Abstract
Sterols have multiple functions in all eukaryotes. In plants, sterol biosynthesis is initiated by the enzymatic conversion of 2,3-oxidosqualene to cycloartenol. This reaction is catalyzed by cycloartenol synthase 1 (CAS1), which belongs to a family of 13 2,3-oxidosqualene cyclases in Arabidopsis thaliana. To understand the full scope of sterol biological functions in plants, we characterized allelic series of cas1 mutations. Plants carrying the weak mutant allele cas1–1 were viable but developed albino inflorescence shoots because of photooxidation of plastids in stems that contained low amounts of carotenoids and chlorophylls. Consistent with the CAS1 catalyzed reaction, mutant tissues accumulated 2,3-oxidosqualene. This triterpenoid precursor did not increase at the expense of the pathway end products. Two strong mutations, cas1–2 and cas1–3, were not transmissible through the male gametes, suggesting a role for CAS1 in male gametophyte function. To validate these findings, we analyzed a conditional CRE/loxP recombination-dependent cas1–2 mutant allele. The albino phenotype of growing leaf tissues was a typical defect observed shortly after the CRE/loxP-induced onset of CAS1 loss of function. In the induced cas1–2 seedlings, terminal phenotypes included arrest of meristematic activity, followed by necrotic death. Mutant tissues accumulated 2,3-oxidosqualene and contained low amounts of sterols. The vital role of sterols in membrane functioning most probably explains the requirement of CAS1 for plant cell viability. The observed impact of cas1 mutations on a chloroplastic function implies a previously unrecognized role of sterols or triterpenoid metabolites in plastid biogenesis.
Keywords: albinism; sterols; triterpenoid metabolites; 2,3-oxidosqualene
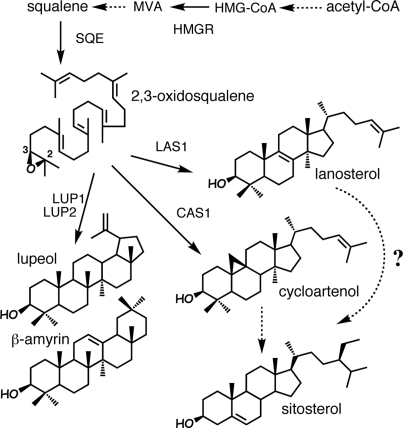
Plant cells use a sterol biosynthetic pathway that is peculiar compared with that of other eukaryotes (1, 2). Animals and fungi cyclize the 30-carbon atom precursor 2,3-oxidosqualene into the tetracyclic triterpene lanosterol, which is metabolized into cholesterol and ergosterol, respectively, whereas plants transform the same precursor into the cyclopropylsterol intermediate cycloartenol, which is converted into sitosterol (Fig. 1). The physiological role of cyclopropylsterol intermediates and the apparent necessity for plants to synthesize sitosterol by the cycloartenol route are not understood. An Arabidopsis thaliana cDNA encoding cycloartenol synthase 1 (CAS1) has been cloned by functional expression in an ergosterol auxotroph of yeast (Saccharomyces cerevisiae) deficient in lanosterol synthesis (3). CAS1 (At2g07050) belongs to a family of 13 triterpene synthases (4), among which At3g45130 has recently been shown to encode a lanosterol synthase 1 (5, 6). Hence, the first committed step in plant sterol biosynthesis may be functionally redundant, as suggested by the possible existence of a lanosterol route besides the major cycloartenol route to sitosterol (7). Other characterized triterpene synthases, such as LUP1 and LUP2 (Fig. 1), catalyze the formation of nonsteroidal polycyclic triterpenes, among which lupeol and β-amyrin are common secondary metabolites (8–12).
Fig. 1.
Sterol and nonsteroidal triterpene biosynthetic pathways in plants. HMG-CoA, 3-hydroxy-3-methylglutaryl-CoA; MVA, mevalonate; SQE, squalene epoxidase; LAS1, lanosterol synthase; LUP1 and LUP2, pentacyclic triterpene alcohol synthases.
Molecular genetic analysis of Arabidopsis mutants affected in the postsqualene segment of sterol metabolism have uncovered a wide spectrum of developmental abnormalities, such as embryo or seedling lethality (13, 14), dwarfism attributable to brassinosteroid deficiency (15–17), and size variation (18–20), and have provided new insights into the physiological functions of membrane sterols and their role as precursors of the brassinosteroid phytohormones (21–23). To better understand the full scope of sterol biological functions in plant cells, allelic series of cas1 mutations were analyzed. The data emphasized an essential role of CAS1 for the plant cell viability, suggested the involvement of this enzyme in the regulation of triterpenoid biosynthesis, and indicated, to our knowledge, a previously unrecognized role of sterols or their precursors in chloroplast differentiation.
Results
The cas1–1 Mutant of Arabidopsis Has an Albino Phenotype Late in Development.
