Abstract
Cancer/testis (CT) antigens—immunogenic protein antigens that are expressed in testis and a proportion of diverse human cancer types—are promising targets for cancer vaccines. To identify new CT antigens, we constructed an expression cDNA library from a melanoma cell line that expresses a wide range of CT antigens and screened the library with an allogeneic melanoma patient serum known to contain antibodies against two CT antigens, MAGE-1 and NY-ESO-1. cDNA clones isolated from this library identified four CT antigen genes: MAGE-4a, NY-ESO-1, LAGE-1, and CT7. Of these four, only MAGE-4a and NY-ESO-1 proteins had been shown to be immunogenic. LAGE-1 is a member of the NY-ESO-1 gene family, and CT7 is a newly defined gene with partial sequence homology to the MAGE family at its carboxyl terminus. The predicted CT7 protein, however, contains a distinct repetitive sequence at the 5′ end and is much larger than MAGE proteins. Our findings document the immunogenicity of LAGE-1 and CT7 and emphasize the power of serological analysis of cDNA expression libraries in identifying new human tumor antigens.
Keywords: human cancer immunology, serological analysis of recombinant cDNA expression libraries
Defining the range of tumor antigens recognized by the immune system of the autologous host has long been a goal of tumor immunology (1). The recent development of a new approach to dissect the humoral immune response to cancer has opened the way to establishing a comprehensive picture of the immune repertoire against human cancer antigens. This approach, called SEREX (serological analysis of recombinant cDNA expression libraries), involves the construction of cDNA expression libraries from primary or metastatic human tumors and immunoscreening these libraries with autologous patient sera (2–4). In this way, two important characteristics of the cloned tumor products are defined simultaneously: immunogenicity in the autologous host and primary sequence of the isolated tumor antigen.
In the past 2 years, SEREX has been applied to a range of tumor types, including melanoma, renal cancer, astrocytoma, and Hodgkin’s disease (2), esophageal cancer (5), lung cancer (6, 7), and colon cancer (8). A large number of novel gene products have been identified, as well as antigens such as MAGE and tyrosinase that had been identified previously as tumor antigens recognized by cytotoxic T lymphocytes (2, 9, 10, 11). The current list of SEREX-defined human tumor antigens fall into several categories, including differentiation antigens, mutational antigens, overexpressed antigens, and retroviral antigens (3, 4). Of particular interest is the category of antigens that we have referred to as cancer/testis (CT) antigens (4, 5). CT antigens share the following characteristics: (i) predominant mRNA expression in testis, but generally not in other normal somatic tissues, (ii) gene activation and mRNA expression in a wide range of human tumor types, (iii) existence of multigene families, and (iv) with one exception, localization of coding genes to chromosome X. Six CT antigens or antigen families have been identified, three of them originally defined by cloning cytotoxic T lymphocyte-recognized antigens expressed by the melanoma cells of a single patient: MAGE (10, 12, 13), BAGE (14), and GAGE (15). The other three CT antigens were identified by SEREX analysis by using sera from patients with melanoma (SSX2) (2, 16), esophageal cancer (NY-ESO-1) (5), and renal cancer (SCP1) (17).
To identify additional members of the CT family, cDNA libraries of testes have been constructed and used as targets for screening, either with sera from cancer patients (17, 18) or with molecular probes identifying known CT antigens (18). Several new CT antigens, e.g., SCP1 (17) and new members of the SSX family (SSX3, 4, and 5) (18), have been uncovered in this way. During SEREX analysis of testicular libraries, however, it became clear that because of the uniquely broad transcriptional repertoire of testis, CT antigens represent only a small fraction of SEREX-identified testicular clones. To enrich CT cDNA species, Türeci et al. (17) used testicular cDNA library subtracted with mRNA from nontesticular tissues. An alternative approach aimed at identifying new CT antigens was pursued in the present study. Melanoma cell lines were screened for expression of known CT antigens, and a cDNA library was constructed from a melanoma cell line (SK-MEL-37) expressing a wide array of known CT antigens. This library was screened with serum from melanoma patient NW38, known to have high-titer antibodies to two CT antigens (19, 20). The rationale for this approach was based on two assumptions: first, SK-MEL-37 has a simpler transcriptional repertoire than testis and CT antigens may be better represented in the SK-MEL-37 cDNA library than in the testicular library; and second, sera from cancer patients with antibodies to one or more known CT antigens might be expected to be a good source of antibodies to other unidentified CT antigens. In addition, the use of cancer cell lines for SEREX analysis has other benefits, including the absence of contaminating normal cell types invariably present in tumor specimens, and the elimination of B cells that give rise to false-positive IgG-expressing clones in the expression library.
