Abstract
An amphipathic α-helical peptide (C5A) derived from the membrane anchor domain of the hepatitis C virus (HCV) NS5A protein is virocidal for HCV at submicromolar concentrations in vitro. C5A prevents de novo HCV infection and suppresses ongoing infection by inactivating both extra- and intracellular infectious particles, and it is nontoxic in vitro and in vivo at doses at least 100-fold higher than required for antiviral activity. Mutational analysis indicates that C5A's amphipathic α-helical structure is necessary but not sufficient for its virocidal activity, which depends on its amino acid composition but not its primary sequence or chirality. In addition to HCV, C5A inhibits infection by selected flaviviruses, paramyxoviruses, and HIV. These results suggest a model in which C5A destabilizes viral membranes based on their lipid composition, offering a unique therapeutic approach to HCV and other viral infections.
Keywords: HCV, amphipathic peptide, antiviral peptide, NS5A, HIV
Hepatitis C virus (HCV), a member of the Flaviviridae family (1), is a single-stranded positive-sense RNA virus that causes acute and chronic hepatitis, cirrhosis, and hepatocellular carcinoma (2, 3). HCV infects >170 million people worldwide and is the most common cause of liver transplantation in the United States (3). There is no vaccine available for HCV, and the only currently approved treatment (combination therapy with IFN and ribavirin) has limited efficacy and serious side effects (4, 5). Thus, development of new classes of antiviral compounds with improved efficacy and toxicity profiles is urgently needed.
The development of HCV replicon technology several years ago (6) greatly accelerated the pace of antiviral drug discovery, leading to the development of HCV protease and polymerase inhibitors that are currently under clinical evaluation (7, 8). The landscape for drug discovery improved further with the establishment of a cell culture model of HCV infection in 2005 (9–11), making it possible to search for inhibitors of every step in the HCV life cycle and agents that target the virus itself. We now report the discovery of several HCV-derived synthetic peptides that inhibit HCV infection in the cell culture infection system. One of those inhibitory peptides, an amphipathic α-helical 18-mer derived from the membrane anchor domain of the HCV nonstructural protein NS5A that was particularly potent against HCV and selected other virus infections, serves as the basis of this report.
Results
Identification of Antiviral Peptides.
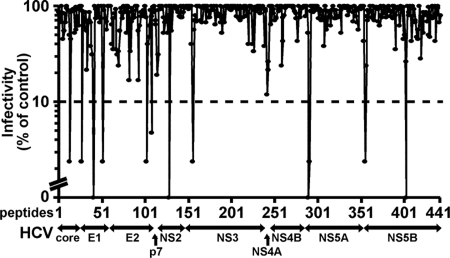
A peptide library of 441 overlapping peptides (18-mers offset by 11 amino acids) covering the entire HCV polyprotein (H77 strain, genotype 1a) was screened (20 μM) for the ability to inhibit HCV infection (JFH-1) in a focus reduction assay using Huh-7.5.1 cells (Fig. 1). Thirteen peptides were shown to inhibit HCV focus formation by >90%. Validation of the antiviral activity of the 13 inhibitory peptides was performed by comparing the ability of each peptide (20 μM) to inhibit the expansion of HCV RNA in Huh-7.5.1 cells 24 and 72 h after infection [multiplicity of infection (moi) 0.1] relative to the solvent control (0.5% DMSO). Most of the peptides inhibited viral expansion 10- to 100-fold in this assay [supporting information (SI) Table 3]. In contrast, the peptide derived from the N terminus of NS5A (SWLRDIWDWICEVLSDFK) inhibited viral expansion by >5 orders of magnitude. Accordingly, that peptide, termed C5A in this report, was studied further. C5A contains residues 3–20 of the amphipathic α-helical N-terminal membrane anchor domain of the genotype 1a HCV NS5A protein (12, 13).
Fig. 1.
Identification of antiviral peptides. A library consisting of 441 overlapping peptides (18-mers) covering the entire HCV polyprotein (H77 strain, genotype 1a) was obtained from the National Institutes of Health AIDS Research and Reference Reagent Program and screened at a concentration of 20 μM for the ability to inhibit HCV infection (JFH-1) in a focus reduction assay in Huh-7.5.1 cells, as described in Materials and Methods. The number of fluorescent HCV-positive foci was counted and expressed as a percentage (%) of foci in cells inoculated with virus and 0.5% DMSO. The peptide that displayed the strongest inhibition (termed C5A in this paper) is highlighted. For all subsequent experiments, this peptide was resynthesized to 95% purity.
C5A Inhibits HCV Infection at Noncytotoxic Concentrations.
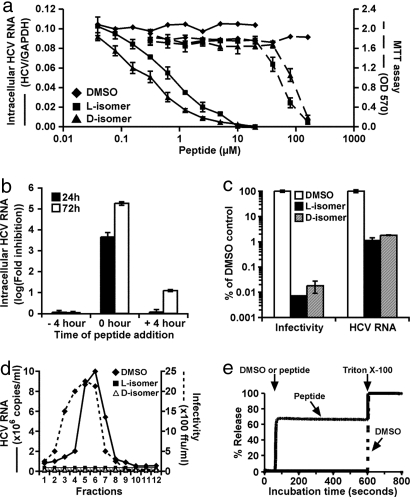
The 50% inhibitory (IC50) and 50% lethal (LC50) concentrations of C5A were determined. As shown in Fig. 2a, both the l- and d-isomers of C5A were highly inhibitory, displaying IC50 values of 0.79 and 0.32 μM, respectively, whereas their LC50 values were ≈100- to 300-fold higher. The slightly lower IC50 of the d-isomer may reflect its increased serum stability, because whereas the antiviral activity of the l-isomer was reduced by ≈2 logarithms after 1 h of preincubation in complete medium the d-isomer was fully active after at least 24 h of incubation (see SI Fig. 5). C5A also displayed a favorable toxicity profile in vivo, because both the l- and d-isomers were nontoxic when administered intravenously at doses as high as 0.5 mg per 25 gm C57BL/6 mouse administered at weekly intervals for 3 consecutive weeks (SI Table 4). Importantly, C5A was not immunogenic after repeated i.v. administration, because anti-C5A antibodies were not detected in the serum of those mice (data not shown).