To identify pigment-deficient mutations in genes that encode nonplastidial proteins, we carried out a phenotypic screening of our collection of 402 sequence-indexed, exon-trap lines generated with a T-DNA vector (24). The line L2–19 had a T-DNA insertion within the CAS1 gene and segregated a characteristic pigment-deficient phenotype of inflorescence shoots (Fig. 2). Albinism affected the flower-carrying parts of the stems. The first 10 to 20 flowers and siliques attached to albino stems had green colored tissues that are normally chlorophyllous in the wild type. Flowers that developed later had either albino or variegated sepals and carpels. Sterility of the purely albino flowers was the probable reason for mutant plants to have, on average, a 2-week longer lifespan than the wild type. Progeny seedlings of homozygous plants showed various developmental abnormalities, such as pigment variegation, polyembryony, fused cotyledons, and multiple cotyledons, reminiscent of phenotypes associated with mutations in sterol biosynthesis (13). Such phenotypes were not observed in the progeny of heterozygous parent plants, suggesting that the maternal tissue provided the CAS1 product to developing embryos.
Fig. 2.
Morphological phenotype of the cas1–1 mutant. (A) Wild-type inflorescence shoot of Arabidopsis ecotype C24. (C) Inflorescence shoot of the cas1–1 mutant. (B and D) Close-ups of inflorescence shoots of wild-type and mutant plants shown in A and C, respectively.
Light and electron microscopy revealed that cells from three cortical layers just below the epidermis of albino stems did not possess fully differentiated chloroplasts (Fig. 3A-D). These albino mutant cells had plastids that were devoid of the organized system of thylakoid membranes (Fig. 3 I–K), which is normally found in fully differentiated plastids from stems of wild types (Fig. 3 E and L) or green stems of cas1–1 (Fig. 3 F–H). Spectral measurements of pigments indicated that albino parts of cas1–1 stems had a reduced carotenoid (10-fold) and chlorophyll (12-fold) content compared with that of the wild type [supporting information (SI) Fig. 7], suggesting an impaired carotenoid biosynthesis in the apical part of the stems. To find out whether the mutant chloroplasts had suffered photooxidative damage, mutant plants were grown first at a light intensity of 40–60 μmol·m−2·s−1, until the flowering shoots reached a height of 10 to 15 cm, and then at a reduced photon fluence of 5–10 μmol·m−2·s−1. Shoots that developed under low-light conditions were able to green, indicating that the albinism of stems was attributable to photooxidative damage of chloroplasts. The albino phenotype cosegregated with the T-DNA in three backcross generations and was genetically complemented by the CAS1 cDNA expressed from a constitutive 35S cauliflower mosaic virus (CaMV) promoter, demonstrating that the mutant phenotype was caused by the deficiency in the CAS1 gene function (SI Methods).
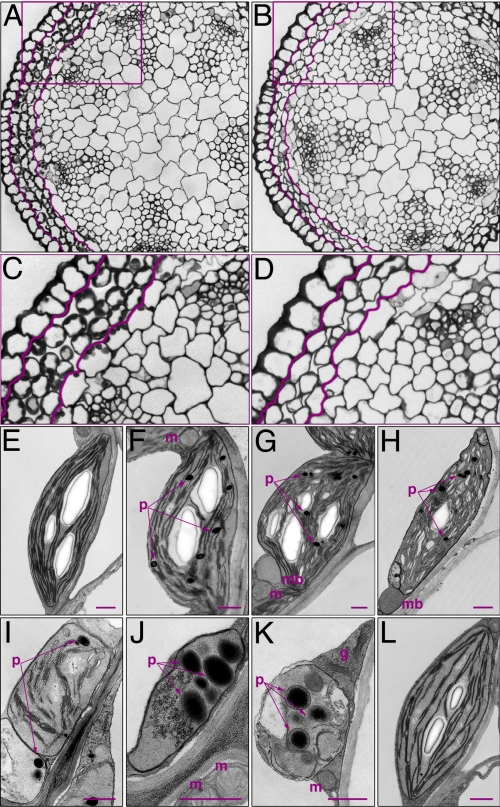
Fig. 3.
Cellular phenotype of the cas1–1 mutant. (A and B) Toluidine blue-stained transverse cross-section of wild-type (A) and albino cas1–1 (B) stems. (C and D) Enlarged parts of images in A and B. Highlighted are the chlorenchyma subepidermal cell layers with chloroplasts (C) and without well differentiated chloroplasts (D). (E) Plastid morphology in cells from the lower part of wild-type stems, ≈3–5 cm above the rosette. The corresponding stem region in the cas1–1 mutant is green as well. (F–K) Progressive acropetal bleaching of stems correlated with plastid morphology. Thin sections of cas1–1 stems collected at ≈2-cm intervals showed plastid types in the developmentally oldest cell positioned at the base of the stem, ≈3.5 cm above the rosette (F), and the youngest albino cell at the top of the shoot (K). Mutant plastids (I–K) lack thylakoid membrane system and starch granules and accumulate electron-dense vesicles that may correspond to plastoglobuli. (L) Chloroplast in wild-type chlorenchyma cell from distal parts of the stem. g, Golgi apparatus; m, mitochondria; mb, microbody; p, plastoglobuli. (Scale bars, 0.5 μm.)
