Abstract
DNA-dependent RNA polymerase (Pol)IV in Arabidopsis exists in two isoforms (PolIVa and PolIVb), with NRPD1a and NRPD1b as their respective largest subunits. Both isoforms are implicated in production and activity of siRNAs and in RNA-directed DNA methylation (RdDM). Deep sequence analysis of siRNAs in WT Arabidopsis flowers and in nrpd1a and nrpd1b mutants identified >4,200 loci producing siRNAs in a PolIV-dependent manner, with PolIVb reinforcing siRNA production by PolIVa. Transposable element identity and pericentromeric localization are both features that predispose a locus for siRNA production via PolIV proteins and determine the extent to which siRNA production relies on PolIVb. Detailed analysis of DNA methylation at PolIV-dependent loci revealed unexpected deviations from the previously noted association of PolIVb-dependent siRNA production and RdDM. Notably, PolIVb functions independently in DNA methylation and siRNA generation. Additionally, we have uncovered siRNA-directed loss of DNA methylation, a process requiring both PolIV isoforms. From these findings, we infer that the role of PolIVb in siRNA production is secondary to a role in chromatin modification and is influenced by chromatin context.
Keywords: RNA polymerase IV, RNA silencing, demethylation
Higher plants encode homologs of DNA-dependent RNA polymerase (Pol) subunits that differ from the canonical PolI, PolII, and PolIII enzymes required for biosynthesis of the major species of cellular RNA. These plant-specific subunits are presumed components of a fourth polymerase (PolIV) that has been implicated in biosynthesis of a 24-nt subclass of short interfering (si)RNAs, although its precise role in siRNA biogenesis is not clear. The largest subunits contain conserved regions that are shared by all Pols, consistent with a role of PolIV in the transcription of a DNA template to generate a long RNA precursor of siRNAs. However, Pol activity has not been shown, and it remains possible that PolIV complexes are RNA-dependent RNA polymerases or that they have a structural rather than an enzymatic role (1).
The link of PolIV with RNA silencing was made first from mutant screens in which the loss of silencing phenotype was associated with loss of RNA-directed DNA methylation (RdDM) at repeated sequence and transgene loci (2, 3). Targets of the PolIV-dependent RdDM include 5S rRNA-encoding DNA (rDNA) arrays; regulatory regions of several protein-coding genes including SUPERMAN, MEDEA, and FLOWERING WAGENINGEN (FWA); transposable elements AtMu1, SIMPLEHAT2, and AtSN1; and a number of unique intergenic regions (2–10). The proteins involved include SNF2 helicases, an RNA-dependent RNA polymerase (RDR2), and a Dicer-like ribonuclease (DCL3) that are involved in siRNA biogenesis (9, 11, 12). An Argonaute protein (AGO4) is an effector protein in these pathways (7), together with DNA methyltransferases (DRM2 and CMT3) (13, 14) and a histone methyltransferase (KYP) (15). In some respects, the mechanism of RdDM may be similar to siRNA-directed heterochromatinization of centromeric repeats and the mating type loci in Schizosaccharomyces pombe (16–19).
There are two genes in Arabidopsis encoding the putative largest subunit of PolIV (NRPD1A and NRPD1B), and it is likely that they share the second largest subunit (NRPD2A) to generate two PolIV isoforms. Mutations in either of the two NRPD1 subunits affect siRNA accumulation, although nrpd1b affects siRNA accumulation at only a subset of the loci affected by nrpd1a (3, 5, 20, 21). This differential effect prompted the proposal that PolIVb acts downstream of PolIVa at repeated sequence loci to amplify siRNA production and methylate DNA, whereas, at less repetitive loci, PolIVa functions without PolIVb in a process that does not involve RdDM (5).