MATERIALS AND METHODS
Cell Lines and Tissues.
Established melanoma cell lines have been described previously (21, 22). Specimens of normal and tumor tissues were obtained from the Departments of Pathology at the New York Hospital–Cornell Medical Center and Memorial Sloan–Kettering Cancer Center.
RNA Extraction and Construction of cDNA Expression Library.
Total RNA was extracted from cultured cell lines and from normal and tumor tissues. A cDNA library was constructed from the SK-MEL-37 melanoma cell line in λZAP Express vector, using a commercial cDNA library kit (Stratagene).
Immunoscreening of the cDNA Library.
The cDNA library was screened with an allogeneic patient’s serum (NW38) at 1:2,000 dilution. This serum has been shown previously to contain high-titer antibody against MAGE-1 and NY-ESO-1 (19, 20). The screening procedure has been described previously (4). Briefly, the serum was diluted 1:10, preabsorbed with transfected Escherichia coli lysate, further diluted to 1:2,000, and incubated overnight at room temperature with the nitrocellulose membranes containing the phage plaques at a density of 4,000–5,000 pfu per 130-mm plate. After washing, the filters were incubated with alkaline phosphatase-conjugated goat anti-human Fcγ secondary antibodies and the reactive phage plaques were visualized by incubating with 5-bromo-4-chloro-3-indolyl-phosphate and nitroblue tetrazolium.
Sequence Analysis of the Reactive Clones.
The reactive clones were subcloned, purified, and in vitro excised to pBK-CMV plasmid forms (Stratagene). Plasmid DNA was prepared by using Wizard Miniprep DNA Purification System (Promega). The inserted DNA was evaluated by EcoRI-XbaI restriction mapping, and clones representing different cDNA inserts were sequenced. The sequencing reactions were performed by DNA Services at Cornell University (Ithaca, NY) by using Applied Biosystems PRISM (Perkin–Elmer) automated sequencers. DNA and amino acid sequences were compared with sequences in the GenBank and other public databases by using the blast program.
Reverse Transcription (RT)-PCR.
To evaluate the mRNA expression pattern of the cloned cDNA in normal and malignant tissues, gene-specific oligonucleotide primers for PCR were designed to amplify cDNA segments of 300–600 bp in length, with the estimated primer melting temperature in the range of 65–70°C. For evaluation of CT antigen expression in melanoma cell lines, primers specific for MAGE-1, MAGE-2, MAGE-3, MAGE-4, BAGE, NY-ESO-1, SSX1, SSX2, SSX4, SSX5, and SCP1 were prepared, either following previously used primer sequences (5, 17, 18, 23) or designed based on published gene sequences (12, 13). All primers were synthesized commercially (Operon Technologies, Alameda, CA). RT-PCR was performed by using 35 amplification cycles in a thermal cycler (Perkin–Elmer) at an annealing temperature of 60°C, and the products were analyzed by 1.5% gel electrophoresis and ethidium bromide visualization.
Genomic Southern Blot Analysis.
Genomic DNA was extracted from normal human tissue. After restriction enzyme digestion, the DNA was separated on a 0.7% agarose gel, blotted onto nitrocellulose filters, and hybridized to a 32P-labeled DNA probe at a high stringency condition (65°C, aqueous buffer).
RESULTS AND DISCUSSION
SK-MEL-37 Expresses a Wide Array of CT Antigens.