Chemical Phenotype of cas1–1.
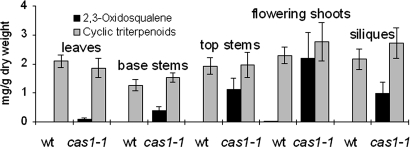
Triterpenoid analysis of wild-type and cas1–1 plants showed that 2,3-oxidosqualene accumulated in cas1–1, a chemical phenotype fully consistent with CAS1 being a functional 2,3-oxidosqualene cyclase in planta. The accumulation of 2,3-oxidosqualene in cas1–1 varied according to organs (Fig. 4 and SI Fig. 8). Rosette leaves contained low amounts, whereas upper stems, flowering shoots, and siliques had the highest quantities of 2,3-oxidosqualene. This accumulation did not occur at the expense of the end products of the cyclic triterpenoid pathway, among which the quantities of total Δ5 sterols were equivalent in the wild-type and in cas1–1 (SI Table 1), as was the case in inflorescences of cas1–1 plants with similar or even larger amounts of cyclic triterpenoids than in the wild type (Fig. 4). To better understand such an apparent increase in the metabolic flux toward total triterpenoids in cas1–1 flowering shoots, we measured the activity of the enzyme 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) (Fig. 1), known to be a rate-limiting step in phytosterol accumulation (25–28). HMGR-specific activity in cas1–1 flowering shoots increased by 30% (5.72 ± 0.1 vs. 4.34 ± 0.25 nmol·mg−1 protein per hour in wild type). These results suggested that a higher activity of an enzyme upstream to CAS1 (Fig. 1) may be responsible for the increase of cyclic triterpenoids and could contribute to the dramatic accumulation of 2,3-oxidosqualene in the cas1–1 mutant.
Fig. 4.
Triterpenoid content of wild-type and cas1–1 tissues. Samples prepared from tissues of at least 15 individual plants with ≈20 siliques were analyzed. The layout for material collection is detailed in SI Fig. 8. Cyclic triterpenoids are metabolites derived from 2,3-oxidosqualene (see SI Table 1 for details). The data are from three independent experiments.
CAS1 Is Essential in Plant Cells.
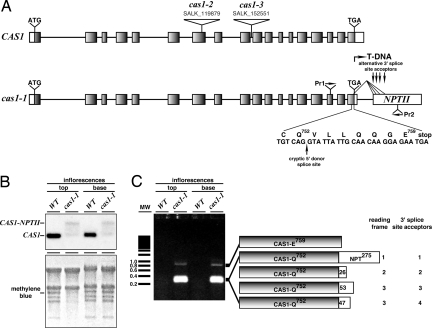
Because CAS1 is thought to be a major 2,3-oxidosqualene cyclase that initiates postsqualene sterol biosynthesis in plants (1, 2, 7) and sterols are known to be essential in yeast, a null mutant allele would be expected to be lethal, given that the CAS1 function is not redundant (2). Indeed, molecular analysis of the cas1–1 mutation showed that it is a weak allele caused by the T-DNA insertion into the 3′ UTR 177 bp downstream from the translation termination codon TGA and ≈50 bp upstream from the polyadenylation site (Fig. 5A), with a lower mRNA expression from the mutated locus as a consequence (Fig. 5B). RT-PCR analysis of the cas1–1 locus suggested that five different CAS1 proteins were synthesized (Fig. 5C and SI Table 2), of which one was a wild-type protein and the four others were mutant isoforms with the last seven amino acid residues of CAS1 replaced by amino acid sequences of different lengths. The developmental pattern of the CAS1 expression was examined by histochemical staining with β-glucuronidase (GUS) in plants that were transformed with a CAS1 promoter driving the expression of the uidA gene of Escherichia coli. The transgene was ubiquitously expressed in Arabidopsis organs (SI Methods and SI Fig. 9). This observation, together with the physiological and chemical phenotypes of cas1–1, mostly restricted to inflorescence shoots (Fig. 4), hinted at a possible redundancy of CAS activity in organs, such as leaves.
Fig. 5.