Structural and biochemical studies have been consistent with this proposal. They have shown that the main difference between NRPD1a and NRPD1b is in the presence of a large carboxyl-terminal region containing 10 copies of a 16-aa repeat (5) that mediates an interaction with AGO4 (22, 23). Based on this finding, it could be envisioned that the difference between PolIVa- and PolIVb-dependent RNA-silencing pathways is attributable to differential interactions of these subunits with the AGO4 effector protein.
However, the previous analyses of the two PolIV isoforms have involved only a few different types of 24-nt siRNA and target loci. Here, to better characterize the function of PolIV proteins, we analyzed the 21- to 25-nt RNA population of WT, nrpd1a, and nrpd1b floral tissue by using high-throughput sequencing technology. Our analysis showed that there were at least 4,600 genomic regions producing siRNAs and micro (mi)RNAs, of which 94% required PolIVa. These PolIVa-dependent loci excluded microRNA (miRNA) genes, and their siRNA products were predominantly 24-nt long. They represented all classes of transposable elements and were most abundant in the pericentromere. Most of these PolIVa-dependent loci were also dependent, to a variable extent, on NRPD1b, indicating that PolIVb enhances production of siRNAs at PolIVa-dependent loci. However, there were some loci at which PolIVb was not required for siRNA accumulation. Our findings also indicate that the relationship between PolIVb dependency and RdDM is more complex than has been appreciated previously (5). We identified two loci, for example, at which RdDM but not siRNA production depends on NRPD1b. Based on this observation, we infer that PolIVb may have separate functions in siRNA biogenesis and RdDM. Also unexpected, we describe a locus at which NRPD1b function was associated with decreased DNA methylation, and we propose that siRNAs may guide DNA demethylation, as well as DNA methylation. At other loci, there was a high level of DNA methylation that was unaffected by loss of PolIV function. Based on these findings, we propose that any effect of PolIV-dependent siRNAs on DNA methylation is determined by locus-dependent interactions with epigenetic mechanisms that may be independent of siRNA.
Results
Databases of PolIV-Dependent Small RNAs.
To discover how PolIV isoforms affect RNA silencing, we used high-throughput pyrosequencing (24) to characterize 15- to 30-nt RNA populations from mixed stage flowers of WT Col-0, nrpd1a-4, and nrpd1b-1 [supporting information (SI) Table 1]. After removing structural RNA sequences (tRNA, rRNA, small nucleolar RNA, and small nuclear RNA) from the datasets, the remaining sequences were predominantly 21- to 24-nt long. We then aligned the sequence of these RNAs with identical regions of the Arabidopsis nuclear genome (SI Table 2) to reveal 10,130 genomic loci with four or more RNA matches aligned <200 bp apart (see SI Methods). However, for repeated sequence loci, it is not clear which copy is a bona fide genomic source of small (s)RNA. To reduce this ambiguity, we focused our analysis on genomic loci with one or more RNAs matching a unique DNA sequence, so that the total number of loci was reduced to 4,685 (SI Table 2). We recognized that this unique sequence filter underestimates the regions in the genome with the potential to generate short RNAs because, of 96 characterized miRNA genes, eight were excluded. However, the less ambiguous identification of sRNA loci is a sound basis for further analysis.
Based on representation of 21- to 24-nt RNA in the three datasets, we infer that most production of these RNAs depends, to some extent, on both PolIVa and PolIVb (Fig. 1A), although there were PolIV-independent loci and loci at which sRNA production depended on PolIVa only (Fig. 1A). The sRNAs from a few (59) loci are less frequent relative to WT in the nrpd1b database, and they are present at WT levels in the nrpd1a dataset. However, we are not confident that these loci are dependent on PolIVb and not on PolIVa because the reduction in nrpd1b was minor and within the likely range of experimental variation.
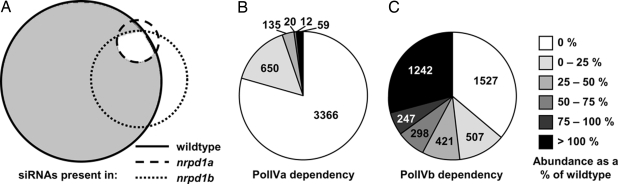
Fig. 1.