Fig. 2.
C5A is virocidal for HCV. (a) To determine the peptide concentration required to inhibit HCV infection by 50% (IC50), >95% pure peptide stock solutions (3.6 mM in DMSO) were serially diluted in DMSO and tested for inhibitory activity as described in Materials and Methods. Peptide cytotoxic activity was measured by 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) cytotoxicity assay according to the manufacturer's instructions (American Type Culture Collection Catalogue no. 30–1010K), as described in SI Text. The peptide concentration that reduced the cell growth by 50% was designated the LC50. (b) C5A was either added to the cells for 4 h and removed before inoculation (−4 h); together with virus at the time of inoculation (0h) or 4 h postinoculation after virus had been removed by washing (+4 h). The −4 h and 0 h cells were washed after 4 h incubation and replaced with virus/peptide-free medium, whereas the peptide remained in the +4 h cultures throughout the experiment. Twenty-four and 72 h postinfection, cells were lysed for HCV RNA quantitation. (c) Peptide (18 μM in 0.5% DMSO) and 0.5% DMSO without peptide were added to complete medium containing 5 × 105 FFU/ml of HCV and incubated at 37°C for 1 h, at which point the virus–peptide and virus–DMSO mixtures were analyzed for total HCV RNA and infectivity. (d) The l- and d-isomers of C5A (18 μM) or 0.5% DMSO were incubated with virus (1 × 105 FFU/ml) for 4 h at 37°C, and 100-μl samples were analyzed by velocity sedimentation ultracentrifugation as described (15). (e) To examine the ability of C5A to permeabilize lipid membranes, a liposome dye release assay was performed as described in Materials and Methods.
C5A Is Virocidal for HCV.
To determine C5A's mechanism of action, the l-isomer was added to the virus or to the cells at different times relative to inoculation. As shown in Fig. 2b, C5A was strongly inhibitory when added to the cells together with the virus, much less inhibitory if added to the cells 4 h after infection, and entirely noninhibitory if added to the cells for 4 h and removed before the virus was added. These results suggest that C5A either blocks a very early step in the HCV life cycle or is virocidal for HCV. To distinguish between these alternatives, viral supernatant was pretreated with the l- or d-isomers or DMSO for 1 h before its HCV RNA content and infectivity were measured. As shown in Fig. 2c, the peptides reduced the HCV RNA content and infectivity by 100- and >10,000-fold, respectively, in contrast to the DMSO control. These results suggest that C5A is virocidal to HCV. To confirm this interpretation, we measured the sedimentation velocity of peptide-treated, and DMSO-treated virus particles by rate zonal ultracentrifugation. As shown in Fig. 2d, the control samples displayed discrete peaks of viral RNA and infectivity, which were virtually abolished by the l- and d-C5A isomers. Collectively, these results strongly suggest that C5A disrupts the structural integrity of HCV; i.e., it is virocidal. Because C5A is derived from the membrane anchor domain of NS5A, which is predicted to be an in-plane amphipathic α-helix (13), it may destabilize the HCV virion by permeabilizing its envelope. To test this hypothesis, we incubated cholesterol-phospholipid liposomes encapsulating a fluorescent dye with peptides or DMSO and measured fluorescence release. As shown in Fig. 2e, the l- and d-isomers of C5A (10 μM) instantly permeabilized 70% of the liposomes, whereas DMSO was inactive. The C5A effect was dose-dependent down to 160 nM, at which point 20% of the liposomes were permeabilized (data not shown). These results suggest that the virocidal effect of C5A reflects its ability to permeabilize cholesterol-rich phospholipid membranes.
C5A Prevents Initiation of HCV Infection and Suppresses Established Infection.
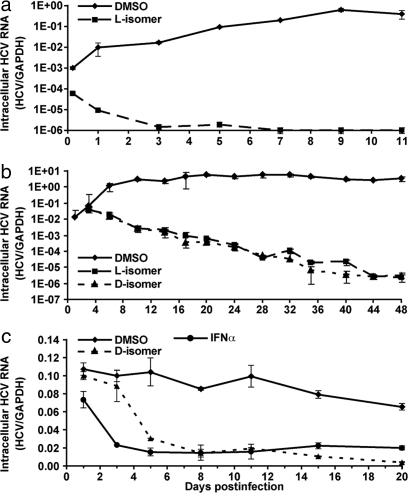
Next, we asked whether C5A can completely and permanently inhibit the establishment of HCV infection. As shown in Fig. 3a, in contrast to the virus spread observed in control cultures, intracellular viral RNA progressively decreased and ultimately disappeared (less than one copy per 1,000 cells) in cells infected with virus that was incubated with C5A before inoculation, and it remained undetectable for at least 11 days. These results suggest that, by virtue of its virocidal activity, C5A prevents initiation and spread of HCV infection. To determine whether C5A can terminate an ongoing HCV infection, it was added to infected cells 3 days after virus inoculation, when 10% of the cells were HCV E2 positive, and it was replenished in the culture medium each time the cells were split. As shown in Fig. 3b, whereas HCV RNA increased in DMSO-treated cells, both the l- and d-forms of C5A abruptly halted viral expansion, and viral RNA gradually decreased to less than one copy in every 1,000 cells by day 45 and remained undetectable for at least 15 days after peptide withdrawal (data not shown). These results suggest that C5A not only prevents HCV infection by destroying the virus and blocking cell-to-cell spread, but it can also terminate ongoing HCV infection in a dividing cell culture. To determine whether C5A can suppress infection in nondividing cells with established infection, d-C5A and DMSO were added to growth-arrested Huh-7 cells (14) 15 days after infection, when >90% of the cells were HCV E2-positive (data not shown). The culture medium was replaced daily with medium containing fresh peptide. For comparison, aliquots of infected cells were treated daily with recombinant human IFNα (100 units/ml). As shown in Fig. 3c, d-C5A suppressed intracellular HCV RNA to the same extent as IFNα by day 5, and it was even more inhibitory (>95%) than IFNα on days 15 and 20 of treatment. These results suggest that C5A can suppress persistent HCV infection in growth-arrested cells.
