Abstract
Aging, defined as a decrease in reproduction rate with age, is a fundamental characteristic of all living organisms down to bacteria. Yet we know little about the causal molecular mechanisms of aging within the in vivo context of a wild-type organism. One of the prominent markers of aging is protein aggregation, associated with cellular degeneracy in many age-related diseases, although its in vivo dynamics and effect are poorly understood. We followed the appearance and inheritance of spontaneous protein aggregation within lineages of Escherichia coli grown under nonstressed conditions using time-lapse microscopy and a fluorescently tagged chaperone (IbpA) involved in aggregate processing. The fluorescent marker is shown to faithfully identify in vivo the localization of aggregated proteins, revealing their accumulation upon cell division in cells with older poles. This accretion is associated with >30% of the loss of reproductive ability (aging) in these cells relative to the new-pole progeny, devoid of parental inclusion bodies, that exhibit rejuvenation. This suggests an asymmetric strategy whereby dividing cells segregate damage at the expense of aging individuals, resulting in the perpetuation of the population.
Keywords: ibpA, inclusion bodies, protein aggregation, small heat-shock protein
Aging is a fundamental characteristic of all living organisms. Recent work in our laboratory has identified and quantified aging in Escherichia coli (1), where cells progressively decline in growth rate and reproductive ability with increasing cell pole age, establishing this organism as a simple experimental model of aging (2). In this outwardly symmetrically dividing bacterium, the cell inheriting the old pole after division grows more slowly and divides less frequently, therefore exhibiting aging (1). Thus, the dividing cell partitions its resources and/or damaged components in a biased fashion, leading to differential growth potential distinguishing the old-pole aging cell and its young-pole counterpart (3, 4).
To shed light on the molecular mechanism underlying aging in E. coli, we focus here on the partitioning of damaged, aggregated proteins in wild-type bacterial cells growing in a nonstressing favorable environment. Aggregated proteins are linked to cellular degeneracy in many age-related diseases [e.g., Huntington's disease, Alzheimer's disease, spongiform encephalopathies, Parkinson's disease, and cataracts (5, 6)]. In addition, numerous reports link protein maintenance and repair functions (e.g., folding and disaggregation-related chaperones and proteases) to aging (7, 8). Consequently, considerable effort has been invested in the study of protein aggregation, resulting in a better understanding of aggregation in vitro (9) and in the identification of a number of genes involved in this process, many of which are widely conserved in all kingdoms of life (10, 11). In contrast, less is known about the aggregation process in vivo, its causes, and its direct consequences on cell fate; such understanding has been hindered by the inability to follow in vivo the formation and outcome of inclusion bodies under native conditions.
The bacterium E. coli has served as an important model for the study of protein aggregation in vivo. These studies focused on specific conditions where proteins readily form insoluble inclusion bodies (e.g., protein overproduction, mutants) (12–14). However, native E. coli proteins can also form inclusion bodies, a process promoted by stress conditions (e.g., oxidative or thermal stress) and certain mutations, as well as by natural transcription and translation errors, resulting in detectable aggregation even under conditions of no external stress (9). Similar inclusion bodies have been found in eukaryotic cells (15). In yeast, in vitro immunostaining of carbonylated proteins (16), correlated with aggregated proteins (17), revealed their retention in mother cells (16) although their in vivo dynamics and influence on aging could not be measured. Recently, the polarized asymmetric inheritance of aggrosomes in Drosophila melanogaster neuronal precursor cells as well as in epithelial crypts of patients suffering from the polyglutamine aggregation-associated ataxia type 3 disease was reported (18). Because the low number of inclusion bodies per bacterial cell (13) may lead to their asymmetric partitioning and because of their potential cellular toxicity, we investigated the hypothesis that asymmetric segregation of damaged proteins and their pole-biased accumulation may explain, at least in part, the observed pattern of E. coli aging.
Results
Reporting Inclusion Bodies in Vivo in Wild-Type Bacteria.