CAS1 mutant alleles. (A) Wild-type (CAS1) and mutated loci cas1–1, cas1–2, and cas1–3. Exon-labeled NPTII encodes a reading frame for neomycin phosphotransferase that lacks the translation initiation codon (ATG). Heterologous splicing events use the cryptic 5′ splice site donor within CAS1 exon 18, as indicated, and any one of the four alternative 3′ splice site acceptors from the T-DNA. The T-DNA insertion mutant alleles cas1–2 (SALK_119879) and cas1–3 (SALK_152551) were reconfirmed by DNA sequencing (SI Fig. 10). (B) RNA gel blot analysis. To verify equal loading and transfer efficiency of RNA, the membrane was stained with methylene blue (Lower). The hybridization signal with a radioactively labeled probe prepared with CAS1 cDNA is presented (Upper). Positions of the wild-type mRNA and chimeric CAS1-NPTII transcript expressed from the cas1–1 locus are indicated. (C) RT-PCR analysis. cDNAs were synthesized with reverse transcriptase and RNA from samples analyzed in B. These cDNAs served as templates in the PCRs with Pr1 and Pr2 shown in A. CAS1 isoforms encoded by alternatively spliced transcripts in the cas1–1 mutant are shown on the right (SI Table 2).
To address this question, we analyzed the knockout mutant alleles cas1–2 (SALK_119879) and cas1–3 (SALK_152551) (29) that were caused by T-DNA insertions in the seventh exon and in the ninth intron of CAS1, respectively (Fig. 5A and SI Fig. 10). No homozygous mutant plants were found among 50 individuals of a self-progeny of either CAS1/cas1–2 or CAS1/cas1–3. Analysis of 50 plants per progeny from reciprocal crosses between heterozygous CAS1/cas1–2(cas1–3) and wild-type CAS1/CAS1 plants revealed that CAS1/cas1–2(cas1–3) progenies were obtained only if CAS1/cas1–2(cas1–3) was used as a female in the cross. Consequently, these two T-DNA insertions caused a male-specific transmission defect, suggesting that the protein is essential for the male gametophyte function.
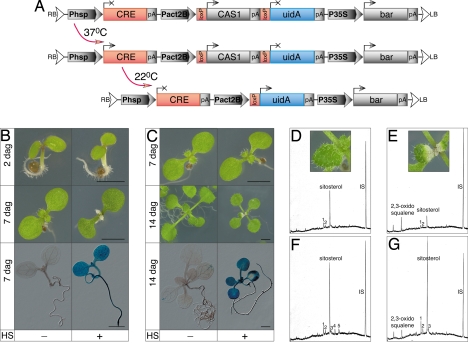
To characterize the function of CAS1 in postgametophytic plant development, we used a CRE/loxP method for a mosaic analysis in the Arabidopsis cas1–2 background (30). The design of the gene construct used, pCRECAS1, is schematically presented in Fig. 6A. Approximately 10% of the primary transformants (T1) generated with pCRECAS1 had abnormal phenotypes that could be categorized into three classes (SI Fig. 11): in one class, plants had albino stems, resembling cas1–1 mutant plants; in a second class, plants were characterized by green leaves with albino bases and petioles; and in the third group, leaf shape and pigmentation were abnormal. Most probably, these mutant phenotypes among the T1 plants were attributable to the DNA homology-dependent cosuppression of the endogenous CAS1 gene (31). T1 lines with abnormal phenotypes as such or in their progeny were excluded from the analysis. In the T2 progeny of the remaining 48 transgenic lines, cas1–2/cas1–2 homozygous individuals were found, demonstrating that the PACT2::CAS1::NOS gene cassette complemented male-specific transmission defect. Five independent cas1–2 homozygous transgenic lines (crecas2–1, crecas 2–4, crecas2–5, cre1, and cre38) were selected because they had constitutive GUS expression patterns after heat shock, indicating efficient CRE recombinase-mediated excision of the CAS1 cDNA.
Fig. 6.
Generation and analysis of the conditional cas1–2 mutant allele. (A) Design of the cas1–2 mutation-complementing vector pCRECAS1 and sequence of events in planta after heat shock. Elements are promoters (Phsp, Pact2B, P35S), protein-coding regions (CRE, CAS1, uidA, bar), mRNA polyadenylation sequences (pA), and loxP sites (loxP); LB and RB are border sequences delineating the DNA transferred (T-DNA) into plant cells from Agrobacterium tumefaciens. Arrows or crosses indicate expression or lack of expression of gene(s). (B) Requirement of CAS1 for plastid differentiation and leaf growth. Two days after germination on a standard synthetic medium, seedlings were heat stressed (HS) at 37°C for 6 h (+) and then grown under standard conditions. Non-HS seedlings (−) are on the left. Phenotypes of seedlings and GUS-staining patterns 7 days after germination (dag) are shown. (C) Assessment of the role of CAS1 in cells with differentiated plastids. Seedlings bearing visible first leaf pairs were HS (+), photographed at 14 dag, and stained for GUS activity, along with control (−) plants. (D–G) Sterol analysis. Post-HS neoformed tissues (i.e., distal half of root, second leaf pair, and proximal half of leaves from first pair) and HS-preformed tissues (i.e., proximal half of root, cotyledons and hypocotyls, and distal half of leaves from first pair) were sampled from 25 seedlings of the wild-type and of the cas1–2 conditional allele in three independent experiments. GC of crude hexanic extracts of nonsaponifiable lipids gave profiles typical of plants with CAS1 (D) and cas1–2 (E) phenotypes in the case of neoformed tissues and profiles typical of plants with CAS1 (F) and cas1–2 phenotypes (G) in the case of preformed tissues. The Insets in D and E demonstrate that HS (+) wild-type and cas1–2 conditional allele had the same growth stage. Chromatograms obtained with the Agilent 6890 GC device show data acquisition after 20 min and until 52 min of the runs. Major peaks are sitosterol and 2,3-oxidosqualene (Fig. 1). Other sterol compounds identified by their mass spectrum are: 1, campesterol; 2, stigmasterol; 3, isofucosterol; 4, cycloartenol; and 5, 24-methylene cycloartenol. IS, internal standard (lupenyl-diacetate) present in identical amount in each sample. (Scale bars, 2 mm.)