PolIVa is required for siRNA accumulation at most endogenous loci. (A) Representative Venn diagram of siRNA-generating loci. Solid line, loci with siRNAs in WT; dotted line, loci with siRNAs at WT or a greater level in nrpd1a; dashed line, loci with siRNAs present at WT or greater level in nrpd1b. (B and C) Pie charts depicting level of siRNA representation at loci requiring at least one PolIV isoform (gray region in A).
Among the loci with reduced siRNA representation in at least one PolIV mutant, PolIVa dependency was stronger than that of PolIVb. The RNAs from most (3,366 of 4,242; 79%) of the PolIV-dependent loci were absent in the nrpd1a dataset, and they were predominantly 24-nt long. Of the 876 remaining loci, there were 650 (74%) from which the 24-nt RNA was present in the nrpd1a dataset at a frequency <25% of WT (Fig. 1B). In contrast, in the nrpd1b dataset, only 1,527 (36%) of these loci were not represented. The remaining loci exhibited various degrees of reduction (1,473 loci; 35%) or were present in the datasets at WT or greater than WT levels (1,242 loci; 29%) (Fig. 1C). The same differential effect of nrpd1a-4 and nrpd1b-1 was evident, even when the analysis was restricted to loci with ≥10, ≥20, ≥50, or ≥100 sRNAs in the WT dataset (SI Fig. 7). We can therefore rule out that the variable effect of PolIVb is an artifact of there being only a few sRNAs represented at certain loci.
From these analyses, we conclude that PolIVb involvement in 24-nt RNA production is more variable than the requirement for PolIVa. We also conclude that PolIVb requirement is almost always tied to PolIVa but that PolIVa can operate independently of PolIVb. Our data therefore support the proposal (5) that PolIVb reinforces or amplifies 24-nt RNA production via PolIVa rather than carrying out a primary role in its biogenesis.
Northern blot analysis with selected loci confirmed that 24-nt RNA representation in the sequence databases reflects their abundance in the RNA sample. The RNAs identified as being PolIVa-dependent in the database were predominantly 24-nt long, they were absent or greatly reduced in samples from nrpd1a, and they were reduced to various degrees in nrpd1b (Fig. 2 and SI Fig. 8). The Northern blot analysis included samples from rdr2–2 plants, and it confirmed the overlap of PolIV- and RDR2 dependency (Fig. 2) (25). In dcl3–1, the 24-nt sRNAs were typically replaced by 21- to 22-nt RNAs, as described previously (9).
Fig. 2.
Genetic dependence of PolIV-dependent loci. siRNA Northern blots of mixed floral tissue confirm that all tested PolIV-dependent loci lose siRNAs in nrpd1a. They also require NRPD2, RDR2, and DCL3 for siRNA accumulation but show various degrees of dependence on NRPD1b. The size markers are estimates based on 20- and 30-nt RNA oligos.
Genomic Features Associated with PolIV-Dependent siRNAs.
There was extensive overlap between the PolIV-independent loci identified here and those described previously from an analysis of nrpd1a nrpd1b and nrpd2a nrpd2b genotypes (25). Our data therefore confirm the previously described enrichment for 21- and 22-nt RNAs and for miRNA and transacting siRNA loci in the datasets of PolIV-independent sRNAs (2, 3, 5, 8). Our data also confirm that the predominantly 24-nt PolIV-dependent sRNAs are siRNAs rather than miRNAs and that they are derived from RDR2- and DCL3-dependent loci.
However, because we used single nrpd1 mutants, we could differentiate the roles of PolIVa and PolIVb. To simplify the analysis of the two PolIV isoforms, we did not consider loci with an intermediate requirement for PolIVb. We focused on the PolIVa-dependent loci with no reduction in siRNAs in nrpd1b databases (PolIVa-dependent only, A type) and those with no siRNAs in nrpd1b databases (PolIVa- and PolIVb-dependent, A+B type). Together, these represented 60% of PolIVa-dependent loci.