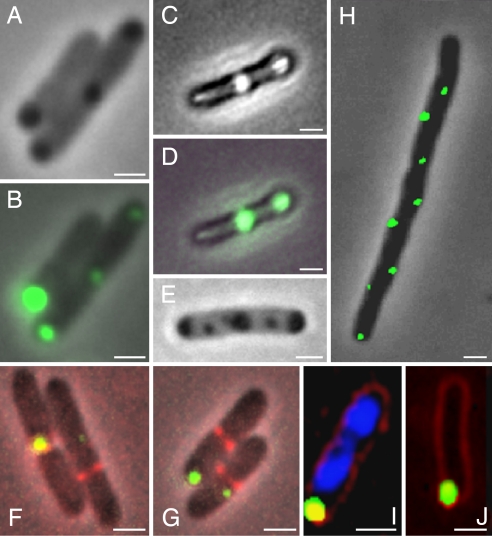
To reveal in vivo the presence and localization of protein aggregates, we followed at the single-cell level the expression and localization of the E. coli inclusion bodies binding small heat-shock protein (sHSP) IbpA (inclusion body protein A) (19). To this end, the endogenous ibpA was replaced by a chromosomal gene fusion to the yellow fluorescent protein (YFP) in the MG1655 sequenced wild-type E. coli strain. IbpA was previously shown to be present in the insoluble cellular fraction of heat-stressed cells (20). To check whether IbpA-YFP can serve as detector for protein aggregates, we exposed the strain expressing the fluorescent gene construct (MGAY) to various aggregating conditions. Indeed, this resulted in strongly fluorescent foci colocalizing with the inclusion bodies, as visualized by fluorescence and phase contrast microscopy [Fig. 1 A–D and supporting information (SI) Fig. 6]. The induced inclusion bodies (one to five per cell) were found to be located at cellular poles, in mid- or quarter-cell positions, independent of the presence of IbpA (Fig. 1 E–G). Under native nonstressed conditions, no inclusion body could be visualized by phase contrast; however, through the increased sensitivity of the fluorescent reporter (SI Fig. 6), small fluorescent foci (mostly one per cell; see below) could be identified. The IbpA-YFP fluorescent foci exhibit the same ordered cellular localization pattern as the above induced inclusion bodies, suggesting a spatial correlation with potential cellular division sites (Fig. 1 F and G). To determine whether this localization is indicative of future septation sites and therefore may be linked to the cell division machinery, we examined aggregate localization in two cell division mutants. We found that these mutations [ftsZ84 at the restrictive temperature (42°C; data not shown) and minCD] did not affect the aggregates' relative positioning in the cell (Fig. 1H). In these mutants, a greater number of foci per cell was observed, corresponding to the increased length of the cells due to filamentation, and these aggregates exhibited a regular distribution along the cells' long axis. Time-lapse microscopy revealed that the foci do indeed colocalize with future septation sites (SI Movie 1). In both the wild-type MGAY (Fig. 1I) and the minCD mutant the inclusion bodies were located within the nucleoid-void cytoplasm. We thus further checked whether nucleoid positioning affects the inclusion bodies' localization. The fluorescent inclusion bodies did not move from their initial position upon treatment of dam mutant strain with 2-aminopurine, which results in chromosome degradation (Fig. 1J). Thus, whereas nucleoid position may determine the initial inclusion body localization, other mechanisms may be responsible for its maintenance (see Discussion).
Fig. 1.
IbpA-YFP in vivo localization. IbpA-YFP overlaps with inclusion bodies and is independent of division machinery or nucleoid occlusion. (A) Representative phase contrast image of MGAY cells after treatment with streptomycin. (B) Overlay of A with the fluorescence image (IbpA-YFP depicted in green). (C) Representative phase contrast image of MGAY cell in the presence of an insoluble protein [antibody single-chain Fv fragment plasmidic expression (38)]. (D) Overlay of C with the fluorescence image. Similar inclusion body and IbpA-YFP colocalization was observed upon heat shock (data not shown). (E) Representative phase contrast image of MG1655(ibp::cat) cell after treatment with streptomycin. (F and G) IbpA-YFP foci are localized in cellular poles or near formed septums, visualized by expression of the ftsZ-CFP fusion (depicted in red) in the MGAY strain. (H) IbpA-YFP foci in the MGAY(minCD) strain lacking the ability to localize the division septum to mid-cell. (I) IbpA-YFP foci in the MGAY strain (blue, DAPI DNA stain; red, FM4-64 membrane stain). (J) IbpA-YFP foci in 2-aminopurine-treated dam− strain where the chromosomes were degraded [blue, DAPI stain (none detected); red, FM4-64). Images were taken using 5% (B, D, and F–H) and 100% (I and J) of available excitation light. (Scale bars: 1 μm.)
Inclusion Bodies Are Formed in Discrete Cellular Positions and Accumulate in Old Poles Under Nonstressing Conditions.
