.
Protein Analysis.
Samples were separated by SDS/PAGE and immunoblotted with antibodies recognizing the following proteins: Tsg101 (4A10, 1:500 dilution; Abcam), EEA1 (ab2900, 1:500 dilution; Abcam), Hrs (ref. 40; 1:200 dilution), or monoubiquitinated and polyubiquitinated conjugates (FK2 antibody, 1:1,000 dilution; Biomol). Horseradish peroxidase-conjugated secondary antibodies were used at 1:10,000 dilution (Jackson Immunochemicals).
Immunofluorescence.
SL2 cells were transferred from 96-well dishes to Con A (Sigma–Aldrich)-coated microscope slides and allowed to adhere for 1 h. RAW cells were plated directly in microscope slides. Cells were fixed in 4% formaldehyde/PBS, washed with PBS plus 0.1% Triton (PBST), blocked in 2% BSA (Sigma)/PBST for 1 h, and incubated overnight at 4°C with monoclonal antibody FK2 (diluted 1:1,000; Biomol). Samples were washed and incubated with anti-mouse Alexa Fluor 488 or anti-mouse Alexa Fluor 594 antibodies (1:200 dilution; Molecular Probes) in 2% BSA/PBST for 1 h. They were stained with 5 μg/ml Hoechst 33342, washed with PBST, and mounted in Antifade (Molecular Probes). Images were acquired on a Leica TCS SP2 AOBS confocal microscope at ×63 or the AutoScope (6).
RAW264.7 Cell Infections.
Bacteria were washed twice in PBS and resuspended in DMEM complete with 10% h.i. FBS. They were centrifuged, and supernatants were harvested as described previously (6). Cells were infected with 4 × 105 bacteria. For experiments with cytochalasin, cells were incubated in 13 μM cytochalasin B or solvent (ethanol) for 5–10 min before adding bacteria. Three hours p.i. the medium was replaced with DMEM 1% h.i. FBS with 10 μg/ml amikacin for 4 h (to kill extracellular bacteria) followed by extensive washing. To determine CFU, either immediately after antibiotic treatment or 22 h later, cells were lysed in 0.2% Triton X-100. Samples were sonicated, and serial dilutions were plated on LB. Colonies were counted after 4 d at 37°C. For microscopy, cells were fixed in 4% formaldehyde/PBS, washed with PBST, and stained with 5 μg/ml Hoechst 33342 and/or TRITC-phalloidin (50 μg/ml; Sigma). Images were acquired on the Autoscope.
S2 Cell Infections.
In 384-well plates, S2 cells were infected with 6.25 × 104 M. smegmatis hsp60::GFP. Before infections, bacteria were washed twice in PBS and resuspended in SM with 10% h.i. FBS. To obtain a single-cell suspension, bacteria were centrifuged and supernatants were harvested as described previously (6). To inhibit growth of extracellular bacteria, 2 h p.i. amikacin was added (2.5 μg/ml). For M. fortuitum, cells were infected as described previously (6). Two days later, cells were fixed with 4% formaldehyde in PBS, washed with PBST, and stained with 5 μg/ml Hoechst 33342. Images (×20) were acquired by using the AutoScope. To quantitate the percentage of infection, the number of bacterial clusters and S2 nuclei were determined by using MetaMorph (Universal Imaging). Automated quantitation closely paralleled manual counting and visual scoring.
Supplementary Material
ACKNOWLEDGMENTS.
We thank C. Cosma and L. Ramakrishnan (University of Washington, Seattle) for map plasmids, M. Komada (Tokyo Institute of Technology, Tokyo) for Hrs antibody, and members of the Harvard University Electron Microscope Facility (Boston) for assistance. We are indebted to N.P. and E.J.R. laboratory members, in particular S. Cherry, M. Markstein, and R. Binari. This work was supported by National Institutes of Health Grants AI061609 and AI57351-01. N.P. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707206105/DC1.