Induction of CAS1 loss of function resulted in a reproducible mutant phenotype. When 2-day-old seedlings were heat stressed, they developed small, narrow albino leaves 5 days after heat shock (DAHS) (Fig. 6B). In another example (Fig. 6C), 7-day-old seedlings with a well emerged first pair of true leaves at the time of heat shock application were analyzed. One week after heat shock, the growth of the first leaf pair that remained green was maintained in the plantlets. In contrast, the third and the fourth leaves were retarded in their development, and their bases were albino, whereas the fifth leaf was very small and purely albino. No other macroscopically discernible leaves appeared during the following 2 weeks of culture, suggesting that the shoot apical meristem activity was terminated. Similarly to leaves, roots of heat-stressed seedlings ceased their growth 5 to 7 DAHS. Albino tissues also turned necrotic and died ≈10–14 DAHS. To verify that the observed mutant phenotypes were caused by the CAS1 loss of function, we compared the composition of hexane extracts of nonsaponifiable lipids in heat-stressed and control plants by GC. Only plantlets from the cas1–2 conditional allele that had experienced heat shock during their growth accumulated 2,3-oxidosqualene (compare chromatograms in Fig. 6 E vs. D and G vs. F). In addition, tissues of the cas1–2 conditional allele that developed after the heat shock and displayed albinism contained a lower amount of the pathway end products than the wild-type counterpart tissues that had experienced the same heat shock. Thus, the induction of the CAS1 loss of function by using the conditional heat shock-inducible cas1–2 allele resulted in cessation of chloroplast differentiation in newly growing leaves, accompanied by growth arrest and followed by the death of bleached tissue.
Discussion
We describe here the defective male-specific transmission of cas1–2 and cas1–3 mutant alleles, as well as abnormal growth of leaves and the arrest of shoot and root meristems in induced loss-of-function cas1–2 seedlings. These phenotypes are most probably a consequence of structural defects in cellular membrane networks caused by the measured depletion of sterols. This conclusion is consistent with studies in yeast and animal cells revealing that sterols are essential components of cell membranes and that their concentration is tightly controlled by a feedback system that operates at transcriptional and posttranscriptional levels (32). Thus, CAS1 is required for the plant cell viability, and its function is not redundant with other 2,3-oxidosqualene cyclases of Arabidopsis.
Analysis of the weak cas1–1 and conditional cas1–2 mutant alleles revealed a role for CAS1 in plastid biogenesis. At the cellular level, albino phenotypes in plants can be caused both by an abortion in biogenesis of the plastidial thylakoid membrane, where chlorophyll–protein complexes accumulate, and by a photooxidative damage to developing thylakoid membranes because of suboptimal production of photoprotective carotenoids. Stability of chlorophyll–protein complexes depends on the availability of lipids (33); however, sterols have not been reported as components of plastid membranes (34). Biogenesis of thylakoids requires extensive exchange, the so-called lipid trafficking, of lipid precursors between the chloroplast and the endoplasmic reticulum (ER) (33). These two cellular compartments develop contact sites between the outer plastid envelope and the sterol-rich ER membranes (35). Unbalanced sterol composition and sterol depletion identified in cas1 mutants could compromise ER–plastid physical contacts.
On the other hand, we found that the cas1–1 albino phenotype is conditionally light-dependent, which correlates with a defect in accumulation of carotenoids. Carotenoids are end products of the plastidial terpenoid biosynthetic pathway, whereas sterols are end products of the cytosolic terpenoid pathway. Albinism of cas1 mutants may represent genetic evidence for a regulatory mechanism that is thought to coordinate activity of compartmentalized terpenoid biosynthesis in the plant cell (36).
Materials and Methods
Plant Growth.
Seeds of Arabidopsis thaliana (L.) Heyhn. were sown in a standard soil compost mixture, and seedlings were grown individually in growth chambers under white fluorescent lamps. Photon fluency was 40–60 μmol·m−2·s−1 at the rosette level and 70–90 μmol·m−2·s−1 at that of inflorescence tips, i.e., ≈20–30 cm above the rosettes. The temperature was 22°C during the day (12 h) and 19°C during the night (12 h).