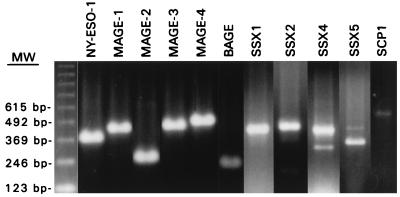
A panel of 12 melanoma cell lines were evaluated for known CT antigen expression by RT-PCR. Of these, SK-MEL-37 was found to have the broadest pattern of CT expression, being positive for MAGE-1, MAGE-2, MAGE-3, MAGE-4, BAGE, NY-ESO-1, SSX1, SSX2, SSX4, SSX5, and SCP1 (Fig. 1).
Figure 1.
RT-PCR analysis of CT antigen expression in the established melanoma cell line SK-MEL-37. SK-MEL-37 showed expression of all CT products tested, i.e., NY-ESO-1, MAGE1, MAGE-2, MAGE-3, MAGE-4, BAGE, SSX1, SSX2, SSX4, SSX5, and SCP1. The minor band of lower molecular mass in SSX4 represents an alternate-spliced variant (18).
SEREX Analysis of SK-MEL-37 cDNA Library with NW38 Serum.
An expression cDNA library of 2.3 × 107 primary clones was established, and immunoscreening was carried out by using absorbed NW38 serum at a 1:2,000 dilution. Sixty-one positive clones were identified after screening of 1.5 × 105 clones. These 61 clones were purified, excised in vitro, and converted to pBK-CMV plasmid forms. cDNA inserts were analyzed and grouped based on a combined strategy of restriction mapping, DNA sequencing, and DNA–DNA hybridization, and the results are summarized in Table 1. Excluding the miscellaneous group, which consisted of 10 clones derived from 9 distinct genes, 4 known and 5 unknown, the remaining 51 clones belonged to 4 distinct groups of tumor products: the KOC family, the MAGE family, the NY-ESO-1 family, and a new CT antigen gene, designated CT7. The isolation of four CT antigen genes—MAGE-4a, NY-ESO-1, LAGE-1, and CT7—after screening only 1.5 × 105 cDNA clones represents a frequency that has not been observed in SEREX analyses to date. For example, a parallel screening of NW38 serum against a testicular library yielded only two MAGE-4a clones after screening of 5.0 × 105 clones, but no other CT-coding clones. This result provides support for our assumption that melanoma cell lines such as SK-MEL-37 may well be a better source than testis for identifying CT cDNA clones.
Table 1.
SEREX-defined genes identified by allogeneic screening of SK-MEL-37 cDNA expression library
| Gene group | No. of clones | Comments |
|---|---|---|
| KOC | 33 | Derived from three related genes, overexpressed antigen |
| MAGE | 11 | Predominantly MAGE-4a (see text) |
| NY-ESO-1 | 5 | Derived from two related genes (NY-ESO-1, LAGE-1) |
| CT7 | 2 | New cancer/testis antigen (see text) |
| Miscellaneous | 2 | S-adenyl homocysteine hydrolase |
| 1 | Glutathione synthetase | |
| 1 | Proliferation-associated protein p38-2G4 | |
| 1 | Phosphoribosyl pyrophosphate synthetase-associated protein 39 | |
| 1 | Unknown gene, identical to ESTs from pancreas, uterus, etc. | |
| 1 | Unknown gene, identical to ESTs from lung, brain, fibroblast, etc. | |
| 1 | Unknown gene, identical to ESTs from multiple tissues | |
| 1 | Unknown gene, identical to ESTs from pancreas and fetus | |
| 1 | Unknown gene, no identical EST sequence, universally expressed |
EST, expressed sequence tags.
The KOC Gene Family.