Fig. 3.
C5A prevents initiation of HCV infection and suppresses established infection. (a) Huh-7.5.1 cells were inoculated with HCV (moi 0.1) and C5A (18 μM) or 0.5% DMSO. After adsorption for 4 h at 37°C, the inocula were removed, the cells were washed two times, overlaid with 120 μl of fresh growth medium, and incubated at 37°C. At the indicated time points, total cellular HCV RNA was measured. (b) Huh-7 cells were infected with HCV (moi 0.1). On day 3 postinfection, when ≈10% of the cells were HCV E2-positive, C5A (18 μM) or 0.5% DMSO were added, and the cells were incubated at 37°C and split 1:6 every 3–4 days when confluent. C5A and DMSO were replenished when the cells were split, at which point total cellular HCV RNA was quantitated. (c) Huh-7 cells were pretreated with 1% DMSO for 10 days to induce differentiation and growth arrest, at which time they were infected by HCV (moi 0.01). Fifteen days after infection, when >90% cells were HCV E2-positive, the l- and d-isomers of C5A (18 μM) were added, and the cells were replenished every day with medium containing fresh peptide. At the indicated time points, total cellular HCV RNA content was measured. For comparison, the infected cells were treated with 100 units/ml of recombinant human IFNα (PBL Biomedical Laboratories) and replenished daily as above.
C5A Enters the Cell and Inactivates Virus Without Blocking HCV Replication.
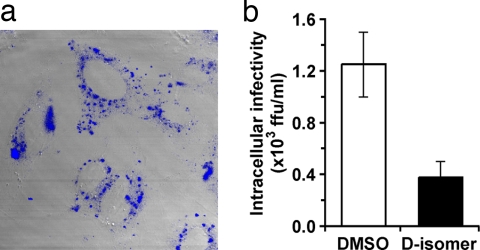
The foregoing results suggest that, in addition to its extracellular virocidal activity, C5A may inactivate HCV intracellularly (15). To test that hypothesis, we asked whether C5A can enter cells and destabilize intracellular virus particles. As shown in Fig. 4a, a fluorescent d-C5A containing a dansyl group at its N terminus efficiently enters Huh-7 cells, accumulating in granular structures in the cytoplasm. Importantly, when infected cells were incubated with the d-isomer of C5A for 6 h, intracellular infectious HCV particles were reduced 3-fold (Fig. 4b), without any change in intracellular HCV RNA (data not shown). These results suggest that, in addition to its extracellular virocidal activity, C5A can enter cells and inactivate intracellular virus, albeit less efficiently.
Fig. 4.
C5A inhibits intracellular HCV particle infectivity. (a) To determine whether C5A enters the cells, a fluorescent C5A containing a dansyl group at its N terminus (18 μM) was incubated with Huh-7 cells for 4 h at 37°C, washed five times with PBS, fixed with 4% paraformaldehyde, immunostained with rabbit polyclonal anti-dansyl antibody (Molecular Probes), and analyzed by confocal fluorescence microscopy. (b) Huh-7 cells previously infected with JFH-1 for 10 days were washed four times, and treated with d-isomers of C5A (18 μM) or DMSO (0.5%). After 6 h, intracellular HCV infectivity, extracellular HCV infectivity, and cellular HCV RNA content were determined as described in Materials and Methods.
Structure–Activity Analysis of C5A Antiviral Activity.
Comparative analysis of the antiviral activity, α-helicity, and membranolytic activity (data not shown) of a series of C- and N-terminal truncation mutants (peptide 3–13) revealed that the 16-mer SWLRDIWDWICEVLSD (peptide 3) retains full antiviral activity and α-helical and membranolytic properties (Table 1). The simultaneous loss of all three activities in the remaining truncation mutants suggests they are probably functionally linked to the ability of the peptide to inhibit HCV infection.
Table 1.
