Abstract
Plants have a sophisticated system for sensing and responding to their light environment. The light responses of populations and species native to different habitats show adaptive variation; understanding the mechanisms underlying photomorphogenic variation is therefore of significant interest. In Arabidopsis thaliana, phytochrome B (PHYB) is the dominant photoreceptor for red light and plays a major role in white light. Because PHYB has been proposed as a candidate gene for several quantitative trait loci (QTLs) affecting light response, we have investigated sequence and functional variation in Arabidopsis PHYB. We examined PHYB sequences in 33 A. thaliana individuals and in the close relative Arabidopsis lyrata. From 14 nonsynonymous polymorphisms, we chose 5 for further study based on previous QTL studies. In a larger collection of A. thaliana accessions, one of these five polymorphisms, I143L, was associated with variation in red light response. We used transgenic analysis to test this association and confirmed experimentally that natural PHYB polymorphisms cause differential plant responses to light. Furthermore, our results show that allelic variation of PHYB activity is due to amino acid rather than regulatory changes. Together with earlier studies linking variation in light sensitivity to photoreceptor genes, our work suggests that photoreceptors may be a common target of natural selection.
Keywords: hypocotyl, linkage disequilibrium, natural variation
Plants use three types of photoreceptors to survey their light environment: phytochromes for red and far-red light and cryptochromes and phototropins for blue light (1). Changes in the light environment sensed through these receptors affect many aspects of plant development. The phytochrome family in Arabidopsis thaliana consists of five genes, PHYA-PHYE, with partially redundant developmental functions (2–10). At the seedling stage, phytochromes regulate emergence from the soil. Seedlings that germinate underground or in the dark cannot photosynthesize and extend their hypocotyl upwards toward the soil surface. Light is a cue that the soil surface has been reached; light perception, therefore, causes inhibition of hypocotyl elongation and the beginning of photoautotrophic growth. The primary photoreceptors for this response are PHYA in far-red light, PHYB in red light, and cryptochromes in blue light. Later in development, plants use PHYB, PHYD, and PHYE to detect neighbor proximity by monitoring the ratio of red to far-red (R/FR) light (4, 5). Because chlorophyll absorbs red but not far-red light, low R/FR ratios indicate close neighbors or canopy shade. Neighbor perception can induce a variety of competitive responses, including stem and leaf petiole elongation and early reproduction, collectively called the shade-avoidance syndrome. Notably, the proper response to light depends on whether or not the plant is native to that environment. For example, the ability of low R/FR to induce shade-avoidance responses is reduced in species and populations normally growing under shady conditions (11), an example of adaptive variation in phytochrome-mediated responses (12, 13).
Phytochromes exist in two photoconvertible forms: Pr, an inactive red-light absorbing form, and Pfr, the active far-red light absorbing form. In sunny conditions, which are characterized by high R/FR ratios, most phytochrome is in the active Pfr form. Shade causes a decrease in Pfr and a concomitant induction of shade-avoidance responses. Phytochromes signal through a web of downstream factors, including a family of related bHLH transcription factors, the PIFs and PILs (14), GIGANTEA (GI) (15), and the bZIP transcription factors HY5 and HYH that integrate signals from multiple photoreceptor pathways (16). Light-regulated protein degradation, often mediated by the E3 ubiquitin ligase COP1 and associated proteins (17), also is important for phytochrome signaling.
An evolutionary question of considerable interest is how selection acts on the different components of metabolic and developmental pathways. For example, in the case of the anthocyanin biosynthesis pathway, which is responsible for producing flower color pigments, it has been found that genes acting later in the pathway evolve more quickly than those upstream (18, 19). Interestingly, for interpretation of light signals, a number of changes have been found at the top of the pathway in the photoreceptors themselves. For example, reduced FR sensitivity of the A. thaliana strain Lm-2 was traced to a single amino acid change in PHYA that reduced photoconversion and autophosphorylation (20). Cloning of an A. thaliana flowering time quantitative trait loci (QTL) revealed that it was caused by an amino acid substitution that stabilized the CRY2 protein, thereby increasing activation of the photoperiod pathway (21). Variation in PHYC is responsible for differences in both flowering time and hypocotyl elongation among A. thaliana accessions (22), and a naturally occurring deletion in PHYD increases stem elongation (3). More generally, phytochrome genes evolve more quickly than the average plant gene (23), and there is evidence for positive selection early in the diversification of PHYA (24).