References
- 1.Vergne I, Chua J, Singh SB, Deretic V. Cell biology of Mycobacterium tuberculosis phagosome. Annu Rev Cell Dev Biol. 2004;20:367–394. doi: 10.1146/annurev.cellbio.20.010403.114015. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen L, Pieters J. The Trojan horse: Survival tactics of pathogenic mycobacteria in macrophages. Trends Cell Biol. 2005;15:269–276. doi: 10.1016/j.tcb.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 3.El-Etr SH, Yan L, Cirillo JD. Fish monocytes as a model for mycobacterial host-pathogen interactions. Infect Immun. 2001;69:7310–7317. doi: 10.1128/IAI.69.12.7310-7317.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dionne MS, Ghori N, Schneider DS. Drosophila melanogaster is a genetically tractable model host for Mycobacterium marinum. Infect Immun. 2003;71:3540–3550. doi: 10.1128/IAI.71.6.3540-3550.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cirillo JD, Falkow S, Tompkins LS, Bermudez LE. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect Immun. 1997;65:3759–3765. doi: 10.1128/iai.65.9.3759-3767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Philips JA, Rubin EJ, Perrimon N. Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science. 2005;309:1251–1253. doi: 10.1126/science.1116006. [DOI] [PubMed] [Google Scholar]
- 7.Ramet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416:644–648. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- 8.Agaisse H, et al. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science. 2005;309:1248–1251. doi: 10.1126/science.1116008. [DOI] [PubMed] [Google Scholar]
- 9.Cheng LW, et al. Use of RNA interference in Drosophila S2 cells to identify host pathways controlling compartmentalization of an intracellular pathogen. Proc Natl Acad Sci USA. 2005;102:13646–13651. doi: 10.1073/pnas.0506461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elwell C, Engel JN. Drosophila melanogaster S2 cells: A model system to study Chlamydia interaction with host cells. Cell Microbiol. 2005;7:725–739. doi: 10.1111/j.1462-5822.2005.00508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorer MS, Kirton D, Bader JS, Isberg R. RNA interference analysis of Legionella in Drosophila cells: Exploitation of early secretor apparatus dynamics. PLoS Pathog. 2006;2:e34. doi: 10.1371/journal.ppat.0020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stroschein-Stevenson SL, Foley E, O'Farrell PH, Johnson AD. Identification of Drosophila gene products required for phagocytosis of Candida albicans. PLoS Biol. 2006;4:e4. doi: 10.1371/journal.pbio.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Da Silva TR, et al. Virulent Mycobacterium fortuitum restricts NO production by a gamma interferon-activated J774 cell line and phagosome-lysosome fusion. Infect Immun. 2002;70:5628–5634. doi: 10.1128/IAI.70.10.5628-5634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramakrishnan L, Federspiel NA, Falkow S. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science. 2000;288:1436–1439. doi: 10.1126/science.288.5470.1436. [DOI] [PubMed] [Google Scholar]
- 15.Chan K, et al. Complex pattern of Mycobacterium marinum gene expression during long-term granulomatous infection. Proc Natl Acad Sci USA. 2002;99:3920–3925. doi: 10.1073/pnas.002024599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Via LE, et al. Arrest of mycobacterial phagosome maturation is caused by a block in vesicle fusion between stages controlled by rab5 and rab7. J Biol Chem. 1997;272:13326–13331. doi: 10.1074/jbc.272.20.13326. [DOI] [PubMed] [Google Scholar]