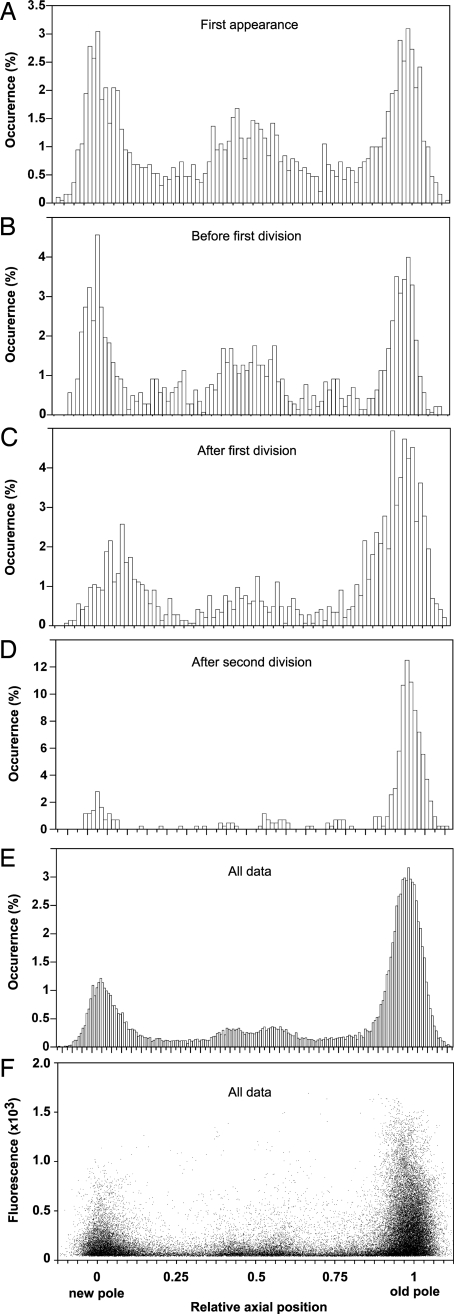
To determine the presence and localization dynamics of damaged proteins marked by the IbpA-YFP fusion in the wild-type MGAY strain under native (37°C; rich media) nonstress, constant growth conditions, time-lapse films of 12 microcolonies (up to 400 cells each) arising from single cells were recorded and analyzed (SI Fig. 7), yielding the relevant physical properties of the cells and the fluorescent foci. Upon the first appearance of foci within a lineage, they are mostly detected at equal frequencies around three discrete positions within the cell: the mid-cell plane (28%), the new pole (30%), and the old pole (31%) (Fig. 2A). The rest of the foci concentrate around the one-quarter and three-quarter cell positions as cells grow and reach their predivision length (Fig. 2B). Because the foci rarely move (<10% incidence) from their initial relative location, their position in the mother cell determines which offspring inherits the inclusion body and where it will be located within it (Fig. 3 and SI Fig. 8). This process results in the systematic accumulation of aggregates (Fig. 2 C–E) with higher fluorescence content (Fig. 2F) in the old poles of the cells (SI Movies 2 and 3) and in a progressively greater aggregate load in older cells (Fig. 4B). The same segregation pattern was observed with large inclusion bodies visualized by phase-contrast microscopy, induced by stress (SI Fig. 6) or by expression of nonsoluble protein (Fig. 1C), indicating a common mechanism irrespective of whether the inclusion body is made of essentially a single protein or an assortment of aggregated polypeptides.
Fig. 2.
Aggregate distribution and associated fluorescence levels along the cell axis. Shown is IbpA-YFP foci localization along the cells' normalized longitude internal coordinate, oriented from the new pole (0) to the old pole (1) of each cell. Binned histograms show foci localization (A–D, 100 bins; E, 200 bins) at first appearance (A), at the last movie frame before first division (B), at the first movie frame after first division (C), at the first movie frame after two consecutive divisions (D), and cumulative over all movies' frames (E). (F) Foci maximal fluorescence intensity (arbitrary gray-level units) as a function of their localization.
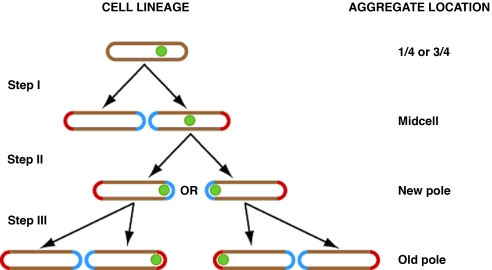
Fig. 3.
Individual aggregates are located to the old pole through cycles of cell divisions. Because movement of foci is rarely observed, the location of an aggregate within the cell after division is determined by its location in the mother cell. Those that were at the one- or three-quarter positions are found concentrated around the mid-cell point after division (Step I). Aggregates at the mid-cell are subsequently located in the new pole, with equal probability to be in either of the two cells (Step II). Those that are found in the new-pole end of the cell immediately before division remain in the same pole; however, that pole, having been formed in the previous division event, is now an old pole in the offspring cell (Step III). Note that aggregates can be initially detected at polar, mid-cell, or quarter-cell positions but are eventually located to an old pole. Once there, they are consistently inherited by the old-pole cell after division. Aggregates are indicated by green dots. Red cell ends are old poles, and blue cell ends are new poles.
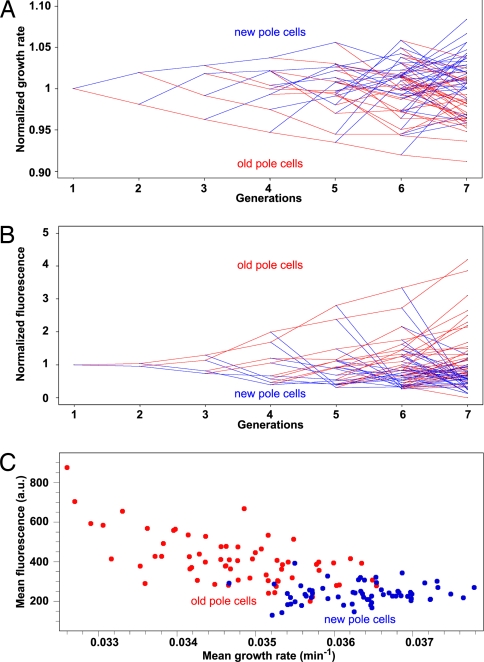
Fig. 4.