Although phenotypic variation due to polymorphisms in A. thaliana PHYA (20), PHYC (22), and PHYD (3) has been described, PHYB plays by far the largest role for white-light responses in standard laboratory strains. In addition, PHYB is a candidate for a white and red light QTL in A. thaliana (25), for a flowering time QTL in tomato (26), and a bud set QTL in poplar (27, 28). Here, we examine the effect of natural variation in PHYB on differential light response in A. thaliana and find that polymorphisms in PHYB proteins contribute to variation in A. thaliana light sensitivity.
Results and Discussion
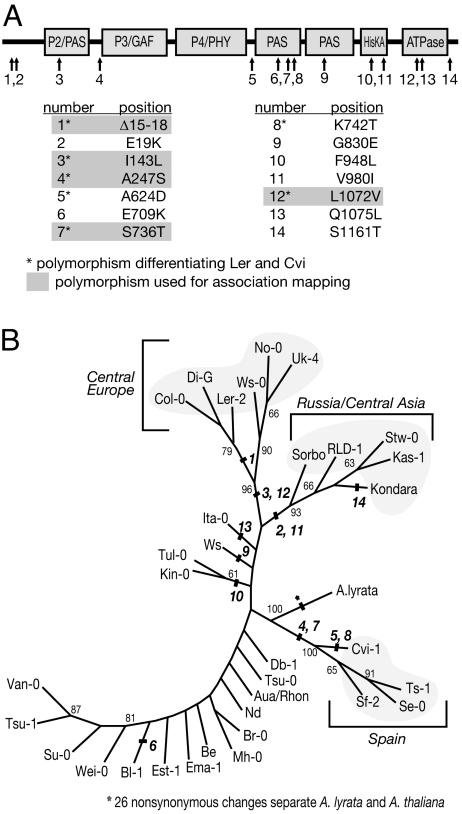
To determine whether polymorphisms in PHYB could contribute to variation in light response, we began by examining PHYB sequence diversity in Arabidopsis. We sequenced the PHYB gene from Ler and Cvi, two accessions known to segregate for LIGHT2, a QTL coincident with PHYB (25); 18 additional A. thaliana accessions; and the sister species A. lyrata. In our sequence analysis, we also included 14 A. thaliana accessions for which PHYB sequences were available in GenBank (29). In A. thaliana, we found 65 synonymous and 14 nonsynonymous polymorphisms. An additional 15 polymorphisms were identified in introns. Of the 14 nonsynonymous sites, 7 differentiated Ler and Cvi (Fig. 1 A and B). A maximum likelihood phylogenetic tree revealed three well supported clades: one clade of six accessions contained the standard laboratory strains Col and Ler, which originate from Central Europe; another one included five Russian and Central Asian accessions; and the third clade comprised of four Spanish accessions (Fig. 1B). The remaining 18 accessions grouped into a large, poorly resolved clade. In general, the pattern seen is consistent with previously reported genomic patterns of isolation by distance (30, 31). A similar tree was obtained by using neighbor-joining methods.
Fig. 1.
Sequence diversity in A. thaliana PHYB. (A) Amino acid changes in A. thaliana PHYB. A schematic diagram of PHYB domains is shown, with arrows indicating each amino acid polymorphism. The table below indicates the specific amino acid change. The amino acid to the left of the number indicates the residue in the reference sequence from the Columbia (Col-0) accession. (B) Maximum likelihood phylogenetic tree, based on nucleotide sequence, showing PHYB divergence in A. thaliana. Bold italic numbers indicate where amino acid variants separate different branches. Plain text numbers in smaller font indicate bootstrap support.
Because the 14 nonsynonymous polymorphisms all fell outside the functionally important GAF and PHY domains (Fig. 1A), we asked whether this pattern could be explained by selection. We modeled sequence evolution, using maximum likelihood methods to assess whether substitution rates differed between the PHY and GAF domains, which are important for chromophore binding and photoconversion (32), and the rest of the protein. For codon position 3, where polymorphisms are usually silent, there was no evidence for different rates (P = 0.95). However, the substitution rate for codon positions 1 and 2, where polymorphisms frequently cause amino acid changes, was significantly lower in the GAF and PHY domains (P = 0.009) compared with the rest of the protein. We only found evidence for depressed substitution rates at positions 1 and 2, suggesting that the reduced sequence diversity in the PHY and GAF domains is due to selection on protein sequence and function, rather than local variation in mutation rate.