Structure–activity analysis of peptide C5A
| No. | Peptides | Description | IC50, μM | Helicity, % |
|---|---|---|---|---|
| Parental C5A | ||||
| 1 | SWLRDIWDWICEVLSDFK | l-isomer | 0.79 | 37.2 |
| 2 | swlrdiwdwicevlsdfk | d-isomer | 0.34 | 39.0 |
| Length series | ||||
| 3 | SWLRDIWDWICEVLSD | ΔC-, 2aa | 0.98 | 37.1 |
| 4 | SWLRDIWDWICEVL | ΔC-, 4aa | 11.3 | 34.2 |
| 5 | SWLRDIWDWICEV | ΔC-, 5aa | >27 | 0 |
| 6 | SWLRDIWDWICE | ΔC-, 6aa | >27 | 0 |
| 7 | SWLRDIWDWI | ΔC-, 8aa | >27 | 0 |
| 8 | SWLRDIWD | ΔC-, 10aa | >27 | 0 |
| 9 | LRDIWDWICEVLSDFK | ΔN-, 2aa | >27 | 4.4 |
| 10 | DIWDWICEVLSDFK | ΔN-, 4aa | >27 | 2.2 |
| 11 | WDWICEVLSDFK | ΔN-, 6aa | >27 | 0 |
| 12 | WICEVLSDFK | ΔN-, 8aa | >27 | 0 |
| 13 | CEVLSDFK | ΔN-, 10aa | >27 | 0 |
| 14 | SGSWLRDIWDWICEVLSDFK | Extend N-, 2aa | 1.7 | 39.8 |
| 15 | GSWLRDIWDWICEVLSDFK | Extend N-, 1aa | 0.51 | 38.0 |
| 16 | SWLRDIWDWICEVLSDFKT | Extend C-, 1aa | 1.7 | 39.8 |
| 17 | SWLRDIWDWICEVLSDFKTW | Extend C-, 2aa | 0.51 | 38.0 |
| Amphipathicity series | ||||
| 18 | SWRLIDWDWICEVLSDFK | Less amphipathic | 4.0 | 22.6 |
| 19 | SWRLDIWDWICESVLDFK | Less amphipathic | >30 | 21.5 |
| 20 | GIGKFLHSAKKFGKAFVGEIMNS | Magainin 2 | >27 | 21.7 |
| 21 | DWLKAFYDKVAEKLKEAF | Apo 18A | >28 | 48.0 |
| 22 | VLDLIYSLHKQINRGLKKIVL | BVDV analogue | >36 | 31.6 |
| Primary sequence series | ||||
| 23 | KFDSLVECIWDWIDRLWS | l- Retro | 0.85 | 40.7 |
| 24 | KFDSLVECIWDWIDRLWS | d- Retro | 0.48 | ND |
| 25 | KWLCRIWSWISDVLDDFE | Hydrophilic scrambled | 0.50 | 38.1 |
| 26 | SIWRDWVDLICEFLSDWK | Hydrophobic scrambled | 0.40 | 37.0 |
| Genotype series | ||||
| 27 | SWLRDVWDWICTVLTDFK | HCV 1b analogue | 3.9 | 50.0 |
| 28 | SWLRDVWDWVCTILTDFK | HCV 2a analogue | 2.1 | 30.2 |
| 29 | DWLRIIWDWVCSVVSDFK | HCV 3a analogue | 0.55 | 42.4 |
| 30 | SWLWEVWDWVLHVLSDFK | HCV 4a analogue | 7.0 | 39.3 |
| 31 | TWLRAIWDWVCTALTDFK | HCV 5a analogue | 7.1 | 42.0 |
| 32 | SWLRDVWDWVCTVLSDFK | HCV 6a analogue | 3.5 | 36.7 |
| Cysteine series | ||||
| 33 | SWLRDIWDWISEVLSDFK | C to S | 13.5 | 31.4 |
| 34 | SWLRDIWDWIREVLSDFK | C to R | 12.5 | 35.7 |
| 35 | SWLRDIWDWIEEVLSDFK | C to E | 13.0 | 31.9 |
| Net-charge series | ||||
| 36 | SWLDDIWDWICEVLSDFE | −6 | 4.7 | 20.2 |
| 37 | SWLRDIWDWICEVLSDFK | −2 | 0.79 | 37.2 |
| 38 | SWLRDIWDWICKVLSDFK | 0 | 6.8 | ND |
| 39 | SWLDRIWRWICKVLSRFE | +2 | 1.7 | 48.9 |
| 40 | SWLRDIWRWICKVLSRFK | +4 | 0.84 | ND |
| 41 | SWLRRIWRWICKVLSRFK | +6 | 0.89 | 56.4 |
The antiviral activity of all peptides (>95% pure) was determined by measuring the effect of serial 2-fold dilutions on intracellular HCV RNA 72 h postinfection and calculating their IC50 values. The helicity of the peptides was determined by CD analysis as described in Materials and Methods. The prototype strains of HCV genotype 3a, 4a, 5a, and 6a selected were K3A (GenBank accession no. D28917), EG. ED43 (Y11604), ZA. SA13 (AF064490), and HK.6a33 (AY859526), respectively. ND, not determined.
As illustrated in the helical wheel diagram shown in SI Fig. 6, C5A is strongly amphipathic. To determine whether amphipathicity is necessary for C5A's antiviral activity, we tested two variants whose amphipathicity was reduced by swapping the positions of its amino acids (underlined in Table 1), while maintaining its amino acid composition and α-helicity (peptides 18 and 19). As shown in Table 1, the antiviral activity of these peptides was reduced 5- to >30-fold. These results contrast with the retention of antiviral activity in C5A variants whose hydrophobic or hydrophilic amino acids were selectively scrambled while retaining their amphipathicity (Table 1, peptides 25 and 26). Collectively, these results suggest that amphipathicity is necessary for C5A's antiviral activity. It is not sufficient, however, because three previously well characterized amphipathic α-helical peptides [Magainin 2 (16), apolipoprotein 18A (17), and the C5A analogue from BVDV (18)] had no antiviral activity against HCV even at high concentrations (Table 1, peptides 20–22).
To determine whether primary amino acid sequence is required for antiviral activity, we tested a series of peptides that have the same amino acid composition as the parental peptides but different primary sequences. As shown in Table 1, C5A analogues containing either reversed (retro-) (peptides 23 and 24) or scrambled sequences (peptides 25 and 26) that maintained amphipathicity and α-helicity were fully active. These results indicate that the antiviral activity of the peptide is independent of its primary amino acid sequence as long as its amphipathic α-helical structure and amino acid composition are maintained.