Growth rate and aggregation accumulation in bacterial lineages. Bacterial lineage representation of average growth rates (A) and fluorescence intensity (B) of new-pole cells (blue) and old-pole cells (red) of the MGAY strain. Summits represent the measured value, and lines follow the lineage propagation. Values of the ordinates are normalized by the generation mean value and averaged over all 12 films. (A) A clear pattern of gradual loss of growth rate of old-pole cells and fast recovery of new-pole cells is discerned. In 90.5% of offspring cell pairs the old-pole cell grows more slowly than the new-pole cell and its mother cell (79.4%) (aging), whereas in 74.6% the new-pole cell grows faster than its mother cell (rejuvenation). (B) Net fluorescence intensity of foci at first time point after division. (C) The above patterns result in a negative correlation between the cells' growth rate and aggregate load (measured as foci fluorescence) whereby the two subpopulations [old-pole cells (red) and new-pole cells (blue)] can be discerned.
Inclusion Body Segregation and Damage Purification.
The majority of MGAY cells followed under the above nonstressed conditions have zero or one inclusion body per cell: 52.3% have no inclusion body, 46.5% of the cells contain only one inclusion body, and only 1.2% carry two inclusion bodies immediately after cell division. No diffuse fluorescence can be seen (Fig. 1), suggesting that soluble IbpA-YFP is too scarce to be detected. In addition, its incorporation time scale may be faster than the maturation of the YFP fluorophore [t1/2 = 2 min (21)]. The recovery of foci detected 2–3 min after photobleaching of the entire cell (data not shown) is in agreement with the latter hypothesis. Importantly, given the observed inclusion body segregation pattern (i.e., eventual accumulation at old pole) (Figs. 2 and 3), late detection would yield a bias where the equal distribution among new pole, middle cell, and old pole positions (Fig. 2A) will be distorted such that the mid-cell population would be relatively decreased and the old pole position would be relatively enriched.
Following the cellular lineages and the history of each of the inclusion bodies enabled the investigation of the laws governing their formation and segregation. The overall probability of a fluorescent IbpA-YFP focus appearing between divisions of a cell devoid of foci at birth is 0.71. The probability of acquiring a second focus during this period is very low (0.037), probably because of a high propensity toward coaggregation of misfolded proteins. The probability of cells developing a focus decreased if there was a focus present in the mother cell that was subsequently passed on to the other offspring cell, as compared with the chance of acquiring one when the mother cell was devoid of aggregates [0.69 (n = 862) vs. 0.85 (n = 202), respectively; Wilcoxon rank test, P < 10−4]. Assuming that the inclusion body represents the majority of aggregated proteins in the cell, this bias suggests a purification effect, such that when an inclusion body is inherited by one offspring it decreases the chance of its appearance in the other. Hence, inclusion bodies appear to act as an intracellular sink for abnormal proteins.
Inclusion Bodies Are Associated with an Age-Related Decrease in Cellular Growth Rate.
We next compared the growth rate pattern within the lineages of the parental wild-type MG1655 strain and the isogenic reporter strain (MGAY) (grown under identical conditions), measured by extracting individual cellular exponential growth rates from microcolony growth followed by time-lapse microscopy. The two strains have similar growth rate (GR) means and distributions [GRMG1655 (n = 5,860 cells) = 0.0353 ± 0.0001 min−1; GRMGAY (n = 2,626) = 0.0356 ± 0.0001 min−1 (± standard errors)] and exhibit the same aging pattern, measured as the mean difference in growth rate between old-pole/new-pole cell pairs (1): [Δ(GRold − GRnew)]mean/GRmean = −3.95 ± 0.5% [t test (≠ 0), P < 10−4] and −3.90 ± 0.5% [± standard errors, t test (≠ 0), P < 10−4] for MG1655 and MGAY, respectively [t test (MG1655 ≠ MGAY), P = 0.67)]. Thus, the presence of the fluorescently tagged IbpA and the exposure to the excitation light did not affect the growth and aging pattern of the cells. In both strains, in the overall majority of the division events in the average lineage of the examined microcolonies, the young pole cell grows faster and the old-pole cell grows more slowly than their mother cell (Fig. 4A). Concurrently, the old-pole cell accumulates increasing levels of the fluorescent marker, whereas the new-pole cell is cleared of the aggregate reporter (Fig. 4B). Because IbpA-YFP foci segregate to the more slowly growing, older pole cells, there is a negative correlation between cellular growth rate and fluorescence intensity (Fig. 4C) (>0.9999 confidence; Spearman test).