The observed distribution of nonsynonymous sites might reflect that the GAF and PHY domains are more highly constrained than the rest of the protein. However, we found no evidence that mutations induced in the laboratory and known to compromise PHYB function unevenly affected the PHY and GAF domains: 12 of 29 missense mutations are in the GAF or PHY domains (32; P = 0.4554, Fisher's exact test, comparing mutation rate in the GAF and PHY domains with the rest of the protein). Thus, with respect to overall activity, the GAF and PHY domains are unlikely to be more highly constrained than the rest of the protein. An alternative explanation is that the observed pattern is a result of adaptive evolution, indicating that domains other than the PHY and GAF are better targets for selection. We used the McDonald–Kreitman test (33) to look for evidence of nonneutral selection. Although there is an excess of nonsynonymous polymorphisms that are fixed between species compared with ones that segregate within species, this excess is not significant [supporting information (SI) Table 1; P > 0.26], so the underlying cause of the observed substitution pattern remains unresolved.
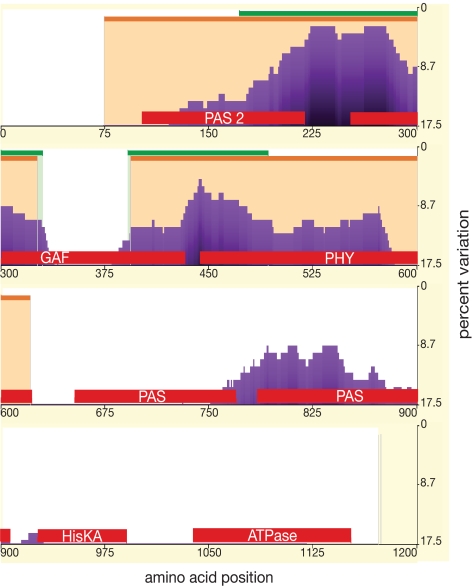
To examine whether the differential distribution of polymorphic sites has persisted over a greater evolutionary distance, a phylogenetic shadowing (34) approach was taken. Using eShadow (35), both divergence threshold (DT) and hidden Markov model islands (HMMI) methods were used to identify conserved regions in four PHYB protein sequences from the Brassicales and 16 further eudicot sequences available in GenBank. Within the Brassicales, the HMMI algorithm predicted conservation in regions spanning most of the P2/PAS, GAF, and PHY domains (Fig. 2), with one interesting exception, an ≈50-aa region in the center of the GAF domain. This less conserved region corresponds to the portion of the GAF domain in bacterial phytochrome that contributes to the light-sensing knot structure (36). Compared with the HHMI method, the DT approach identified a slightly smaller conserved region from the C-terminal third of the P2/PAS domain to approximately the first 50 aa of the PHY domain, again excluding the light-sensing knot region in the GAF domain. Using eudicot sequences, similar patterns of conservation were seen with both the DT and HMMI models (SI Fig. 6). In these more divergent sequences, conservation is limited to the GAF domain excluding the light-sensing knot and the N-terminal end of the PHY domain. In summary, the patterns over longer time scales are predictive for the variation seen within A. thaliana, suggesting that the PHY and GAF domains are the most slowly-evolving domains during evolution. That the GAF contribution to the phytochrome knot is not conserved suggests that primary sequence may not be a particularly important determinant for this structural feature.
Fig. 2.
Phylogenetic shadowing. Shown is the percentage variation by amino acid position for four Brassicales PHYB sequences. Regions determined to be slow-evolving are indicated by orange bars (HMMI) or green bars (DT). Functional domains are marked with red bars.