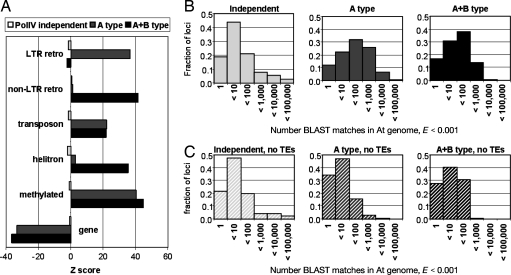
Both A type and A+B type loci over-represent transposable elements relative to PolIV-independent loci, but they were distinct from each other in that LTR retrotransposons were prevalent only at A type loci, whereas non-LTR retrotransposon and helitrons were significantly represented only at A+B type loci (Fig. 3A). Both A and A+B type loci were more repetitive than the independent loci, with A type loci more frequently matching the genome <1,000 or <10,000 times when compared with A+B type loci (Fig. 3B). When loci overlapping transposable elements were removed from the analysis, A and A+B type loci exhibited repetitiveness similar to the independent loci (Fig. 3C). It is likely, therefore, that the increased repetitiveness of A type relative to A+B type loci is largely attributable to the transposons associated with these loci.
Fig. 3.
PolIV-dependent loci exhibit differential association with transposable elements but no inherent repetitive character. (A) Over- or underrepresentation of genomic features at PolIV-independent, A type, and A+B type loci. The locus positions were randomized 100 times, and the associated genomic elements were counted to calculate Z scores: Z = (observed − averagerand)/SDrand. (B and C) Histograms of loci with respect to their genomic repetitiveness. Each locus was BLASTed against the Arabidopsis genome, and the resulting matches (E < 0.001) were counted and plotted.
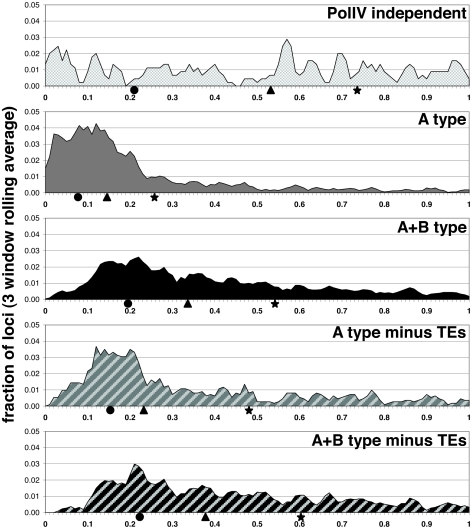
The PolIV-dependent loci of both A and A+B type are abundant in the pericentromeric region, as reported by others for siRNA loci (26, 27) (Fig. 4). This pericentromeric pattern was more pronounced with the A type loci (Fig. 4), and it could be a consequence of the abundance of transposons in pericentromeric regions of Arabidopsis chromosomes (28, 29). The pericentromeric bias persisted even when transposons-related loci were eliminated from the analysis, although to a slightly reduced extent with A type loci (Fig. 4). However, because prediction of transposable elements is imprecise, we could not rule out that unannotated elements influence the pericentromeric bias of these classes. In distal chromosomal regions, 41.5% of PolIV-dependent loci in distal chromosomal regions overlapped a transposable element compared with 4.4% of random loci.
Fig. 4.
PolIV-dependent loci are pericentromerically localized independent of transposable element character. Histograms of relative centromeric bias of sRNA-generating loci. 0, centromere; 1, telomere. The rolling average of the fraction of loci in three 1% windows is plotted. The short arms of chromosomes 2 and 4 were removed for clarity (33). Circles, triangles, and stars mark the first, second, and third quartiles, respectively.