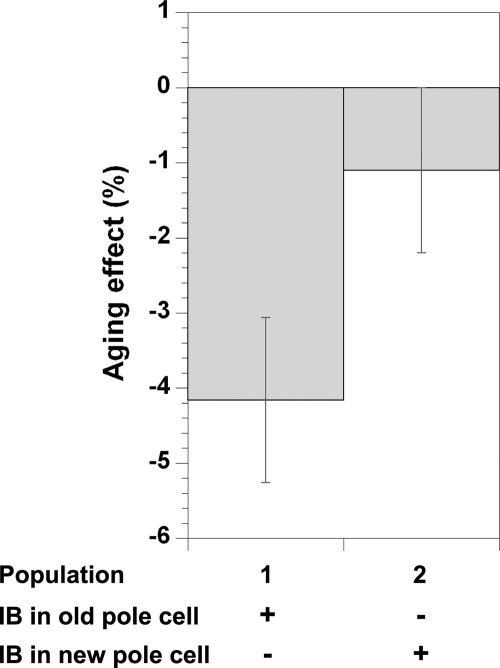
To determine whether the presence of the inclusion bodies is actually associated with some of the fitness loss in these cells, as opposed to passively marking the old pole, we chose an age-homogeneous subgroup consisting of all newborn cells (those that had inherited the new pole from their parent and had yet to divide themselves) and followed them through their first division. The pairs of cells produced from this division were divided into two discrete populations: in the first population, the focus present in the mother cell is retained in the old-pole cell after division (population 1), whereas in the second population, the focus is inherited by the new-pole cell (population 2). Newborn mother cells containing two foci (1% of the cases) were sorted according to retention of the brighter focus in their offspring. Aging, as reflected from the negative mean of the differences in growth rate between the old-pole and new-pole offspring cells is manifested in population 1 but not in population 2 (where the difference is smaller and not statistically significant), indicating that the presence of inclusion bodies negatively correlates with the cell growth rate (Fig. 5 and Table 1). However, retention of the inclusion body in the new-pole cell (population 2) does not completely reverse the growth rate difference between the offspring cells, suggesting that aging cannot be explained solely by presence of inclusion bodies. By comparing the average growth rates of the cells in the two populations, the aging effect associated with the presence of the aggregates (Agg) can be determined and separated from other aggregate-independent old-pole-associated effects (Pole). Subtracting the average growth rate of old-pole cells that inherit an inclusion body from that of new-pole cells that do not (both classes seen in population 1) yields the total aging effect on growth rate (Agg + Pole = 1.5 × 10−3 min−1). Comparing this with the difference in growth rate between same pole cells (of the two populations) that inherit an inclusion body and those that do not (Agg = 5 × 10−4 and 6 × 10−4 min−1 for old-pole and new-pole cells, respectively) suggests that the fraction of the growth rate decrease (aging) associated with the presence of the aggregate, Agg/(Agg + Pole) is ≈30–40% (see Table 1). Thus, independent of the old-pole effect, the mere presence of aggregates, as reported by IbpA-YFP, is significantly correlated with diminished growth rates. Moreover, in population 1, where the inclusion body is inherited by the old pole, the new-pole offspring grows significantly more rapidly than the mother cell (rejuvenation), and the old-pole cell grows more slowly than before division (aging), as expected from the internal asymmetric division of damaged cellular components. In population 2, where the inclusion body is inherited by the new-pole cell, the old-pole offspring still grows more slowly than before division (albeit to a lesser extent), whereas the new-pole cell no longer exhibits an increased growth rate relative to its mother cell. Thus, when inheriting the inclusion body, the new-pole cell no longer experiences rejuvenation (Table 1). This strongly supports the interpretation that the aggregate itself is the cause of the reduced growth rate seen in these cells.
Fig. 5.
Aging correlation with presence of protein aggregation. The aging effect calculated from the relative growth rate (GR) difference between old-pole and new-pole offspring [e.g., (GRold − GRnew)/GRoffspring] of newborn mother cells where inclusion body is inherited by the old-pole cell (population 1) or the new-pole cell (population 2). See Table 1 for actual values. IB, inclusion bodies.
Table 1.
Quantifying the contribution of protein aggregation to E. coli aging
| Offspring with inclusion body | Mean growth rate (×10−2 min−1) |
|
|---|---|---|
| Population 1 (332 cell pairs) | Population 2 (257 cell pairs) | |
| Overall offspring (GRoffspring) | 3.61 ± 0.01 | 3.61 ± 0.01 |
| Old-pole offspring (GRold) | 3.54 ± 0.02 | 3.59 ± 0.02 |
| New-pole offspring (GRnew) | 3.69 ± 0.02 | 3.63 ± 0.02 |
| Mother cell (GRmother) | 3.62 ± 0.02 | 3.66 ± 0.02 |
Exponential growth rates (GR) of the old-pole (population 1) and new-pole (population 2) offspring cells from all new-pole mother cells containing inclusion bodies, subdivided into two populations as described. Statistical significance values (t test) for the GR differences across the obtained data are as follows: GRold ≠ GRnew, P < 10−4 for population 1 and n.s. (not significant; P > 0.1) for population 2; GRmother ≠ GRnew, P = 2.3 × 10−3 for population 1 and n.s. for population 2; GRmother ≠ GRold, P = 3.9 × 10−4 for population 1 and 1.7 × 10−3 for population 2. Difference in aging between the two populations: (GRold − GRnew)population1 ≠ (GRold − GRnew)population2, P = 3.4 × 10−4. Effect of inclusion body presence on new-pole offspring: (GRnew)population1 ≠ (GRnew)population2, P = 0.019. Effect of inclusion body presence on old-pole offspring: (GRold)population1 ≠ (GRold)population2, P = 0.035.