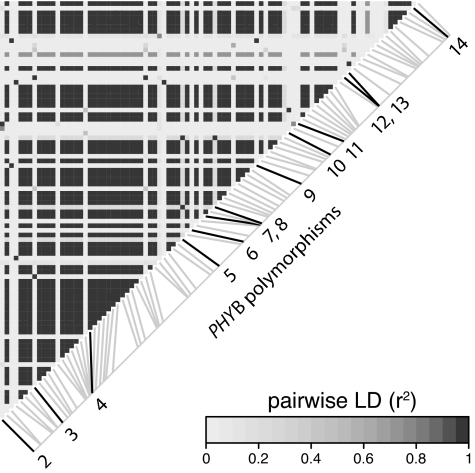
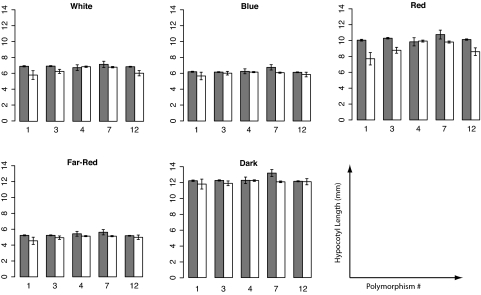
We next asked whether any of the observed sequence variation could impact PHYB activity. Association or linkage disequilibrium (LD) mapping uses historical recombination events in natural populations to associate polymorphisms with phenotypic variation (37). We used this technique to address whether any of the amino acid polymorphisms between A. thaliana accessions Ler and Cvi are associated with natural variation in light response. A panel of 140 accessions that had been phenotyped for seedling light responses (20) was genotyped for the five nonsingleton polymorphisms that distinguish Ler and Cvi. A whole-genome survey of LD in A. thaliana has shown that haplotype blocks are typically gene-sized (38). Consistent with this finding, there is strong LD across the PHYB gene (Fig. 3). Polymorphisms 1, 3, and 12 were specifically found to be associated with differences in hypocotyl elongation in red light (P < 0.01, P < 0.005, and P < 0.05, respectively; Fig. 4). Red light is precisely the condition where variation in PHYB would be most easily detected, suggesting that these associations are meaningful.
Fig. 3.
PHYB linkage disequilibrium. Pairwise linkage disequilibrium (r2) between single nucleotide polymorphisms in the A. thaliana PHYB gene. Polymorphic sites are arranged along the diagonal from lower left (5′) to upper right (3′). Numbered polymorphisms highlighted by darker lines correspond to the numbered amino acid polymorphisms in Fig. 1. To read the LD between any two sites, trace a column upwards from the 5′ site and a row leftward from the 3′ site; the square in which the column and row intersect indicates the LD.
Fig. 4.
Phenotypic association with PHYB polymorphisms. For each condition and polymorphism the bar height indicates average hypocotyl length of accessions with Ler (gray) or Cvi (white) alleles. Hypocotyl data are from ref. 21 and represent an average of 14 hypocotyls per condition for each of 140 accessions. Error bars indicate the SEM.
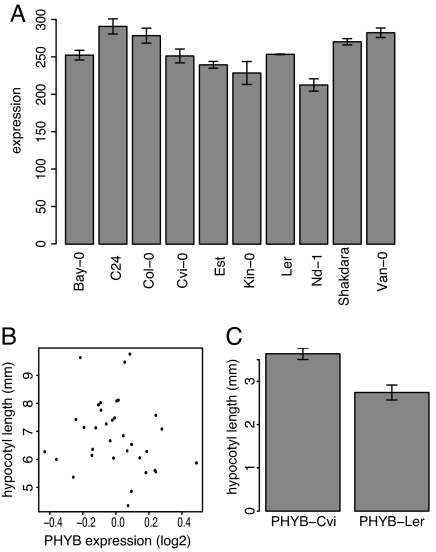
Because of the potentially confounding effects of population structure (39), the PHYB association can only be suggestive of being causal for differential light response across accessions. Therefore, we decided to compare the function of PHYB-Ler and PHYB-Cvi experimentally. We first asked whether differences in PHYB activity (if any) would be more likely due to coding or regulatory changes. Three findings suggested that regulatory differences might not be important. First, protein blots did not show any obvious differences in PHYB levels among accessions (data not shown). Second, an analysis of published microarray data (40) revealed that PHYB mRNA levels are similar across accessions, with no significant difference between Ler and Cvi (P > 0.8), and that there was no correlation between PHYB mRNA levels and hypocotyl elongation (Fig. 5 A and B). Third, an eQTL study that used microarrays to examine expression differences in Ler, Cvi, and a derived mapping population found no evidence for differential PHYB expression (41). Therefore, we concluded that if PHYB was a source of variation in light sensitivity, then amino acid changes, rather than regulatory polymorphisms, between PHYB-Ler and PHYB-Cvi were most likely the cause.
Fig. 5.