A further difference between A and A+B type loci was in nucleotide composition. The A+B type loci had a lower mean G+C content (35.9%) than either the A type (39.3%) or the PolIV-independent loci (41.8%) (SI Fig. 9). These differences may indicate that loci with fewer potential methylation sites require PolIVb reinforcement of siRNA production. Alternatively, it may be a footprint of ancestral genomes in which T residues were introduced by deamination of methyl C at PolIV-dependent loci. Consistent with this possibility, there is under-representation of CG dinucleotides and corresponding over-representation of TG dinucleotides at A type loci (SI Fig. 9) but not at A+B type loci.
PolIV-Dependent Effects on RNA-Directed DNA Methylation.
It has been hypothesized that PolIVb-dependent siRNAs target RdDM, whereas PolIVb-independent siRNAs do not (5). Consistent with this idea, there was over-representation of methylated DNA at the A+B loci (Fig. 3A). However, there was also over-representation of methylated DNA and hallmarks of CG methylation at the A type loci (Fig. 3A and SI Fig. 9). This pattern suggested to us that PolIV could be recruited to DNA that is methylated by an siRNA-independent mechanism. Alternatively it could be that RNA-directed DNA methylation occurs without the involvement of PolIVb.
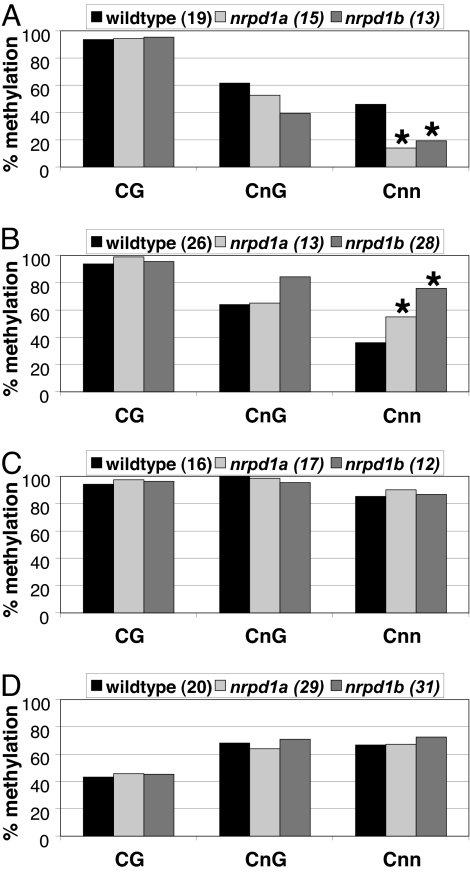
To assess these alternatives, we used bisulfite sequencing to monitor DNA methylation at four PolIV-dependent loci in WT and nrpd mutants (Fig. 5 and SI Fig. 10 and Table 3). The results in Fig. 5 represent the methylation status at C residues in either symmetrical (CnG, CG) context or at Cnn motifs where C methylation is diagnostic of RNA-directed DNA methylation (6, 7). All four loci exhibited unexpected effects on the DNA methylation status. Locus 00687, for example, exhibited loss of methylation at asymmetric Cnn in the nrpd1a and nrpd1b mutants (Fig. 5A), consistent with the predicted involvement of PolIVb in RdDM. However, the siRNAs were lost from this mutant only in the nrpd1a mutant: it was an A type locus (Fig. 2). From this analysis, we conclude that PolIVb can influence target DNA methylation independent of its role in siRNA biogenesis.
Fig. 5.
RNA-directed DNA methylation at PolIVa-dependent loci. Bisulfite sequencing of PolIV-dependent loci 00687 (A), 08002 (B), 10102 (C), and 04138 (D). Data from two biological replicates were combined. Asterisks mark changes that were statistically significant (two-tailed t test, P < 0.01) in both replicates. The total number of clones analyzed is in parenthesis.