The first and by far the predominant group, consisting of 33 clones, was related to KOC (KH-domain containing gene overexpressed in cancer) gene, a gene shown to be overexpressed in pancreatic cancer and mapped to chromosome 7p11.5 (24). Among the 33 clones, 2 were derived from the KOC gene, whereas the other 31 clones were derived from two previously unidentified closely related genes, indicating that KOC belongs to a gene family with at least three expressed members. The KOC gene contains an ORF of 1740 bp, encoding a protein of 579 aa (Mr 65 kDa). The two other homologous genes encode proteins slightly different in size, with 60–70% amino acid homology among the three gene products. Alternate splicing forms were observed in one of the two KOC-like genes, but not in the others. In the original study by Müller-Pillasch et al. (24), Northern blot analysis showed that the KOC expression was restricted to placenta and was not found in heart, brain, lung, liver, kidney, pancreas, or skeletal muscle. By RT-PCR analysis, however, we have observed significant levels of KOC mRNA expression in testis and in nontesticular normal tissues, including liver, colon, kidney, and brain. In this regard, the expression pattern of KOC resembles the unrelated cytotoxic T lymphocyte-defined antigen PRAME (25), i.e., restricted expression by Northern blotting and ubiquitous expression by the more sensitive RT-PCR assay. A detailed description of these findings regarding KOC family genes will be reported elsewhere.
The MAGE Family.
The second group consisting of 11 clones were derived from genes belonging to the MAGE family. Sequencing of five representative clones revealed overlapping sequences, all of them derived from the MAGE-4a gene (ref. 14, GenBank accession no. U10687). The other six clones showed positive dot blot hybridization to a MAGE-4a probe derived from the 5′ sequences, indicating that they belonged to the MAGE family (data not shown). Restriction mapping further suggested that these clones probably were all derived from MAGE-4a, as they all shared the same EcoRI site, which is present at the 3′ end of the MAGE-4a cDNA (nucleotide position 10932, GenBank accession no. U10687).
It is of interest that MAGE-4a, but not other MAGE genes, were isolated in the present study. Although MAGE-1 has been isolated by SEREX (2), and NW38 serum reacts with MAGE-1 recombinant protein in vitro (19), MAGE-4a appears to be more frequently detected by SEREX. MAGE-4a has been isolated from an ovarian cancer library by autologous screening (3) and also from testicular and SK-MEL-37 libraries with NW38 serum. In contrast, MAGE-1 has only been isolated once (2), and the products of other MAGE family genes have not been detected by SEREX. Although one can speculate that MAGE-4a mRNA may be more abundant than other MAGE family genes, the fact that MAGE-4a has been isolated from cDNA libraries derived from different tissue sources—testis, ovarian cancer, and a melanoma cell line—makes this simple explanation unlikely. It is therefore possible that MAGE-4a is significantly more immunogenic to the humoral immune system than other MAGE members.
The NY-ESO-1 Family.
The third group consisted of five clones from the NY-ESO-1 family. Two clones were identical to the NY-ESO-1 gene that we described previously (5). The other three clones were derived from a second gene of the NY-ESO-1 family. This gene, sharing 94% nucleotide and 87% amino acid homology to NY-ESO-1, previously has been identified by Léthe et al. (26) by using representational difference analysis comparing testicular vs. nontesticular mRNA and has been designated as LAGE-1. This NY-ESO-1-related gene has also been isolated by nucleotide hybridization with a NY-ESO-1 probe (unpublished data).
Although the LAGE-1 protein shares strong homology to NY-ESO-1, there has been no direct evidence that LAGE-1 is immunogenic in tumor-bearing patients. Isolation of LAGE-1 by SEREX in the present study documents that the LAGE-1 product, similar to NY-ESO-1, is recognized by the humoral immune system. However, because NY-ESO-1 was also identified in this screening, it is still possible that only one of the two NY-ESO-1 genes was primarily responsible for eliciting the antibody response in patient NW38, and that the other gene was isolated as a consequence of antibody cross-reactivity.
The CT7 Gene.
The fourth group consists of two clones derived from a novel gene and this gene has been designated CT7 (4).¶
Two SEREX-reactive CT7 clones, MNW16b and MNW25c, were identified. MNW25c contained a cDNA insert of 2,184 bp, 219 bp longer than MNW16b. An ORF of 543 aa was identified in MNW25c, extending to the 5′ end of the cloned sequence, indicating that this is a partial cDNA clone. To obtain complete cDNA sequences and to seek related gene members, a human testicular cDNA library was screened with probes derived from MNW25c. Eleven positive clones were identified, and sequencing data from the six longest clones indicated that they were all derived from the same gene. The full-length CT7 transcript, excluding poly(A) tail, consists of 4,265 nt, i.e., 286 bp of 5′ untranslated region, 550 bp of 3′ untranslated region, and a coding region of 3,429 bp (GenBank accession no. AF056334). The predicted protein, 1,142 residues in length, has a predicted molecular mass of 123,872 Da.