To determine whether the antiviral activity strictly depends on amino acid composition, we compared the activity of several C5A analogues from six HCV genotypes whose amino acid sequence and composition are not strictly conserved. As shown in Table 1, the antiviral activity of the peptides was found to vary over a 10-fold range, with genotypes 2a and 3a (peptides 28 and 29) being most active and genotypes 4a and 5a (peptides 30 and 31) least active. These results imply that the amino acid composition of the peptide appears to influence the efficiency of its antiviral activity. It is noteworthy that the cysteine residue in C5A, which could lead to disulfide bond formation between peptide monomers, is essential for full antiviral activity (peptides 33–35), and that positively charged congeners of C5A (peptides 39–41) display strong antiviral activity.
Antiviral Specificity of C5A.
To determine whether the antiviral activity of C5A is specific to HCV, we asked whether it inhibited other viruses (Table 2). As described in SI Text, 18 μM C5A had no significant impact on the infectivity of adenovirus 5, Borna disease virus, coronavirus, coxsackie virus, influenza A virus, lymphocytic choriomeningitis virus, rhinovirus, rotavirus, vaccinia virus, or vesicular stomatitis virus, or on the antigenicity and DNA content of hepatitis B virus. In contrast, it strongly inhibited the infectivity of chimeric viruses containing the envelope proteins of HCV1a and 1b genotypes, and of other human Flaviviridae members, including West Nile virus and dengue 2 virus. Surprisingly, we found that infectivity of the paramyxoviruses, measles and respiratory syncytial virus, and HIV-1 were also inhibited by C5A. The IC50 values of C5A against these viruses are presented in Table 2. Detailed descriptions of those results will be reported separately.
Table 2.
Antiviral specificity and spectrum of C5A
| Virus | Enveloped | Genome | IC50, μM |
|---|---|---|---|
| HCV (JFH-1) genotype 2a | Yes | RNA+ | 0.6 |
| HCV (H77 envelope) genotype 1a | Yes | RNA+ | 3.9 |
| HCV (Con1 envelope) genotype 1b | Yes | RNA+ | 1.6 |
| HCV (J6CF envelope) genotype 2a | Yes | RNA+ | 1.1 |
| Dengue virus | Yes | RNA+ | 2.0 |
| West Nile virus | Yes | RNA+ | 4.5 |
| Measles virus | Yes | RNA− | 2.7 |
| Respiratory syncytial virus | Yes | RNA−xs | 4.5 |
| Human immunodeficiency virus | Yes | RNA+ | 1.3 |
| Adenovirus | No | DNA | >18 |
| Borna disease virus | Yes | RNA− | >18 |
| Coronavirus 229E | Yes | RNA+ | >18 |
| Coxsackie virus | No | RNA+ | >18 |
| Hepatitis B virus | Yes | DNA | >18 |
| Influenza virus | Yes | RNA− | >18 |
| Lymphocytic choriomeningitis virus | Yes | RNA− | >18 |
| Rhinovirus | No | RNA+ | >18 |
| Rotavirus WISC2 | No | dsRNA | >18 |
| Vaccinia virus | Yes | DNA | >18 |
| Vesicular stomatitis virus | Yes | RNA− | >18 |
To determine its antiviral specificity, C5A was added to virus stocks of predetermined infectivity (1–105 FFU or TCID50/ml) by using 2-fold serial dilutions starting from 18 to 20 μM. The virus–peptide and virus–DMSO mixtures were incubated at 37°C for at least 1 h before addition to susceptible cells or as indicated. In parallel, peptide and HCV (10,000 FFU/ml) were added to Huh-7.5.1 cells as a positive control for antiviral activity. Viral infections were assessed either by detection of a cytopathic effect (CPE) or by immunostaining with antibody against the corresponding viral protein, respectively and described in supporting text. HCV infections were carried out with JFH-1 (genotype 2a) and with chimeric viruses containing the structural region of prototype isolates from genotypes 1a (H77), 1b (con1) and an additional genotype 2a molecular clone (J6CF) as described in SI Text.
Discussion
Synthetic peptides derived from the functional domains of viral gene products have been shown to inhibit the corresponding viruses (19–21). For example, a 36-aa peptide, corresponding to a heptad repeat sequence in the HIV-1 transmembrane glycoprotein (gp41), is a potent inhibitor of HIV-1 membrane fusion and virus entry (20, 22, 23) and is now in clinical use in HIV-1 infected patients (24). Interestingly, the N-terminal proteolytic cleavage product of a peptide that spans the NS5A-NS5B cleavage site in HCV inhibits the HCV NS3/4A protease, and a peptidomimentic macrocyclic compound based on that structure has potent antiviral activity in vitro and in humans (19). These and similar results in other virus systems (21,25) served as the impetus for the studies reported herein.
Using an infectious focus-reduction assay to screen a 441-member synthetic peptide library covering the HCV polyprotein, we identified 13 peptides that inhibited HCV infection at least 10-fold, the most potent of which blocked infection >100,000-fold. Several of these peptides have potentially diverse mechanisms of action, based on their location in the corresponding protein (Fig. 1, SI Table 3). The most potent of these peptides, C5A, an 18-mer derived from the membrane anchor domain of the HCV NS5A protein whose IC50 is in the nanomolar range, which is at least 100-fold lower than the toxic dose observed in vitro and in vivo in mice (SI Table 4), was selected for further analysis. Importantly, C5A's d-isomer is at least as potent as the l-isomer (Fig. 2), is less cytotoxic in vitro (Fig. 2), is stable in serum-containing medium (SI Fig. 5), and is nonimmunogenic in vivo (data not shown). Moreover, C5A displays an entirely unique antiviral mechanism of action: it destabilizes viral structural integrity and has viral membranolytic activity, i.e., it is virocidal. Thus, it is not surprising that C5A completely and permanently prevents de novo HCV infection and suppresses ongoing HCV infection in dividing and nondividing cells. Collectively, these results suggest that C5A inhibits HCV infection by destabilizing HCV virions extracellularly and within infected cells.