The A type locus 08002 (Fig. 2) also exhibited an unexpected pattern of DNA methylation in the WT and mutant plants. There were siRNAs in nrpd1b but an increase in asymmetric DNA methylation in both mutants (Fig. 5B). As with the 00687 data, this result implies an activity of PolIVb that is independent of its role in siRNA biogenesis or amplification. However, unlike locus 00687, this role would be in loss of DNA methylation. This effect could be direct if a PolIV complex targeted DNA demethylation enzymes. It could also be indirect if loci with the potential to target RdDM in a PolIV-independent manner were themselves silenced by PolIV-dependent siRNAs.
The two A+B type loci (10102 and 04138) illustrate how PolIVb-dependent siRNA production is not always associated with methylation of the target DNA. There was reduced siRNA accumulation from these loci in nrpd1b but no changes of DNA methylation in either of the mutants (Fig. 5 C and D). One interpretation of these data is that PolIVb-dependent siRNA is not always associated with RNA-directed DNA methylation. However, we cannot formally rule out that there is functional redundancy, so that loss of PolIVb is compensated by other RNA directed DNA methylation pathways.
The unexpected patterns of DNA methylation and siRNA accumulation at these four loci show that DNA methylation is not an inevitable consequence of targeting by PolIVb-generated siRNAs. There can be PolIVb-dependent changes in DNA methylation that do not require PolIVb-generated siRNAs and PolIVb-generated siRNAs with no effect on DNA methylation. PolIVb, therefore, has separable roles in DNA methylation and siRNA generation.
Discussion
Until now, PolIV was considered a cofactor in siRNA biogenesis. Various models have proposed that PolIVa would play a role in siRNA biogenesis either alone or together with PolIVb and that, when both isoforms are involved, PolIVa acts upstream of PolIVb (5, 22, 23). RNA-directed DNA methylation has been presumed to be an effect of siRNAs generated by PolIVb. However, from the data presented here, we must revise that view. At two of the four loci inspected in detail, there was an effect of nrpd1b on DNA methylation without an effect on siRNA accumulation, and at two other loci, PolIVb was required for siRNA accumulation without affecting DNA methylation (Figs. 2 and 5).
The simplest explanation of these results is that PolIVb has a role as an effector of RNA silencing independent of any function in siRNA biogenesis. We envision that a complex including the PolIV subunit NRPD1b and its interaction partner AGO4 is targeted to genomic loci by siRNAs associated with AGO4. The recruitment of this large complex may cause structural perturbation to allow access for DNA methyltransferases, DNA demethylation enzymes, or other cofactors of silencing. Alternatively, this complex could have an enzymatic function that influences the structure or biochemical activity of silencing cofactors either directly or indirectly, perhaps by influencing their localization, compartmentalization, or chemical modification. Presumably, the characteristics of chromatin adjacent to the targeted region would influence whether PolIVb affected methylation or loss of methylation.
This proposal that PolIVb has a role that is independent of siRNA biogenesis may mean that it has an effect at many more loci than those at which it influences siRNA accumulation and perhaps at all PolIVa-dependent loci. This proposal may also be interpreted to indicate that PolIVb does not have a direct involvement in siRNA biogenesis at any loci. It could be that PolIVb somehow allows access for PolIVa in siRNA biogenesis at the loci where there is a high degree of PolIVb dependency.
To explain the different degrees of PolIVb-dependent siRNA accumulation, we invoke self-reinforcing silencing pathways, as proposed by Pontier et al. (5), in which PolIVa would generate primary siRNAs at loci with preexisting epigenetic marks (2). This requirement for preexisting epigenetic marks may explain the predominant pericentromeric localization of PolIV-dependent loci. These regions are enriched in epigenetic marks as a consequence of proximity to the centromere, whereas more distant regions may have epigenetic marks that are too weak for PolIVa recruitment.