DNA and protein sequence homology analysis indicated strongest homology with MAGE family genes, MAGE-10 in particular (13). The region of homology, however, was limited strictly to the ≈210 aa stretch at the carboxyl ends of these two gene products, specifically, amino acids 908-1115 of CT7 and 134–342 of MAGE-10 (GenBank accession no. P43363). Despite the 56% amino acid homology (75% including conservative changes) in this region, no homology was identified 5′ to this sequence, with the predicted CT7 protein (1,142 aa residues) much larger than the MAGE-10 protein (369 residues).
A unique feature of CT7, dissimilar to MAGE or any other known genes, resides in the 5′ region. Examination of the nucleotide and amino acid sequences in this region revealed a strikingly repetitive pattern, as illustrated in Fig. 2. The repeats, although inexact, appear to be rich in serine, proline, glutamine, and leucine residues, with an almost invariable (P)QS(P)LQ(I) core. The most consistent repetitive element is located in the middle of the molecule, where 10 almost-exact repeats of 35 aa residues were observed. Overall the repetitive portion of this molecule comprises ≈70% of the entire sequence, initiating shortly after the translational initiation codon (≈amino acid position 15) and ending shortly before the MAGE-homologous region. A highly repetitive coding structure previously has been found by us in a gene coding for CDR34, a cerebellar-degeneration-related 34-kDa protein that we isolated by antibody screening of a cerebellar library, with serum from a patient with paraneoplastic cerebellar degeneration (27). The CDR34 gene contains a highly repetitive element consisting of 34 inexact tandem repeats of 6 aa. CT7 and CDR34 are structurally unrelated, and the observation that they both contain tandem repeats and were both isolated by antibody screening suggests that molecules with this tandem-repeat feature may be highly immunogenic to the humoral immune system.
Figure 2.
Predicted amino acid sequence of CT7, illustrating the repetitive structure encoded by the 5′ sequences of this gene. The sequence has a number of repeating elements, most of them containing a (P)QS(P)LQ(I) core sequence. The most highly conserved repeating element consisted of a 35-aa unit that was repeated 10 times in tandem (amino acid positions 125–475). The carboxyl-end sequence of 208 aa (908–1115), which is homologous to MAGE-10 gene, is underlined.
Restricted CT7 Expression in Normal and Tumor Tissues.
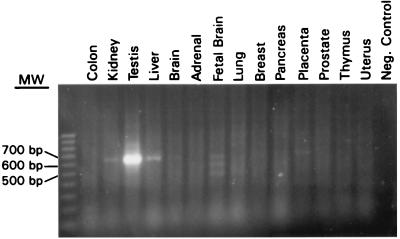
RT-PCR assays were used to evaluate CT7 mRNA expression in normal tissues, tumor cell lines, and tumor samples. Of 14 normal tissues tested, strong expression was identified only in testis, and no expression was detected in colon, brain, adrenal, lung, breast, pancreas, prostate, thymus, or uterus. Trace amounts of RT-PCR products on ethidium bromide-stained gels, were seen in liver, kidney, placenta, and fetal brain (Fig. 3). Fetal brain showed three transcripts of different sizes; the two additional bands of lower molecular weight, however, were proven to be nonspecific amplification products. The level of mRNA expression in somatic tissues, estimated from the intensities of signals, was at least 20- to 50-fold lower than that in the testis.
Figure 3.
RT-PCR analysis of CT7 expression in normal tissues. High-level expression is seen only in testis. Trace amounts of PCR products were detected in kidney, liver, placenta, and fetal brain. Two additional bands of lower molecular mass also were seen in fetal brain; by sequencing, these products were proven to be nonspecific amplification products.
Of 12 melanoma cell lines examined, 7 showed strong expression (NW38, SK-MEL-13, 19, 23, 30, 37, 179), one showed weak expression (SK-MEL-33), and 4 were negative (MZ2-MEL-3.1, MZ2-MEL-2.2, SK-MEL-29, SK-MEL-31).