C5A is composed of amino acids 3–20 at the N terminus of NS5A, a phosphorylated zinc metalloprotein that is an essential component of the HCV replication complex (26, 27). The N-terminal 30 amino acids of NS5A serve as its membrane anchor and target the protein to the endoplasmic reticulum (ER) (28). Structural analysis indicates that this region forms an amphipathic α-helix that is embedded in-plane in the cytosolic leaflet of the membrane bilayer (13, 31). As expected, CD analysis reveals that C5A displays α-helical structure in aqueous solution. Similarly, helical wheel projections showed that C5A residues are distributed in a perfect amphipathic pattern (SI Fig. 6). Nonetheless, C5A's antiviral activity is not a general property of all membrane association domains, because peptides from the membrane anchor domains of other HCV nonstructural proteins [e.g., NS4A, NS4B, and NS5B (29)] present in our peptide screening panel do not inhibit HCV infection.
Structure–activity analysis revealed that the antiviral specificity of C5A correlates with its α-helical structure and its ability to permeabilize liposome membranes. These results suggest that C5A interacts with the viral membrane to disrupt its integrity, release viral capsids, and expose the viral genome to exonucleases for degradation. In addition, mutational analysis revealed that amphipathicity is necessary for C5A's antiviral activity. This unique mechanism of action distinguishes C5A from peptides that interfere with viral ligand–host receptor interaction [e.g., Fuzeon and Virus Inhibitory Peptide (VIRIP) for HIV; EB peptide for influenza] (20, 30, 31) and viral escape from endosomes (e.g., HNP1 and HD5 for human papillomaviruses) (32). Although amphipathicity and α-helicity are both necessary for C5A's antiviral activity, they are not sufficient, because several other amphipathic α-helical peptides that have antimicrobial activity in other systems (33, 34) are not active against HCV.
Structure–activity analysis also revealed that C5A's antiviral activity is sequence-independent, because retropeptides or peptides with scrambled hydrophobic or hydrophilic amino acids were fully active, suggesting that its virocidal activity is independent of specific peptide–membrane protein interactions. This is supported by the full antiviral activity of the d-isomer of C5A, which is not likely to participate in the same specific peptide–membrane protein interactions as its l-isomer counterpart. In contrast, amino acid substitution analysis revealed that C5A's antiviral activity depends on its amino acid composition, that its internal cysteine residue appears to be essential and that its positively charged congeners are active as well. Synthetic analogues of C5A derived from the prototype sequence of six independent HCV genotypes displayed variable antiviral activity against HCV despite retention of amphipathic α-helical structure, presumably reflecting their different amino acid composition.
Interestingly, the antiviral activity of C5A is not limited to HCV, because it displays potent antiviral activity against other members of the Flaviviridae (West Nile virus and dengue virus), paramyxoviruses (measles and respiratory syncytial virus), and HIV, the details of which will be reported separately. Importantly, preliminary analysis suggests that C5A is virocidal for Dengue and HIV, similar to HCV. These results support the notion that C5A targets cellular components of virus membranes rather than virus-encoded proteins. Based on its in-plane orientation in the cytosolic leaflet of the ER membrane (13), we suggest that C5A recognizes cellular components of virus membranes, most likely their lipid composition, which would reflect that of the cellular compartment within which their morphogenesis occurs. Because HCV, West Nile virus, and dengue virus bud into the ER, their membranes may reflect the lipid composition required for insertion of the membrane anchor of HCV NS5A. In contrast, measles, RSV, and HIV bud from the surface of infected cells, so the basis for their inactivation by C5A remains to be determined. Theoretically, they could bud through lipid rafts, which are rich in cholesterol and sphingolipids (35), and therefore could contain similar membrane lipid composition to HCV, which has also been shown to associate with detergent-resistant membranes in the cell (36, 37). Further studies are needed to address these possibilities, as well as the basis for C5A resistance of the other viruses tested in this study.
In conclusion, these studies have defined the viral membrane as target for intervention in certain virus infections. In addition, they establish C5A as the prototype of a class of antiviral agents that specifically focus on that target. As such, membrane-targeting agents like C5A could be combined with agents that target intracellular aspects of the viral life cycle. Importantly, such agents should not select for escape variants and, if combined with drugs that do select resistance mutations, their extracellular virocidal mode of action should prevent the spread of those variants as well.
Materials and Methods
Peptide Synthesis.
Highly purified peptides (>95% purity) were synthesized with either l- or d-amino acids using fluorenylmethoxycarbonyl (Fmoc) chemistry by A & A Labs or Mimotopes or at The Scripps Research Institute.
HCV Focus Reduction Assay.
Peptides were reconstituted in 100% DMSO at a concentration of 10 mg/ml and stored at −20°C. The peptide stock solution was diluted 1:200 to a final concentration of ≈20 μM in complete DMEM growth medium containing 50 focus-forming units (FFU) (11) of HCV. The virus–peptide mixture was transferred to Huh-7.5.1 cells plated at a density of 8,000 cells per well in a 96-well plate. After adsorption for 4 h at 37°C, the inocula were removed, and the cells were washed two times, overlaid with 120 μl of fresh growth medium, and incubated at 37°C for 3 days when the cells were fixed with paraformaldehyde and immunostained with antibody against HCV nonstructural protein NS5A. The number of HCV-positive foci was counted by fluorescence microscopy.