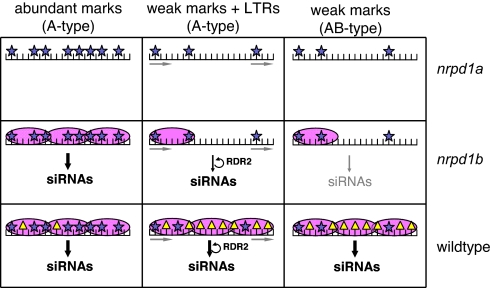
At loci with a high degree of PolIVb dependency for siRNA accumulation (Fig. 1C), we envision that the primary PolIVa-mediated siRNAs would bind to AGO4 and, indirectly, to PolIVb. This AGO4/PolIVb effector complex would then facilitate DNA or histone methyltransferases, so that PolIVa would be recruited for production of additional secondary siRNAs. PolIVb would influence siRNA accumulation at these loci, but its effect would be indirect. We envision that these loci would include those at which the initial signal for recruitment of PolIVa is weak or sparse (Fig. 6).
Fig. 6.
Model of PolIV dependency of siRNA-generating loci. PolIV-independent epigenetic marks (blue stars) recruit PolIVa (pink ovals) to generate siRNAs. In the presence of PolIVb, additional epigenetic marks (yellow triangles) can reinforce recruitment of PolIVa. Additionally, RDR2-mediated amplification of siRNAs can occur when direct repeats are present in the transcript, as in LTR retroelement RNAs.
At loci with a low degree of PolIVb dependency for siRNA biogenesis (Fig. 1C), including the A type loci, we envision that the initial signal for recruitment for PolIVa would be strong. The primary PolIVa-dependent siRNAs would guide an AGO4/PolIVb effector complex, and there could be epigenetic modification of DNA at the target loci, such as at loci 00687 and 08002 (Fig. 5 A and B), but any additional secondary siRNA biogenesis would be negligible compared with the abundant primary siRNAs (Fig. 6).
There may also be A type loci with a weak initial signal for recruitment of PolIVa at which primary siRNA production is amplified in an RDR2-dependent mechanism. We envision that LTR retrotransposons that are significantly over-represented in A-type but not A+B type loci could be in this category (Fig. 3A) because they have direct transcribed repeats. RNA molecules with direct transcribed repeats would be good substrates for an RDR-dependent amplification mechanism of siRNA production (30). The RDR2-generated siRNAs would guide AGO4/PolIVb, but, as at the loci with a strong primary signal for recruitment of PolIVa, any secondary siRNA biogenesis following from epigenetic modification would be negligible in comparison with the RDR2-generated siRNAs (Fig. 6).
A test of this hypothesis would be through an extension of analyses in which siRNA accumulation is monitored in mutant plants with mutations at DDM1 and MET1. These loci control epigenetic modification at many loci, including those producing siRNAs. We predict that in single mutants, the primary epigenetic marks responsible for PolIV recruitment would be lost and that A type loci would be converted into A+B type loci or they would lose the potential for siRNA production completely.
The next challenge, having defined the overlapping PolIVa and PolIVb transcriptomes in Arabidopsis, is to elucidate the biological roles of PolIV-generated siRNAs. Loci generating PolIV-dependent siRNAs comprise at least 1% of the Arabidopsis genome, and, in addition to transposons and pericentromeric sequences, they are derived from thousands of regions adjacent to protein coding genes. Although strongly under-represented at A and A+B type loci, genes are known targets of PolIV (Fig. 3). Based on the analysis of PolIVa-dependent natural antisense siRNAs (31, 32), it is possible that there are additional genic siRNAs that are induced during growth, development, and stress and that they influence responses of the plant to various stimuli. Alternatively, intergenic siRNAs may directly influence gene expression because a slight but significant correlation between siRNA production and gene expression has been shown (27). Intergenic siRNAs may influence transcription through altered chromatin organization or the creation of boundary elements or nuclear subdomains.