PHYB protein variants confer different light sensitivities. (A) PHYB mRNA expression levels in 4-day-old seedlings, as determined on replicated microarrays (40). (B) Correlation between mean-centered PHYB mRNA expression levels and hypocotyl length. This set includes both replicated and unreplicated microarray measurements. (C) Hypocotyl length of phyB-9 T2 plants transgenic with either phyB-Ler or phyB-Cvi. An average of 56 plants from each of 40 (phyB-Ler) or 44 (phyB-Cvi) independent single-insertion T1 lines was measured (4,711 plants total). Error bars indicate SEM.
To examine possible differences in protein activity, we linked PHYB cDNAs from both Ler and Cvi to the constitutive Cauliflower Mosaic Virus 35S promoter (35S), uncoupling PHYB function from any possible promoter differences. We transformed the null mutant phyB-9, which has greatly reduced light sensitivity and tall hypocotyls. Based on the allelic effects of the LIGHT2 QTL (25) and our association mapping results, 35S::PHYB-Ler was expected to produce a more active PHYB than 35S::PHYB-Cvi. Indeed, hypocotyls of phyB-9 35S::PHYB-Ler plants were significantly shorter than those of phyB-9 35S::PHYB-Cvi (P < 0.008 for T1 generation, and P < 0.0008 for T2 generation plants; Fig. 5C), indicating that PHYB-Cvi confers less light responsiveness than PHYB-Ler in a phyb-9 (Col) background. It is possible that this difference is due to genetic interactions between the Col background and the different PHYB alleles. However, because PHYB-Cvi is associated with reduced light response across accessions and no loci were found to be epistatic with LIGHT2 in Ler X CviQTL analysis, it is more likely that the reduced response of PHYB-Cvi is inherent to the protein. In summary, PHYB polymorphisms are likely responsible for the significant associations across accessions and at least partially responsible for the LIGHT2 QTL.
We used regression analysis to determine which of the three polymorphisms identified as significant in association mapping are most likely to be responsible for differences in PHYB activity. We regressed the polymorphisms both individually and in combination against the light responses. This process identified polymorphism 3 as the strongest candidate for the causative change. Specifically, models that included polymorphism 3 in combination with either 1 or 12 fit the data significantly better than models with polymorphism 1 or 12 alone. However, the combination of polymorphisms 3 plus 1 or 3 plus 12 did not provide any improvement over polymorphism 3 as the only explanatory variable (SI Table 2). Polymorphism 3 falls within a moderately conserved region of Brassicaceae PHYB. Comparison of the A. thaliana PHYB and PHYA sequences with the bacterial phytochrome DrBphP sequence indicated that this polymorphism lies within the P2/PAS domain of the photosensory core (36). By superimposing secondary structural elements of bacterial phytochrome on Arabidopsis PHYB sequence, we found that polymorphism 3 was predicted to be on the surface in a span of three α-helices, away from the chromophore binding pocket. This surface is speculated to be involved in protein–protein interactions (36), suggesting a possible mode-of-action for polymorphism 3.
Determining the genes responsible for natural variation and identifying the underlying QTL remains an important challenge (42). We have shown that amino acid polymorphisms in the major photoreceptor for red light contribute to variation in photomorphogenesis across A. thaliana accessions. There is a long-standing debate whether developmental variation is primarily due to changes in gene expression or in protein activity. Our findings with PHYB, together with those for PHYA, PHYC, and CRY2 (20–22), weigh in on the side of protein changes. Second, it has been proposed that in some pathways variants are more likely to be found downstream, rather than upstream, because of relatively relaxed constraints on downstream genes (18, 19) or selective sweeps acting on upstream genes (43). Here, we report that there is functional variation at the top of the PHYB pathway, complementing previous studies of other light-response pathways that also pointed to the photoreceptors themselves being responsible for variation in light sensing (20–22, 44). Similarly, another study has shown evidence for selection upstream in a floral development pathway (43).
Changes in PHYA and CRY2 are limited to individual accessions (20, 21), making it difficult to know whether they confer adaptive advantages or are simply only mildly deleterious mutations that have not yet been purged from nature. In contrast, alternative variants that cause differential light sensitivity are more common in the case of PHYC (22) and PHYB (this work), suggesting more strongly that they may be important in adaptation. If any of these changes are indeed adaptive, it could indicate that overall light sensitivity rather than a particular downstream process is more prone to selection. It will therefore be interesting to study the molecular evolution of the different light-sensing pathways as comprehensive whole-genome variation data become available.