In Vitro HCV RNA Inhibitory Assay.
To quantify the inhibitory effect of peptides on HCV infection, the intracellular HCV RNA content of peptide-treated and untreated cells was quantitated by real-time RT-QPCR. The peptide stock solution and DMSO solvent were diluted 1:100 and mixed with an equal volume of stock viral supernatant to yield a final peptide concentration of 18 μM and a final DMSO concentration of 0.5%. The virus–peptide and virus–DMSO mixtures were then used to infect Huh-7.5.1 cells at a moi of 0.1. After 3-day incubation at 37°C, total cellular RNA was isolated by the guanidine thiocyanate method, and the HCV RNA level was measured by real-time RT-QPCR, as described in SI Text. Inhibitory activity was detected by comparing the normalized intracellular HCV RNA levels of the peptide- and solvent-treated inocula. To determine IC50, serial 2-fold dilutions of the peptide stock solution were prepared in 100% DMSO, and aliquots of each dilution were then diluted 1:100 in complete growth medium and mixed with an equal volume of virus supernatant. The peptide concentration that inhibited infection by 50% relative to the DMSO control was designated the IC50 of that peptide. All results are presented as mean ± SD of a representative experiment.
Time of Addition Assay.
Huh-7.5.1 cells were seeded at 8,000 cells per well in a 96-well plate, and the next day they were infected with 800 FFU per well of HCV. Peptide was added at a final concentration of 18 μM under the following conditions: (i) preinoculation: peptide was added to cells for 4 h at 37°C followed by washing four times with growth medium before virus infection; (ii) coinoculation: peptide was added to cells together with virus for 4 h, at which time the cells were washed as above and replenished with complete media without peptide; (iii) postinoculation: cells were infected for 4 h at which point the virus was removed and the peptide was added and left on the cells for the duration of the experiment without washing. At 24 and 72 h postinfection, cells were lysed, and the HCV RNA level was measured as described above.
HCV Virocidal Assay.
Peptide was diluted in complete growth medium containing 5 × 105 FFU/ml of HCV to a final concentration of 18 μM and incubated at 37°C for 1 h, at which point the virus–peptide mixture was analyzed for HCV RNA and infectivity and compared with a comparably prepared virus–DMSO control. HCV RNA content was measured by real-time RT-QPCR and normalized to the level of GAPDH RNA that was added to the RNA samples to control for RNA extraction efficiency. HCV infectivity was measured by diluting 25 μl of each sample in growth medium 200-fold (i.e., below the inhibitory concentration of the diluted peptide), and residual infectivity was determined by incubating the diluted samples with Huh-7.5.1 cells and counting the number of HCV E2-positive foci 3 days later.
Liposome Dye Release Assay.
Peptides were examined for their ability to permeabilize lipid membranes in a liposome dye release assay, as described in SI Text.
CD Spectroscopy.
The CD spectra of peptides were recorded by using an Aviv model 62DS CD spectrometer (Aviv Associates), as described in SI Text.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Takaji Wakita (National Institute of Infectious Diseases, Tokyo) for providing JFH-1; Michael Houghton (Chiron, Emeryville, CA) for providing anti-NS5A antibodies; Dennis Burton and Juan Carlos de la Torre (TSRI, La Jolla, CA) for providing recombinant human anti-E2, lymphocytic choriomeningitis virus, and Borna disease virus respectively; Adolfo Garcia-Sastre (Mt. Sinai School of Medicine, New York) for providing influenza virus A/WSN/33; Jin Zhong and Sharookh Kapadia for assistance, comments, and discussion; Angelina Eustaquio, Josan Chung, and Bryan Boyd for technical support; the National Institutes of Health AIDS Research and Reference Reagent Program for the HCV synthetic peptide library; the Molecular Biology Service Laboratory of the Department of Molecular Medicine, supported by the Sam and Rose Stein Endowment Fund, for oligonucleotide preparation; and Drs. Jens Bukh (University of Copenhagen, Copenhagen, Denmark) and Richard Whitley (University of Alabama, Birmingham) for helpful critical comments. G.C., P.A.G., C.W.-B., S.F.W., M.I., and F.V.C. were supported by National Institutes of Health Grant CA108304 and by a gift from Mr. Clifford Evans; A.M. and M.R.G. were supported by Grant GM52190. B.F. was supported by Grant 1K22AI06302801. S.S. and P.A.G. were supported by Grant AI071952.
Footnotes
Conflict of interest statement: Francis V. Chisari has a financial interest in Viriome, Inc., which has licensing rights to the information provided in this paper.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712380105/DC1.