Table 2 summarizes mRNA expression pattern of CT7 in malignant tumors of various types. Of 70 specimens tested, CT7 transcripts were detected in 26 of the cases (37%). Similar to our experience with other CT antigens (ref. 28 and unpublished results), the level of transcript varied substantially among positive samples, with low-level expression (signal intensities estimated <1/10 of the stronger expressers using two different primer pairs) seen in 12 of the 26 positive specimens: 2 of 7 positive melanomas, 1 of 3 breast cancers, 1 of 3 lung cancers, 3 of 5 head and neck cancers, 1 of 4 transitional cell carcinomas, 1 of 1 leiomyosarcoma, 2 of 2 synovial sarcomas, and 1 of 1 colon cancer. The overall frequency of tumors with strong CT7 expression is thus 20% (14/70) in this group.
Table 2.
CT7 mRNA expression in various human tumors by RT-PCR
| Tumor type | mRNA, positive/total |
|---|---|
| Melanoma | 7/10 |
| Breast cancer | 3/10 |
| Lung cancer | 3/9 |
| Head/neck cancer | 5/14 |
| Bladder cancer | 4/9 |
| Colon cancer | 1/10 |
| Leimyosarcoma | 1/4 |
| Synovial sarcoma | 2/4 |
| Total | 26/70 |
Southern Blot Analysis of CT7.
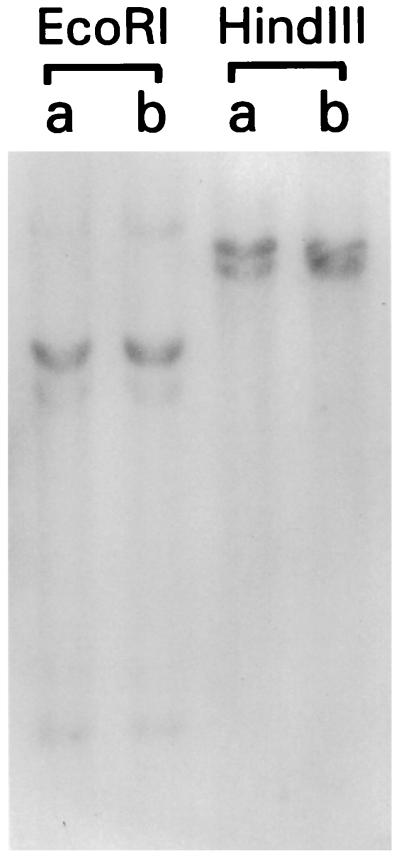
Genomic Southern blot analysis with CT7 probe showed two to four bands in EcoRI, and HindIII digests, suggesting the possibility of two genes (Fig. 4). However, sequencing of six testicular cDNA clones showed identical sequences in overlapping regions, with the only variation at nucleotide position 360 (GenBank accession no. AF056334). Of four testicular clones containing this region, this position was adenine in two clones, and guanine in two other clones. This difference, most likely representing alleleic polymorphism, also would result in a corresponding variation in the amino acid sequence, i.e., tyrosine (TAT) versus cysteine (TGT).
Figure 4.
Southern blot analysis of CT7 gene. Genomic DNA extracted from normal tissues of two individuals were digested with EcoRI and HindIII and analyzed with a CT7 probe derived from the MAGE-unrelated 5′ sequences. Two bands of similar intensity were seen in HindIII digests, whereas EcoRI digests showed one strong band and three weaker bands, indicating that CT7 does not belong to a multigene family.
CT7 and MAGE-C1.
Using representational difference analysis to identify genes selectively expressed in testis and melanoma, Lucas et al. (29) recently defined a gene with >99% identity to CT7 in coding sequences, which they designated as MAGE-C1. CT7 sequences differed from MAGE-C1 in having 30 additional nucleotides in the 5′ untranslated region, and also in 14 single nucleotide differences in the coding region, resulting in 11 corresponding amino acid differences. Ten of 11 different amino acid residues clustered in 2 of 10 35-aa core repeating units, and CT7 and MAGE-C1 probably represent different alleles of the same gene. MAGE-C1 has been mapped to chromosome Xq26 (29) and therefore joins the other CT-coding genes that also have been mapped to the X chromosome: MAGE, GAGE, SSX, and NY-ESO-1.