Materials and Methods
DNA Sequencing and Assembly.
Detailed sequencing methods are described in SI Text. All polymorphisms were confirmed in multiple sequencing reads. PHYB sequence for Arabidopsis lyrata subspecies lyrata was assembled from whole genome shotgun reads, using the Staden package (45). Sequences have been deposited in GenBank (29) as accession nos. EU352775–EU352793.
Phylogenetic Analysis.
PHYB sequences were obtained either from the above sequencing or from GenBank (SI Table 3) and were aligned with ClustalX (46, 47). PHYLIP (48) was used to bootstrap the dataset 100 times, determine maximum likelihood and neighbor joining trees, and find majority rule consensus trees. PAUP* software, Version 4.0b10 (49), was used to determine whether there were different rates of nucleotide substitution in the P2/PAS, GAF, and PHY domains relative to the rest of the protein within A. thaliana by comparing different substitution models, as described in SI Text.
Phylogenetic Shadowing.
Sequences obtained from GenBank (SI Table 3) were used as input for eShadow (http://eshadow.dcode.org). HMMI analysis of Brassicales used the following probabilities: eS = 0.85, eF = 0.80, and T = 0.2. Brassicales DT analysis used a maximum percent variation of 5% and a minimum length of 80 aa. For eudicot sequences, HMMI analysis used probabilities of eS = 0.75, eF = 0.60, and T = 0.1, whereas DT analysis used a maximum percent variation of 20% and a minimal length of 80 aa.
Association Mapping.
Simple sequence length polymorphism, cleavable amplified polymorphic sequence (CAPS) and derived CAPS assays were designed and used to genotype >100 A. thaliana accessions for the five nonsingleton nonsynonymous polymorphisms between Ler and Cvi (SI Tables 4 and 5). A permutation-based approach was used to determine association with phenotypic differences. First, a t statistic for correlation between genotype and phenotype was calculated for each polymorphism. To establish the significance of these associations, 10,000 permuted datasets were analyzed, and, for each permutation, the highest t statistic across all polymorphisms was recorded. An association was deemed significant if its t statistic was larger than the appropriate quantile t statistic from the permuted dataset.
LD Analysis.
Ninety-two SNPs were identified from a MAFFT multiple sequence alignment of 33 genomic PHYB sequences (SI Tables 3 and 6). One was removed from the analysis because of incomplete data. The LDheatmap package (50) in the R statistical environment (51) was used to determine and plot pairwise linkage disequilibrium, using allelic correlation (r2).
Transgenic Analysis.
The null phyB-9 allele was transformed with PHYB-Cvi and PHYB-Ler as described in SI Text. Two independent transformations were performed with a total of 483 PHYB-Cvi and 662 PHYB-Ler T1 transformants assayed in nine independent experiments. Significance was determined by using a linear mixed-effects model with PHYB construct as a fixed effect and transformation plus experiment (within transformation) as random effects. To confirm these results, kanamycin resistant T2 progeny from 40 PHYB-Cvi and 44 PHYB-Ler T1 plants were analyzed in two independent experiments. Experiment was not a significant factor, so the mixed-effects model contained PHYB construct (fixed effect), plate (random effect), and T1 parent (random effect).
Microarray Analysis.
Microarray data (40) were imported into R (51) and Bioconductor (52) and normalized by RMA (53). Because we were only querying a single gene, a simple t test was used to determine whether there was evidence for differential PHYB expression between Ler and Cvi (n = 2 and 3 replicates, respectively).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Andreah Wallace and Gabriel Trainer for technical assistance. This work was funded by National Science Foundation Grant DBI-0227103 (to J.N.M and Cynthia Weinig), National Insitutes of Health Grant GM62932 (to J.C. and D.W.), The Howard Hughes Medical Institute, and National Insitutes of Health Training Grant T32 GM070377 (to D.L.F.).
Footnotes
The authors declare no conflict of interest.
Data Deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU352775–EU352793).
This article contains supporting information online at www.pnas.org/cgi/content/full/0712174105/DC1.