Discussion
Reconstructing the exponential growth phenotype of a bacterial population from its individuals revealed a steady state between aging and rejuvenation that is maintained in part by segregation of damage (e.g., protein aggregates). This may indicate that evolution has selected an asymmetric distribution of damage, prevailing over unbiased dilution of damage. Recent in silico models suggest that under most conditions such asymmetry may be selected (4, 22, 23). Interestingly, in the asymmetrically dividing bacteria Caulobacter crescentus, where aging was previously observed (24), the abundance of heat-shock proteins involved in disaggregation (e.g., Lon and DnaK) is biased toward the aging stalked cell (25). Previous hypotheses for the deleterious effects of aggregates in disease are generally based on either a direct toxic effect of aggregates (or their intermediates) or an indirect loss of function via depletion of freely diffused key proteins trapped with the aggregate (10). The loss-of-function model predicts that the draining of essential diffusible components should be shared by both siblings and would thus result in slower rejuvenation with mother cell age. This is inconsistent with the observed pattern where the growth rate difference between sibling pairs tends to increase with mother cell age (figures 2A and 3 in ref. 1), suggesting that rejuvenation is linked to loss of dysfunction as opposed to regaining of function. Our results thus imply that the apparent toxicity of the aggregates (as revealed by a decreased growth rate in their presence) may be due to a gain of dysfunction, most likely caused by the presence of the aggregate.
The observed aggregates' segregation pattern may be the result of a passive mechanism whereby nucleoid occlusion determines the initial positioning of the stochastically appearing inclusion bodies (Figs. 1 and 2) followed by stabilization of their position. This in turn may be a result of limited diffusion of the large molecular assembly in a crowded cytoplasmic environment (26) or perhaps of anchorage. An alternative mechanism may rely on the existence of an upstream symmetry break that directs the inclusion body segregation via active machinery targeting nascent aggregates to polar and mid-cell positions. Although there exists no evidence for an active mechanism in E. coli, it has been previously proposed for other organisms. Examples include carbonylated protein sorting in yeast, mediated by actin filaments in a sir2p-dependent manner (27), the Shigella outer membrane protein IcsA (28), and other bacterial surface structure localization (29) as well as the microtubule-dependent aggrosome localization in mammalian cells (15). Intriguing is the similarity between aggrosome localization to the eukaryotic centrosome (18) and the inclusion bodies to the bacterial division plane. In the former model, putative stem cells exhibited a lower aggregation level as compared with differentiated cells. It remains to be seen whether, as in bacteria, germ-line cells use the same mechanism to ensure their optimal survival. Is the unknown segregation mechanism common across kingdoms? Is the bacterial mechanism a precursor (possibly “passive,” e.g., non-energy consuming?) mechanism or rather evolutionarily converged to the same optimal solution? These are some of the challenges that follow these works. Previous works on asymmetrically dividing unicellular organisms suggest similar strategies of damage retention (16, 30, 31), although their direct effect on cellular fitness or fate could not be assessed or quantified. Nondividing cells cannot segregate damage and may therefore need to invest in other mechanisms to deal with damage accumulation (e.g., in more effective disaggregation machinery). Indeed, RpoH expression-dependent chaperone genes (including ibpA) were shown to be induced in stationary-phase bacteria (32). This suggests that, although cells can potentially invest more in maintenance and repair during growth phase, this may not be cost-effective, prevailing in cycles of aging and rejuvenation by damage segregation. Interestingly, in terminally differentiated postmitotic cells such as neurons, aggregate accumulation (33) correlates with diseases (10). Because all organisms face challenges of protein folding and aggregation, it is unsurprising that the genes coding for the machinery for protein folding quality control are highly conserved across the domains of life. Given the newly apparent universality, E. coli may provide a powerful system for studying the formation and prevention of toxic aggregates, such as those responsible for a number of degenerative diseases.
Materials and Methods
Strains.
The sequenced wild-type strain of E. coli, MG1655 (34), was modified to express an improved version of the YFP fused to the C terminus of IbpA under the control of the endogenous chromosomal ibpA promoter resulting in the strain MGAY. The ftsZ84(TS) {from DRC14 [MC4100 ftsZ84(TS) leu::Tn10]}, ftsZ-ecfp under a LacI-controlled promoter {JOE521 [JOE309 D(λattL-lom)::bla lacIq P208-ftsZ-ecfp]} (kindly provided by J. Beckwith (Harvard Medical School, Boston) (35)], minB::kan [minCD locus mutant kindly provided by R. d'Ari (Institut Jacques Monod, Paris) (36)], and dam [constructed by direct gene knockout in the MG1655 strain (37)] loci were introduced separately into the MGAY strain by P1vir transductions. For overexpression of insoluble protein, the pET-28-based D2.3–4 single-chain Fv (scFv) antibody fragment expression plasmid (38) was introduced into the MGAY strain. Bugbuster kit (Novagen) protocol was used for inclusion body purification. The resulting insoluble fraction was spread on a glass slide and was visualized by using a fluorescence microscope. Additional cloning information is provided in SI Methods.