CONCLUSION
As illustrated in this study, the serological screening of cDNA expression libraries prepared from established human cancer cell lines (rather than tumor specimens) offers new opportunities for analyzing the repertoire of the humoral immune response to human cancer. Selection of the appropriate cell line and the source of serum of course is critical in this approach, and in the present effort to define additional CT antigens we focused on a melanoma cell line known to be rich in CT antigen expression and a human serum known to have antibodies to at least two CT antigens. This search for new CT antigens is being extended to other cancer cell lines with a different profile of CT antigen expression and sera from other patients with a different serological reactivity pattern to CT antigens. The development of ELISA methodology for broad scale screening of human sera for antibodies to known CT antigens (19) facilitates the identification of patients with high-titered serological responses to CT antigens.
ABBREVIATIONS
- CT
cancer/testis
- SEREX
serological analysis of recombinant cDNA expression libraries
- RT-PCR
reverse transcription–PCR
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF056334).
Because the function of only one CT antigen, SCP1, is known, a standardized nomenclature for these antigens has not been established. We suggest that new CT antigens be numbered in the order of their discovery, e.g., CT7 for the seventh CT antigen or antigen family to be identified. In the case of CT antigens belonging to a multigene family, each member would be distinguished by a number after the CT designation, e.g., CT7.1, CT7.2, etc.
References
- 1.Oettgen H F, Old L J. In: Biologic Therapy of Cancer. DeVita V T Jr, Hellman S, Rosenberg S A, editors. Philadelphia: Lippincott; 1991. [Google Scholar]
- 2.Sahin U, Türeci Ö, Schmitt H, Cochlovius B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I, Pfreundschuh M. Proc Natl Acad Sci USA. 1995;92:11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sahin U, Türeci Ö, Pfreundschuh M. Curr Opin Immunol. 1997;9:709–716. doi: 10.1016/s0952-7915(97)80053-2. [DOI] [PubMed] [Google Scholar]
- 4.Old L J, Chen Y-T. J Exp Med. 1998;187:1163–1167. doi: 10.1084/jem.187.8.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y-T, Scanlan M, Sahin U, Türeci Ö, Gure A O, Tsang S, Williamson B, Stockert E, Pfreundschuh M, Old L J. Proc Natl Acad Sci USA. 1997;94:1914–1918. doi: 10.1073/pnas.94.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brass N, Heckel D, Sahin U, Pfreundschuh M, Sybrecht G W, Meese E. Hum Mol Genet. 1997;6:33–39. doi: 10.1093/hmg/6.1.33. [DOI] [PubMed] [Google Scholar]
- 7.Güre A O, Altorki N K, Stockert E, Scanlan M J, Old L J, Chen Y-T. Cancer Res. 1998;58:1034–1041. [PubMed] [Google Scholar]
- 8.Scanlan, M. J., Chen, Y.-T., Williamson, B., Gure, A. O., Stockert, E., Gordan, J. D., Türeci, Ö., Sahin, U., Pfreundschuh, M. & Old, L. J. (1998) Int. J. Cancer, in press. [DOI] [PubMed]
- 9.van der Bruggen P, Traversari C, Chomez P, Lurquin C, Deplaen E, Van den Eynde B, Knuth A, Boon T. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 10.Brichard V A, Van Pel A, Wölfel T, Wölfel C, De Plaen E, Léthe B, Coulie P, Boon T. J Exp Med. 1993;178:489–495. doi: 10.1084/jem.178.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robbins P F, El-Gamil M, Kawakami Y, Rosenberg S A. Cancer Res. 1994;54:3124–3126. [PubMed] [Google Scholar]