References
- 1.Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Annu Rev Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- 2.Reed JW, Nagatani A, Elich TD, Fagan M, Chory J. Phytochrome A, phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 1994;104:1139–1149. doi: 10.1104/pp.104.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aukerman MJ, et al. A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell. 1997;9:1317–1326. doi: 10.1105/tpc.9.8.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Devlin PF, Patel SR, Whitelam GC. Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell. 1998;10:1479–1487. doi: 10.1105/tpc.10.9.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devlin PF, et al. Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation growth and flowering time. Plant Physiol. 1999;119:909–915. doi: 10.1104/pp.119.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monte E, et al. Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling pathways. Plant Cell. 2003;15:1962–1980. doi: 10.1105/tpc.012971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halliday KJ, Whitelam GC. Changes in photoperiod or temperature alter the functional relationships between phytochromes and reveal roles for phyD and phyE. Plant Physiol. 2003;131:1913–1920. doi: 10.1104/pp.102.018135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clack T, Mathews S, Sharrock RA. The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol. 1994;25:413–427. doi: 10.1007/BF00043870. [DOI] [PubMed] [Google Scholar]
- 9.Franklin KA, et al. Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol. 2003;131:1340–1346. doi: 10.1104/pp.102.015487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franklin KA, Davis SJ, Stoddart WM, Vierstra RD, Whitelam GC. Mutant analyses define multiple roles for phytochrome C in Arabidopsis photomorphogenesis. Plant Cell. 2003;15:1981–1989. doi: 10.1105/tpc.015164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maloof JN, Borevitz JO, Weigel D, Chory J. Natural variation in phytochrome signaling. Semin Cell Dev Biol. 2000;11:523–530. doi: 10.1006/scdb.2000.0198. [DOI] [PubMed] [Google Scholar]
- 12.Dorn LA, Pyle EH, Schmitt J. Plasticity to light cues and resources in Arabidopsis thaliana: Testing for adaptive value and costs. Evolution. 2000;54:1982–1994. doi: 10.1111/j.0014-3820.2000.tb01242.x. [DOI] [PubMed] [Google Scholar]
- 13.Donohue K, Messiqua D, Pyle EH, Heschel MS, Schmitt J. Evidence of adaptive divergence in plasticity: Density- and site-dependent selection on shade-avoidance responses in Impatiens capensis. Evolution. 2000;54:1956–1968. doi: 10.1111/j.0014-3820.2000.tb01240.x. [DOI] [PubMed] [Google Scholar]
- 14.Duek PD, Fankhauser C. bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plants Sci. 2005;10:51–54. doi: 10.1016/j.tplants.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Huq E, Tepperman JM, Quail PH. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc Natl Acad Sci USA. 2000;97:9789–9794. doi: 10.1073/pnas.170283997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holm M, Ma LG, Qu LJ, Deng XW. Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 2002;16:1247–1259. doi: 10.1101/gad.969702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serino G, Deng XW. The COP9 signalosome: regulating plant development through the control of proteolysis. Annu Rev Plant Biol. 2003;54:165–182. doi: 10.1146/annurev.arplant.54.031902.134847. [DOI] [PubMed] [Google Scholar]
- 18.Lu Y, Rausher MD. Evolutionary rate variation in anthocyanin pathway genes. Mol Biol Evol. 2003;20:1844–1853. doi: 10.1093/molbev/msg197. [DOI] [PubMed] [Google Scholar]
- 19.Rausher MD, Miller RE, Tiffin P. Patterns of evolutionary rate variation among genes of the anthocyanin biosynthetic pathway. Mol Biol Evol. 1999;16:266–274. doi: 10.1093/oxfordjournals.molbev.a026108. [DOI] [PubMed] [Google Scholar]
- 20.Maloof JN, et al. Natural variation in light sensitivity of Arabidopsis. Nat Genet. 2001;29:441–446. doi: 10.1038/ng777. [DOI] [PubMed] [Google Scholar]
- 21.El-Din El-Assal S, Alonso-Blanco C, Peeters AJ, Raz V, Koornneef M. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat Genet. 2001;29:435–440. doi: 10.1038/ng767. [DOI] [PubMed] [Google Scholar]
- 22.Balasubramanian S, et al. The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat Genet. 2006;38:711–715. doi: 10.1038/ng1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alba R, Kelmenson PM, Cordonnier-Pratt MM, Pratt LH. The phytochrome gene family in tomato and the rapid differential evolution of this family in angiosperms. Mol Biol Evol. 2000;17:362–373. doi: 10.1093/oxfordjournals.molbev.a026316. [DOI] [PubMed] [Google Scholar]