References
- 1.Miller RH, Purcell RH. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses and members of two plant virus supergroups. Proc Natl Acad Sci USA. 1990;87:2057–2061. doi: 10.1073/pnas.87.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36:S21–S29. doi: 10.1053/jhep.2002.36227. [DOI] [PubMed] [Google Scholar]
- 3.Alter HJ, Seeff LB. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin Liver Dis. 2000;20:17–35. doi: 10.1055/s-2000-9505. [DOI] [PubMed] [Google Scholar]
- 4.Di Bisceglie AM, McHutchison J, Rice CM. New therapeutic strategies for hepatitis C. Hepatology. 2002;35:224–231. doi: 10.1053/jhep.2002.30531. [DOI] [PubMed] [Google Scholar]
- 5.Tan SL, Pause A, Shi Y, Sonenberg N. Hepatitis C therapeutics: current status and emerging strategies. Nat Rev Drug Discov. 2002;1:867–881. doi: 10.1038/nrd937. [DOI] [PubMed] [Google Scholar]
- 6.Lohmann V, et al. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 7.Lin C, et al. In vitro studies of cross-resistance mutations against two hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061. J Biol Chem. 2005;280:36784–36791. doi: 10.1074/jbc.M506462200. [DOI] [PubMed] [Google Scholar]
- 8.Stuyver LJ, et al. Inhibition of hepatitis C replicon RNA synthesis by beta-D-2′-deoxy-2′-fluoro-2′-C-methylcytidine: a specific inhibitor of hepatitis C virus replication. Antivir Chem Chemother. 2006;17:79–87. doi: 10.1177/095632020601700203. [DOI] [PubMed] [Google Scholar]
- 9.Lindenbach BD, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 10.Wakita T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong J, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elazar M, et al. Amphipathic helix-dependent localization of NS5A mediates hepatitis C virus RNA replication. J Virol. 2003;77:6055–6061. doi: 10.1128/JVI.77.10.6055-6061.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Penin F, et al. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J Biol Chem. 2004;279:40835–40843. doi: 10.1074/jbc.M404761200. [DOI] [PubMed] [Google Scholar]
- 14.Sainz B, Jr, Chisari FV. Production of infectious hepatitis C virus by well-differentiated, growth-arrested human hepatoma-derived cells. J Virol. 2006;80:10253–10257. doi: 10.1128/JVI.01059-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gastaminza P, Kapadia SB, Chisari FV. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J Virol. 2006;80:11074–11081. doi: 10.1128/JVI.01150-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bechinger B, Zasloff M, Opella SJ. Structure and orientation of the antibiotic peptide magainin in membranes by solid-state nuclear magnetic resonance spectroscopy. Protein Sci. 1993;2:2077–2084. doi: 10.1002/pro.5560021208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung BH, Anatharamaiah GM, Brouillette CG, Nishida T, Segrest JP. Studies of synthetic peptide analogs of the amphipathic helix. Correlation of structure with function. J Biol Chem. 1985;260:10256–10262. [PubMed] [Google Scholar]
- 18.Sapay N, et al. NMR structure and molecular dynamics of the in-plane membrane anchor of nonstructural protein 5A from bovine viral diarrhea virus. Biochemistry. 2006;45:2221–2233. doi: 10.1021/bi0517685. [DOI] [PubMed] [Google Scholar]
- 19.Lamarre D, et al. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature. 2003;426:186–189. doi: 10.1038/nature02099. [DOI] [PubMed] [Google Scholar]
- 20.Kilby JM, et al. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 21.Lambert DM, et al. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc Natl Acad Sci USA. 1996;93:2186–2191. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan DC, Kim PS. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 23.Wild C, Oas T, McDanal C, Bolognesi D, Matthews T. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc Natl Acad Sci USA. 1992;89:10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenberg ML, Cammack N. Resistance to enfuvirtide, the first HIV fusion inhibitor. J Antimicrob Chemother. 2004;54:333–340. doi: 10.1093/jac/dkh330. [DOI] [PubMed] [Google Scholar]
- 25.Pastey MK, Gower TL, Spearman PW, Crowe JE, Jr, Graham BS. A RhoA-derived peptide inhibits syncytium formation induced by respiratory syncytial virus and parainfluenza virus type 3. Nat Med. 2000;6:35–40. doi: 10.1038/71503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tellinghuisen TL, Marcotrigiano J, Rice CM. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature. 2005;435:374–379. doi: 10.1038/nature03580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang L, et al. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J Biol Chem. 2005;280:36417–36428. doi: 10.1074/jbc.M508175200. [DOI] [PubMed] [Google Scholar]
- 28.Brass V, et al. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J Biol Chem. 2002;277:8130–8139. doi: 10.1074/jbc.M111289200. [DOI] [PubMed] [Google Scholar]
- 29.Gosert R, et al. Characterization of nonstructural protein membrane anchor deletion mutants expressed in the context of the hepatitis C virus polyprotein. J Virol. 2005;79:7911–7917. doi: 10.1128/JVI.79.12.7911-7917.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones JC, Turpin EA, Bultmann H, Brandt CR, Schultz-Cherry S. Inhibition of influenza virus infection by a novel antiviral peptide that targets viral attachment to cells. J Virol. 2006;80:11960–11967. doi: 10.1128/JVI.01678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munch J, et al. Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide. Cell. 2007;129:263–275. doi: 10.1016/j.cell.2007.02.042. [DOI] [PubMed] [Google Scholar]
- 32.Buck CB, et al. Human alpha-defensins block papillomavirus infection. Proc Natl Acad Sci USA. 2006;103:1516–1521. doi: 10.1073/pnas.0508033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Srinivas RV, et al. Antiviral effects of apolipoprotein A-I and its synthetic amphipathic peptide analogs. Virology. 1990;176:48–57. doi: 10.1016/0042-6822(90)90229-k. [DOI] [PubMed] [Google Scholar]
- 34.Chinchar VG, et al. Inactivation of viruses infecting ectothermic animals by amphibian and piscine antimicrobial peptides. Virology. 2004;323:268–275. doi: 10.1016/j.virol.2004.02.029. [DOI] [PubMed] [Google Scholar]
- 35.Booth AM, et al. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J Cell Biol. 2006;172:923–935. doi: 10.1083/jcb.200508014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi ST, Lee KJ, Aizaki K, Hwang SB, Lai MM. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J Virol. 2003;77:4160–4168. doi: 10.1128/JVI.77.7.4160-4168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matto M, Rice CM, Aroeti B, Glenn JS. Hepatitis C virus core protein associates with detergent-resistant membranes distinct from classical plasma membrane rats. J Virol. 2004;78:12047–12053. doi: 10.1128/JVI.78.21.12047-12053.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.