Growth Conditions.
Cells were inoculated from exponentially growing cultures onto a solid matrix of LB-agarose (LB from DIFCO, Becton Dickinson; agarose from Qbiogene) in microscope cavity slides as previously described (1) at 37°C. Induction of aggregates in MG1655, MG1655(ibp::cat), and MGAY strains was achieved by addition of streptomycin (10 μg/ml; Sigma) to exponentially growing cultures 30 min before inoculation on the slide. IPTG (50 μM) was added to exponentially growing culture of the MGAY(ftsZ-ecfp) strain 60 min before inoculation on the slide. 2-Aminopurine (200 μg/ml; Sigma) was added to the MGAY(dam) strain during exponential phase 2 h before inoculation on the slide (39). For live cell DNA and membrane staining, DAPI (0.3 μg/ml; Sigma) and FM4-64 (2 μg/ml; Molecular Probes, Invitrogen) were added to the culture 20 min before inoculation as well as to the LB-agarose support.
Microscopy.
The previously described experimental protocol (1) was followed with minor changes: phase contrast as well as fluorescent images (6-second exposure) were taken every 90 seconds.
Image Analysis.
The custom analysis program [Bacterial Home Vision, BHV (1)] was used with the following introduced changes: phase contrast images, treated with the “flatten background” filter of Metamorph software (Roper Scientific), were used to automatically segment cells; where necessary (5–10% of cells), manual correction of the segmentation was implemented. Fluorescent foci were detected through their maximal intensity pixel, recording their intensity, coordinates, and affiliation to its containing cell, enabling their tracking throughout the cellular lineages.
Data Analysis.
Growth rates were calculated by exponential fit to the length change of individual bacteria as a function of time, as implemented in the BHV software (1), taking into account cells with at least eight time point measurements (R > 0.95). No correlation was found between the geographical location of cells within colonies and their growth rate (SI Fig. 9), suggesting that there is no nutrient limitation during colony growth (1). Indeed, the overall colony growth rate (as measured by colony area increase over time) remained unchanged throughout the experiments (data not shown).
Statistical t tests were made on the normally distributed growth rate values by using the t test for unpaired data with equal variance (as judged by an F test value; F > 0.05) implemented in Kaleidagraph 4.0 software.
Supplementary Material
ACKNOWLEDGMENTS.
We thank M. F. Bredeche and A. Babic for help in cloning; G. Paul for help in preparing Fig. 4; and M. Radman, B. Bukau, and M. Meselson for discussions and comments. A.B.L. was supported by European Molecular Biology Organization and Marie Curie postdoctoral fellowships. E.J.S. was supported as an Institut National de la Santé et de la Recherche Médicale Young Investigator fellowship. This work was supported by an Action Concertée Incitative grant from the French Ministry of Research, a European Young Investigator award, an Agence Nationale de la Recherche systems biology grant, and a Human Frontier Science Program grant.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. EU446025).
This article contains supporting information online at www.pnas.org/cgi/content/full/0708931105/DC1.
References
- 1.Stewart EJ, Madden R, Paul G, Taddei F. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol. 2005;3:e45. doi: 10.1371/journal.pbio.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirkwood TB. Understanding the odd science of aging. Cell. 2005;120:437–447. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 3.Kirkwood TB. Asymmetry and the origins of ageing. Mech Ageing Dev. 2005;126:533–534. doi: 10.1016/j.mad.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Ackermann M, Chao L, Bergstrom CT, Doebeli M. On the evolutionary origin of aging. Aging Cell. 2007;6:235–244. doi: 10.1111/j.1474-9726.2007.00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobson CM. The structural basis of protein folding and its links with human disease. Philos Trans R Soc London B. 2001;356:133–145. doi: 10.1098/rstb.2000.0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harding JJ. Viewing molecular mechanisms of ageing through a lens. Ageing Res Rev. 2002;1:465–479. doi: 10.1016/s1568-1637(02)00012-0. [DOI] [PubMed] [Google Scholar]
- 7.Soti C, Csermely P. Aging and molecular chaperones. Exp Gerontol. 2003;38:1037–1040. doi: 10.1016/s0531-5565(03)00185-2. [DOI] [PubMed] [Google Scholar]
- 8.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 9.Mogk A, Deuerling E, Vorderwulbecke S, Vierling E, Bukau B. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol Microbiol. 2003;50:585–595. doi: 10.1046/j.1365-2958.2003.03710.x. [DOI] [PubMed] [Google Scholar]
- 10.Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- 11.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 12.Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol. 2004;22:1399–1408. doi: 10.1038/nbt1029. [DOI] [PubMed] [Google Scholar]