- 24.Mathews S, Burleigh JG, Donoghue MJ. Adaptive evolution in the photosensory domain of phytochrome A in early angiosperms. Mol Biol Evol. 2003;20:1087–1097. doi: 10.1093/molbev/msg123. [DOI] [PubMed] [Google Scholar]
- 25.Borevitz JO, et al. Quantitative trait loci controlling light and hormone response in two accessions of Arabidopsis thaliana. Genetics. 2002;160:683–696. doi: 10.1093/genetics/160.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiménez-Gómez JM, et al. Quantitative genetic analysis of flowering time in tomato. Genome. 2007;50:303–315. doi: 10.1139/g07-009. [DOI] [PubMed] [Google Scholar]
- 27.Ingvarsson PK, García MV, Hall D, Luquez V, Jansson S. Clinal variation in phyB2, a candidate gene for day-length-induced growth cessation and bud set, across a latitudinal gradient in European aspen (Populus tremula). Genetics. 2006;172:1845–1853. doi: 10.1534/genetics.105.047522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frewen BE, et al. Quantitative trait loci and candidate gene mapping of bud set and bud flush in populus. Genetics. 2000;154:837–845. doi: 10.1093/genetics/154.2.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank Nucleic Acids Res. 2007;35:D21–D25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nordborg M, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharbel TF, Haubold B, Mitchell-Olds T. Genetic isolation by distance in Arabidopsis thaliana: biogeography and postglacial colonization of Europe. Mol Ecol. 2000;9:2109–2118. doi: 10.1046/j.1365-294x.2000.01122.x. [DOI] [PubMed] [Google Scholar]
- 32.Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 34.Boffelli D, et al. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- 35.Ovcharenko I, Boffelli D, Loots GG. eShadow: a tool for comparing closely related sequences. Genome Res. 2004;14:1191–1198. doi: 10.1101/gr.1773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner JR, Brunzelle JS, Forest KT, Vierstra RD. A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature. 2005;438:325–331. doi: 10.1038/nature04118. [DOI] [PubMed] [Google Scholar]
- 37.Flint-Garcia SA, Thornsberry JM, Buckler ES. Structure of linkage disequilibrium in plants. Annu Rev Plant Biol. 2003;54:357–374. doi: 10.1146/annurev.arplant.54.031902.134907. [DOI] [PubMed] [Google Scholar]
- 38.Kim S, et al. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet. 2007;39:1151–1155. doi: 10.1038/ng2115. [DOI] [PubMed] [Google Scholar]
- 39.Zhao K, et al. An Arabidopsis example of association mapping in structured samples. PLoS Genet. 2007;3:e4. doi: 10.1371/journal.pgen.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lempe J, et al. Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet. 2005;1:109–118. doi: 10.1371/journal.pgen.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keurentjes JJ, et al. Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. Proc Natl Acad Sci USA. 2007;104:1708–1713. doi: 10.1073/pnas.0610429104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weigel D, Nordborg M. Natural variation in Arabidopsis. How do we find the causal genes? Plant Physiol. 2005;138:567–568. doi: 10.1104/pp.104.900157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olsen KM, Womack A, Garrett AR, Suddith JI, Purugganan MD. Contrasting evolutionary forces in the Arabidopsis thaliana floral developmental pathway. Genetics. 2002;160:1641–1650. doi: 10.1093/genetics/160.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olsen KM, Halldorsdottir SS, Stinchcombe JR, Weinig C, Schmitt J, Purugganan MD. Linkage disequilibrium mapping of Arabidopsis CRY2 flowering time alleles. Genetics. 2004;167:1361–1369. doi: 10.1534/genetics.103.024950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- 46.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Felsenstein J. PHYLIP (Phylogeny Inference Package), Version 3.6. 2005 Available at http://evolution.genetics.washington.edu/phylip.html.
- 49.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinaur Associates; 2003. [Google Scholar]
- 50.Shin J-H, Blay S, McNeney B, Graham J. LDheatmap: An R Function for Graphical Display of Pairwise Linkage Disequilibria Between Single Nucleotide Polymorphisms. J Stat Soft. 2006;16 Code Snippet 3. [Google Scholar]
- 51.R Development Core Team. R: A Language and Environment for Statistical Computing. 2006 Available at www.r-project.org.
- 52.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.