Molecular Analysis of Nucleic Acids.
Total RNA was isolated and analyzed by RNA gel blot hybridization (37). For RT-PCR analysis, total RNA was treated with DNaseI and additionally purified with an RNeasy kit (Qiagen). The oligo(dT)-primed first-strand cDNA was synthesized on 5 μg of total RNA with thermoscript reverse polymerase (Invitrogen). Chimeric cDNAs of CAS1 and the neomycin phosphotransferase II-encoding gene (NPTII) were amplified by PCR with Taq DNA polymerase (Roche Molecular Biochemicals) and the CAS1-specific primer Pr1 (primer 1; 5′-GGCTATGCTCGCACTCATTGGT-3′), and NPTII-specific primer Pr2 (primer 2; 5′-CCCCTGCGCTGACAGCCGGAACACG-3′) (Fig. 5A). Amplified DNA fragments were subcloned into pCRTOPO (Invitrogen) for DNA sequence analysis.
Construction of a Conditional cas1–2 Allele.
The details of the preparation of the binary vector pCRECAS1 are available upon request. Elements incorporated into this vector were the vector backbone from a binary vector pPZP200 (38); a plant selectable marker conferring resistance to glufosinate-ammonium (BASTA) as a CaMV 35S promoter driving the expression of the BAR gene (39); the heat shock promoter from AtHSP18.2 (40) and octopine synthase polyadenylation sequences to drive the expression of the CRE-intron gene (41); the promoter of the At3g18780 that encodes ACTIN2 (42); and the nopaline synthase polyadenylation sequences used to drive the expression of the CAS1 cDNA. Progenies of transformed CAS1/cas1–2 plants were screened for individuals resistant to BASTA that were genotyped by PCR for plants homozygous for the SALK_119879 cas1–2 knockout mutant allele (SI Methods).
Triterpenoid Analysis.
Samples were collected, freeze-dried, and ground with an Ultra-Turrax blender (Janke und Künkel, IKA Labortechnik) in 15–25 ml of 6% KOH in MeOH at 80°C for 2 h. The nonsaponifiable compounds were extracted with three volumes of n-hexane and separated on silica TLC plates (0.25 mm thickness; Merck) with two runs of dichloromethane. 2,3-Oxidosqualene was scraped off the plate (Rf = 0.6) and identified by comparison with an authentic standard of 2,3-oxidosqualene (racemic) (6E,10E,14E,18E)-2,3-epoxy-2,6,10,15,19,23-hexamethyl-6,10,14,18,22-tetracosapentaene (C30H50O) (Echelon Biosciences Laboratories) and sterols as acetate derivatives with a 8200 gas chromatograph (Varian) equipped with a DB-1 column as described (4, 14, 26). Lupenyl-3,28-diacetate was added to the extract of nonsaponifiable compounds as an internal standard for quantification of 2,3-oxidosqualene and total triterpenoids (sterols and triterpenes). Identification of compounds was confirmed by GC/MS with a 6890 gas chromatograph (Agilent) equipped with a DB-5MS column (J&W; 30 m long, 0.32 mm i.d., 0.25-μm film thickness) coupled with a 5973 mass selective detector (Agilent). The temperature program included a steep ramp from 60 to 220°C at 30°C per minute, followed by a 2°C per minute ramp from 220 to 300°C and a plateau at 300°C (He as carrier gas, 2 ml/min).
Measurement of HMGR Activity.
Membranes from Arabidopsis tissues were isolated as described (43). Microsomal proteins were quantified by the Bio-Rad protein assay with BSA as a standard. The standard HMGR assay consisted of 100 mM K2PO4, pH 7.5, 4.2 mM EDTA, 8.4 mM DTE, 100 μg BSA, 600 μM NADPH, and 30 μM (0.1 μCi) (R,S)-[3-14C]3-hydroxy-3-methylglutaryl-CoA in a final volume of 60 μl, incubated, and processed as described (25, 26) (SI Methods).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Professors Pierre Benveniste and Dirk Inzé for encouragement; Yves Poirier, Michel Rohmer, Thomas Bach, and Jacques-Henry Weil for comments and advice; Màrta Ramel for horticultural work; Riet De Rycke for help with the microscopical analysis; Karel Spruyt and Pascal Disdier for photographic work; and Martine De Cock for help in preparing the manuscript. The Arabidopsis Biological Resource Center (Ohio State University) is acknowledged for providing T-DNA insertional mutant lines. This work was supported by Centre National de la Recherche Scientifique and Agence Nationale de la Recherche Grant TERPENE ANR-05-BLAN-0217-02 (to V.C.) and by European Union Grant EXOTIC QLG2-CT-1999-000351 (to S.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712190105/DC1.