- 13.Carrio MM, Villaverde A. Role of molecular chaperones in inclusion body formation. FEBS Lett. 2003;537:215–221. doi: 10.1016/s0014-5793(03)00126-1. [DOI] [PubMed] [Google Scholar]
- 14.Ignatova Z, Gierasch LM. Monitoring protein stability and aggregation in vivo by real-time fluorescent labeling. Proc Natl Acad Sci USA. 2004;101:523–528. doi: 10.1073/pnas.0304533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 16.Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- 17.Dukan S, et al. Protein oxidation in response to increased transcriptional or translational errors. Proc Natl Acad Sci USA. 2000;97:5746–5749. doi: 10.1073/pnas.100422497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rujano MA, et al. Polarised asymmetric inheritance of accumulated protein damage in higher eukaryotes. PLoS Biol. 2006;4:e417. doi: 10.1371/journal.pbio.0040417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen SP, Polazzi JO, Gierse JK, Easton AM. Two novel heat shock genes encoding proteins produced in response to heterologous protein expression in Escherichia coli. J Bacteriol. 1992;174:6938–6947. doi: 10.1128/jb.174.21.6938-6947.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laskowska E, et al. Aggregation of heat-shock-denatured, endogenous proteins and distribution of the IbpA/B and Fda marker-proteins in Escherichia coli WT and grpE280 cells. Microbiology. 2004;150:247–259. doi: 10.1099/mic.0.26470-0. [DOI] [PubMed] [Google Scholar]
- 21.Nagai T, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 22.Watve M, Parab S, Jogdand P, Keni S. Aging may be a conditional strategic choice and not an inevitable outcome for bacteria. Proc Natl Acad Sci USA. 2006;103:14831–14835. doi: 10.1073/pnas.0606499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans SN, Steinsaltz D. Damage segregation at fissioning may increase growth rates: A superprocess model. Theor Popul Biol. 2007;71:473–490. doi: 10.1016/j.tpb.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ackermann M, Stearns SC, Jenal U. Senescence in a bacterium with asymmetric division. Science. 2003;300:1920. doi: 10.1126/science.1083532. [DOI] [PubMed] [Google Scholar]
- 25.Reuter SH, Shapiro L. Asymmetric segregation of heat-shock proteins upon cell division in Caulobacter crescentus. J Mol Biol. 1987;194:653–662. doi: 10.1016/0022-2836(87)90242-7. [DOI] [PubMed] [Google Scholar]
- 26.Elowitz MB, Surette MG, Wolf PE, Stock JB, Leibler S. Protein mobility in the cytoplasm of Escherichia coli. J Bacteriol. 1999;181:197–203. doi: 10.1128/jb.181.1.197-203.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erjavec N, Nystrom T. Sir2p-dependent protein segregation gives rise to a superior reactive oxygen species management in the progeny of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2007;104:10877–10881. doi: 10.1073/pnas.0701634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janakiraman A, Goldberg MB. Evidence for polar positional information independent of cell division and nucleoid occlusion. Proc Natl Acad Sci USA. 2004;101:835–840. doi: 10.1073/pnas.0305747101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shapiro L, McAdams HH, Losick R. Generating and exploiting polarity in bacteria. Science. 2002;298:1942–1946. doi: 10.1126/science.1072163. [DOI] [PubMed] [Google Scholar]
- 30.Cox B, Ness F, Tuite M. Analysis of the generation and segregation of propagons: Entities that propagate the [PSI+] prion in yeast. Genetics. 2003;165:23–33. doi: 10.1093/genetics/165.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinclair DA, Guarente L. Extrachromosomal rDNA circles: A cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 32.Saint-Ruf C, Taddei F, Matic I. Stress and survival of aging Escherichia coli rpoS colonies. Genetics. 2004;168:541–546. doi: 10.1534/genetics.104.028704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stroikin Y, Dalen H, Brunk UT, Terman A. Testing the “garbage” accumulation theory of ageing: Mitotic activity protects cells from death induced by inhibition of autophagy. Biogerontology. 2005;6:39–47. doi: 10.1007/s10522-004-7382-y. [DOI] [PubMed] [Google Scholar]
- 34.Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 35.Chen JC, Beckwith J. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Mol Microbiol. 2001;42:395–413. doi: 10.1046/j.1365-2958.2001.02640.x. [DOI] [PubMed] [Google Scholar]
- 36.Jaffe A, Boye E, D'Ari R. Rule governing the division pattern in Escherichia coli minB and wild-type filaments. J Bacteriol. 1990;172:3500–3502. doi: 10.1128/jb.172.6.3500-3502.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindner AB, Kim SH, Schindler DG, Eshhar Z, Tawfik DS. Esterolytic antibodies as mechanistic and structural models of hydrolases: A quantitative analysis. J Mol Biol. 2002;320:559–572. doi: 10.1016/s0022-2836(02)00418-7. [DOI] [PubMed] [Google Scholar]
- 39.Matic I, Ekiert D, Radman M, Kohiyama M. Generation of DNA-free Escherichia coli cells by 2-aminopurine requires mismatch repair and nonmethylated DNA. J Bacteriol. 2006;188:339–342. doi: 10.1128/JB.188.1.339-342.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.