- 12.Gaugler B, Van den Eynde B, van der Bruggen P, Romero P, Gaforio J, De Plaen E, Lethé B, Brasseur F, Boon T. J Exp Med. 1994;179:921–930. doi: 10.1084/jem.179.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Plaen E, Arden K C, Traversari C, Gaforio J, Szikora J-P, De Smet C, Brasseur F, van der Bruggen P, Lethé B, De Backer O, et al. Immunogenetics. 1994;40:360–369. doi: 10.1007/BF01246677. [DOI] [PubMed] [Google Scholar]
- 14.Boel P, Wildmann C, Sensi M L, Brasseur R, Renauld J, Coulie P, Boon T, van der Bruggen P. Immunity. 1995;2:167–175. doi: 10.1016/s1074-7613(95)80053-0. [DOI] [PubMed] [Google Scholar]
- 15.Van den Eynde B, Peeters O, De Backer O, Gaugler B, Lucas S, Boon T. J Exp Med. 1995;182:689–698. doi: 10.1084/jem.182.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Türeci Ö, Sahin U, Schobert I, Koslowski M, Schmitt H, Schild H J, Stenner F, Seitz G, Rammensee H G, Pfreundschuh M. Cancer Res. 1996;56:4766–4772. [PubMed] [Google Scholar]
- 17.Türeci, Ö., Sahin, U., Zwick, C., Koslowski, M., Seitz, G. & Pfreundschuh, M. (1998) Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 18.Güre A O, Türeci Ö, Sahin U, Tsang S, Scanlan M J, Jäger E, Knuth A, Pfreundschuh M, Old L J, Chen Y-T. Int J Cancer. 1997;72:965–971. doi: 10.1002/(sici)1097-0215(19970917)72:6<965::aid-ijc8>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 19.Stockert E, Jäger E, Chen Y-T, Scanlan M J, Gout I, Karbach J, Arand M, Knuth A, Old L J. J Exp Med. 1998;187:1349–1354. doi: 10.1084/jem.187.8.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jäger E, Chen Y-T, Drijfout J W, Karback J, Ringhoffer M, Jäger D, Arand M, Wada H, Noguchi Y, Stockert E, Old L J, Knuth A. J Exp Med. 1998;187:265–270. doi: 10.1084/jem.187.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carey T E, Lloyd K O, Takahashi T, Travassos L R, Old L J. Proc Natl Acad Sci USA. 1979;76:2898–2902. doi: 10.1073/pnas.76.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van den Eynde B, Hainaut P, Herin M, Knuth A, Lemoine C, Weynants P, van der Bruggen P, Fauchet R, Boon T. Int J Cancer. 1989;44:634–640. doi: 10.1002/ijc.2910440413. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y-T, Stockert E, Chen Y, Garin-Chesa P, Rettig W J, van der Bruggen P, Boon T, Old L J. Proc Natl Acad Sci USA. 1994;91:1004–1008. doi: 10.1073/pnas.91.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller-Pillasch F, Lacher U, Wallrapp C, Micha A, Zimmerhackl F, Hameister H, Varga G, Freiss H, Büchler M, Beger H G, Vila M R, Adler G, Gress T M. Oncogene. 1997;14:2729–2733. doi: 10.1038/sj.onc.1201110. [DOI] [PubMed] [Google Scholar]
- 25.Ikeda H, Lethé B, Lehmann F, Van Baren N, Baurain J-F, De Smet C, Chambost H, Vitale M, Moretta A, Boon T, Coulie P G. Immunity. 1997;6:199–208. doi: 10.1016/s1074-7613(00)80426-4. [DOI] [PubMed] [Google Scholar]
- 26.Lethé, B., Lucas, S., Michaux, L., De Smet, C., Godelaine, D. & Boon, T. (1998) Int. J. Cancer, in press. [DOI] [PubMed]
- 27.Dropcho E J, Chen Y-T, Posner J B, Old L J. Proc Natl Acad Sci USA. 1987;84:4552–4556. doi: 10.1073/pnas.84.13.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Türeci, Ö., Chen, Y.-T., Sahin, U., Güre, A. O., Zwick, C., Villena, C., Tsang, S., Seitz, G., Old, L. J. & Pfreundschuh, M. (1998) Int. J. Cancer, in press. [DOI] [PubMed]
- 29.Lucas S, De Smet C, Arden K C, Viars C S, Lethé B, Lurquin C, Boon T. Cancer Res. 1998;58:743–752. [PubMed] [Google Scholar]