References
- 1.Benveniste P. Sterol biosynthesis. Annu Rev Plant Physiol. 1986;37:275–308. [Google Scholar]
- 2.Benveniste P. Biosynthesis and accumulation of sterols. Annu Rev Plant Biol. 2004;55:429–457. doi: 10.1146/annurev.arplant.55.031903.141616. [DOI] [PubMed] [Google Scholar]
- 3.Corey EJ, Matsuda SPT, Bartel B. Isolation of an Arabidopsis thaliana gene encoding cycloartenol synthase by functional expression in a yeast mutant lacking lanosterol synthase by the use of a chromatographic screen. Proc Natl Acad Sci USA. 1993;90:11628–11632. doi: 10.1073/pnas.90.24.11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Husselstein-Muller T, Schaller H, Benveniste P. Molecular cloning and expression in yeast of 2,3-oxidosqualene-triterpenoid cyclases from Arabidopsis thaliana. Plant Mol Biol. 2001;45:75–92. doi: 10.1023/a:1006476123930. [DOI] [PubMed] [Google Scholar]
- 5.Kolesnikova MD, Xiong Q, Lodeiro S, Hua L, Matsuda SPT. Lanosterol biosynthesis in plants. Arch Biochem Biophys. 2006;447:87–95. doi: 10.1016/j.abb.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki M, et al. Lanosterol synthase in dicotyledonous plants. Plant Cell Physiol. 2006;47:565–571. doi: 10.1093/pcp/pcj031. [DOI] [PubMed] [Google Scholar]
- 7.Phillips DR, Rasbery JM, Bartel B, Matsuda SPT. Biosynthetic diversity in plant triterpene cyclization. Curr Opin Plant Biol. 2006;9:305–314. doi: 10.1016/j.pbi.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Herrera JBR, Bartel B, Wilson WK, Matsuda SPT. Cloning and characterization of the Arabidopsis thaliana lupeol synthase gene. Phytochemistry. 1998;49:1905–1911. doi: 10.1016/s0031-9422(98)00366-5. [DOI] [PubMed] [Google Scholar]
- 9.Kushiro T, Shibuya M, Ebizuka Y. β-Amyrin synthase. Cloning of oxidosqualene cyclase that catalyzes the formation of the most popular triterpene among higher plants. Eur J Biochem. 1998;256:238–244. doi: 10.1046/j.1432-1327.1998.2560238.x. [DOI] [PubMed] [Google Scholar]
- 10.Segura MJR, Meyer MM, Matsuda SPT. Arabidopsis thaliana LUP1 converts oxidosqualene to multiple triterpene alcohols and a triterpene diol. Org Lett. 2000;2:2257–2259. doi: 10.1021/ol006016b. [DOI] [PubMed] [Google Scholar]
- 11.Ebizuka Y, Katsube Y, Tsutsumi T, Kushiro T, Shibuya M. Functional genomics approach to the study of triterpene biosynthesis. Pure Appl Chem. 2003;75:369–374. [Google Scholar]
- 12.Fazio GC, Xu R, Matsuda SPT. Genome mining to identify new plant triterpenoids. J Am Chem Soc. 2004;126:5678–5679. doi: 10.1021/ja0318784. [DOI] [PubMed] [Google Scholar]
- 13.Schrick K, et al. FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev. 2000;14:1471–1484. [PMC free article] [PubMed] [Google Scholar]
- 14.Kim HB, et al. Arabidopsis cyp51 mutant shows postembryonic seedling lethality associated with lack of membrane integrity. Plant Physiol. 2005;138:2033–2047. doi: 10.1104/pp.105.061598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- 16.Klahre U, et al. The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell. 1998;10:1677–1690. doi: 10.1105/tpc.10.10.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choe S, et al. The Arabidopsis dwf7/ste1 mutant is defective in the Δ7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell. 1999;11:207–221. [PMC free article] [PubMed] [Google Scholar]
- 18.Schaeffer A, Bronner R, Benveniste P, Schaller H. The ratio of campesterol to sitosterol that modulates growth in Arabidopsis is controlled by STEROL METHYLTRANSFERASE 2;1. Plant J. 2001;25:605–615. doi: 10.1046/j.1365-313x.2001.00994.x. [DOI] [PubMed] [Google Scholar]
- 19.Carland FM, Fujioka S, Takatsuto S, Yoshida S, Nelson T. The identification of CVP1 reveals a role for sterols in vascular patterning. Plant Cell. 2002;14:2045–2058. doi: 10.1105/tpc.003939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasbery JM, et al. Arabidopsis thaliana squalene epoxidase 1 is essential for root and seed development. J Biol Chem. 2007;282:17002–17013. doi: 10.1074/jbc.M611831200. [DOI] [PubMed] [Google Scholar]
- 21.Clouse SD. Arabidopsis mutants reveal multiple roles for sterols in plant development. Plant Cell. 2002;14:1995–2000. doi: 10.1105/tpc.140930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindsey K, Pullen ML, Topping JF. Importance of plant sterols in pattern formation and hormone signalling. Trends Plants Sci. 2003;8:521–525. doi: 10.1016/j.tplants.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 23.Schaller H. The role of sterols in plant growth and development. Prog Lipid Res. 2003;42:163–175. doi: 10.1016/s0163-7827(02)00047-4. [DOI] [PubMed] [Google Scholar]