Materials and Methods
All RNA and DNA samples were collected from mixed-stage flower buds grown under 16 h of light. Fifteen- to 30-nucleotide siRNAs were ligated with adapters, reverse transcribed, and amplified before being sent to 454 Life Sciences for pyrosequencing. SI Table 4 lists the adapters and primers used. Perl scripts were generated to trim adapters, filter structural RNAs, match siRNAs to the genome, and analyze siRNA loci. For Northern blot transfer, 50 μg of total RNA was separated on an 8% acrylamide gel. Labeled riboprobes were synthesized from T7-tailed PCR products before hybridization in PerfectHyb (Sigma). DNA was bisulfite-converted for methylation analysis by using the EZ DNA Methylation-Gold kit (Zymo Research). For detailed description of methods and analysis, see SI Methods.
Supplementary Material
ACKNOWLEDGMENTS.
This work was supported by the Gatsby Charitable Foundation to the Sainsbury Laboratory, Biotechnology and Biological Sciences Research Council Grant BB/E004091/1, the European Union Sixth Framework Programme Integrated Project SIROCCO (Grant LSHG-CT-2006-037900), and postdoctoral fellowships from the Marshall Sherfield Foundation and the National Science Foundation (R.A.M.). D.C.B. is funded as a Royal Society Research Professor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.C.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709632105/DC1.
References
- 1.Herr AJ. FEBS Lett. 2005;579:5879–5888. doi: 10.1016/j.febslet.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 2.Herr AJ, Jensen MB, Dalmay T, Baulcombe D. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 3.Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ. Nat Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- 4.Chan SW, Henderson IR, Zhang X, Shah G, Chien JS, Jacobsen SE. PLoS Genet. 2006;2 doi: 10.1371/journal.pgen.0020083. 0791–0797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T. Genes Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qi Y, He XH, Wang X, Kohany O, Jurka J, Hannon GJ. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- 7.Zilberman D, Cao X, Jacobsen SE. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 8.Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zilberman D, Cao X, Johansen LK, Xie Z, Carrington JC, Jacobsen SE. Curr Biol. 2004;14:1214–1220. doi: 10.1016/j.cub.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 11.Kanno T, Mette F, Kreil DP, Aufsatz W, Matzke AJM, Matzke M. Curr Biol. 2004;14:801–805. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 12.Smith LM, Pontes O, Searle I, Yelina NE, Yousafzai FK, Herr AJ, Pikaard C, Baulcombe D. Plant Cell. 2007;19:1507–1521. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao X, Jacobsen SE. Proc Natl Acad Sci USA. 2002;99:16491–16498. doi: 10.1073/pnas.162371599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindroth AM, Cao XF, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 15.Jackson JP, Lindroth AM, Cao XF, Jacobsen SE. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 16.Djupedal I, Portoso M, Spahr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K. Genes Dev. 2005;19:2301–2306. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 18.Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. Science. 2005;309:467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- 19.Volpe T, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen R. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 20.Chan SW, Zhang X, Bernatavichute YV, Jacobsen SE. PLoS Biol. 2006;4:e363. doi: 10.1371/journal.pbio.0040363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJM, Matzke M. EMBO J. 2006;25:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pontes O, Li CF, Nunes PC, Haag JR, Ream T, Vitins A, Jacobsen SE, Pikaard CS. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 23.Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SWL, Lagrange T, Pikaard C, Jacobsen SE. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 24.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen Y, Chen Z, et al. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE. Proc Natl Acad Sci USA. 2007;104:4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The Arabidopsis Genome Initiative. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Martienssen RA. Nat Genet. 2003;35:213–214. doi: 10.1038/ng1252. [DOI] [PubMed] [Google Scholar]
- 31.Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu J-K. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Jr, Zhu JK, Staskawicz BJ, Jin H. Proc Natl Acad Sci USA. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pereira V. Genome Biol. 2004;5:R79. doi: 10.1186/gb-2004-5-10-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.