- 17.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 18.Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- 19.Sevrioukov EA, Moghrabi N, Kuhn M, Kramer HA. Mutation in dVps28 reveals a link between a subunit of the endosomal sorting complex required for transport-I complex and the actin cytoskeleton in Drosophila. Mol Biol Cell. 2005;16:2301–2312. doi: 10.1091/mbc.E04-11-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson BJ, et al. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev Cell. 2005;9:711–720. doi: 10.1016/j.devcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 21.Vaccari T, Bilder D. The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating Notch trafficking. Dev Cell. 2005;9:687–698. doi: 10.1016/j.devcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 22.Moberg KH, Schelble S, Burdick SK, Hariharan IK. Mutations in erupted, the Drosophila ortholog of mammalian tumor susceptibility gene 101, elicit non-cell-autonomous overgrowth. Dev Cell. 2005;9:699–710. doi: 10.1016/j.devcel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 23.Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4 AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perrin AJ, Jiang X, Birmingham CL, Brummell JH. Recognition of bacteria in the cytosol of mammalian cells by the ubiquitin system. Curr Biol. 2004;14:806–811. doi: 10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 25.Bache KG, et al. The growth-regulatory protein HCRP1/hVps37A is a subunit of mammalian ESCRT-I and mediates receptor down-regulation. Mol Biol Cell. 2004;15:4337–4346. doi: 10.1091/mbc.E04-03-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cosma CL, Humbert O, Ramakrishnan L. Superinfecting mycobacteria home to established tuberculous granulomas. Nat Immunol. 2004;5:828–835. doi: 10.1038/ni1091. [DOI] [PubMed] [Google Scholar]
- 27.Chau J, Deretic V. Mycobacterium tuberculosis reprograms waves of phosphatidylinositol 3-phosphate on phagosomal organelles. J Biol Chem. 2004;279:36982–36992. doi: 10.1074/jbc.M405082200. [DOI] [PubMed] [Google Scholar]
- 28.Vergne I, et al. Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2005;102:4033–4038. doi: 10.1073/pnas.0409716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purdy GE, Owens RM, Bennett L, Russell DG, Butcher BA. Kinetics of phosphatidylinositol-3-phosphate acquisition differ between IgG bead-containing phagosomes and Mycobacterium tuberculosis-containing phagosomes. Cell Microbiol. 2005;7:1627–1634. doi: 10.1111/j.1462-5822.2005.00580.x. [DOI] [PubMed] [Google Scholar]
- 30.Fratti RA, Backer JM, Gruenberg J, Corvera S, Deretic V. Role of phosphatidylinositol 3-kinase and Rab5 effectors in phagosomal biogenesis and mycobacterial phagosome maturation arrest. J Cell Biol. 2001;154:631–644. doi: 10.1083/jcb.200106049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vergne I, Chua J, Deretic V. Tuberculosis toxin blocking phagosome maturation inhibits a novel Ca2+/calmodulin-PI3K hVPS34 cascade. J Exp Med. 2003;198:653–659. doi: 10.1084/jem.20030527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vieira OV, et al. Acquisition of Hrs, an essential component of phagosomal maturation, is impaired by Mycobacteria. Mol Cell Biol. 2004;24:4593–4604. doi: 10.1128/MCB.24.10.4593-4604.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Razi M, Futter CE. Distinct roles for Tsg101 and Hrs in multivesicular body formation and inward vesiculation. Mol Biol Cell. 2006;17:3469–3483. doi: 10.1091/mbc.E05-11-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci USA. 2007;104:6031–6036. doi: 10.1073/pnas.0700036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mira MT, et al. Susceptibility to leprosy is associated with PARK2 and PACRG. Nature. 2004;427:636–640. doi: 10.1038/nature02326. [DOI] [PubMed] [Google Scholar]
- 36.Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 37.Valiathan RR, Resh MD. Expression of human immunodeficiency virus type 1 Gag modulates ligand-induced downregulation of EGF receptor. J Virol. 2004;78:12386–12394. doi: 10.1128/JVI.78.22.12386-12394.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teitelbaum R, et al. Mycobacterial infection of macrophages results in membrane-permeable phagosomes. Proc Natl Acad Sci USA. 1999;96:15190–15195. doi: 10.1073/pnas.96.26.15190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boutros M, et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- 40.Komada M, Kitamura N. Growth factor-induced tyrosine phosphorylation of Hrs, a novel 115-kilodalton protein with a structurally conserved putative zinc finger domain. Mol Cell Biol. 1995;15:6213–6221. doi: 10.1128/mcb.15.11.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.