- 24.Babiychuk E, Fuangthong M, Van Montagu M, Inzé D, Kushnir S. Efficient gene tagging in Arabidopsis thaliana using a gene trap approach. Proc Natl Acad Sci USA. 1997;94:12722–12727. doi: 10.1073/pnas.94.23.12722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gondet L, Weber T, Maillot-Vernier P, Benveniste P, Bach TJ. Regulatory role of microsomal 3-hydroxy-3-methylglutaryl-coenzyme A reductase in a tobacco mutant that overproduces sterols. Biochem Biophys Res Commun. 1992;186:888–893. doi: 10.1016/0006-291x(92)90829-a. [DOI] [PubMed] [Google Scholar]
- 26.Schaller H, et al. Expression of the Hevea brasiliensis (HBK.) Müll. Arg. 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1 in tobacco results in sterol overproduction. Plant Physiol. 1995;109:761–770. doi: 10.1104/pp.109.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chappell J, Wolf F, Proulx J, Cuellar R, Saunders C. Is the reaction catalyzed by 3-hydroxy-3-methylglutaryl coenzyme A reductase a rate-limiting step for isoprenoid biosynthesis in plants? Plant Physiol. 1995;109:1337–1343. doi: 10.1104/pp.109.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki M, et al. Loss of function of 3-hydroxy-3-methylglutaryl coenzyme A reductase 1 (HMG1) in Arabidopsis leads to dwarfing, early senescence and male sterility, and reduced sterol levels. Plant J. 2004;37:750–761. doi: 10.1111/j.1365-313x.2004.02003.x. [DOI] [PubMed] [Google Scholar]
- 29.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 30.Sieburth LE, Drews GN, Meyerowitz EM. Non-autonomy of AGAMOUS function in flower development: Use of a Cre/loxP method for mosaic analysis in Arabidopsis. Development. 1998;125:4303–4312. doi: 10.1242/dev.125.21.4303. [DOI] [PubMed] [Google Scholar]
- 31.Qin H, Dong Y, von Arnim AG. Epigenetic interactions between Arabidopsis transgenes: Characterization in light of transgene integration sites. Plant Mol Biol. 2003;52:217–231. doi: 10.1023/a:1023941123149. [DOI] [PubMed] [Google Scholar]
- 32.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 33.Awai K, Xu C, Lu B, Benning C. Lipid trafficking between the endoplasmic reticulum and the chloroplast. Biochem Soc Trans. 2006;34:395–398. doi: 10.1042/BST0340395. [DOI] [PubMed] [Google Scholar]
- 34.Melkonian M, Robenek H, Steup M. Occurrence and distribution of filipin-sterol complexes in chloroplast envelope membranes of algae and higher plants as visualized by freeze-fracture. Protoplasma. 1981;109:349–358. [Google Scholar]
- 35.Andersson MX, Goksör M, Sandelius AS. Optical manipulation reveals strong attracting forces at membrane contact sites between endoplasmic reticulum and chloroplasts. J Biol Chem. 2007;282:1170–1174. doi: 10.1074/jbc.M608124200. [DOI] [PubMed] [Google Scholar]
- 36.Laule O, et al. Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2003;100:6866–6871. doi: 10.1073/pnas.1031755100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Babiychuk E, Kushnir S, Belles-Boix E, Van Montagu M, Inzé D. Arabidopsis thaliana NADPH oxidoreductase homologs confer tolerance of yeasts toward the thiol-oxidizing drug diamide. J Biol Chem. 1995;270:26224–26231. doi: 10.1074/jbc.270.44.26224. [DOI] [PubMed] [Google Scholar]
- 38.Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- 39.Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- 40.Matsuhara S, Jingu F, Takahashi T, Komeda Y. Heat-shock tagging: A simple method for expression and isolation of plant genome DNA flanked by T-DNA insertions. Plant J. 2000;22:79–86. doi: 10.1046/j.1365-313x.2000.00716.x. [DOI] [PubMed] [Google Scholar]
- 41.Joubès J, De Schutter K, Verkest A, Inzé D, De Veylder L. Conditional, recombinase-mediated expression of genes in plant cell cultures. Plant J. 2004;37:889–896. doi: 10.1111/j.1365-313x.2004.02004.x. [DOI] [PubMed] [Google Scholar]
- 42.An Y-Q, et al. Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 1996;10:107–121. doi: 10.1046/j.1365-313x.1996.10010107.x. [DOI] [PubMed] [Google Scholar]
- 43.Du L, Lykkesfeldt J, Olsen CE, Halkier BA. Involvement of cytochrome P450 in oxime production in glucosinolate biosynthesis as demonstrated by an in vitro microsomal enzyme system isolated from jasmonic acid-induced seedlings of Sinapis alba L. Proc Natl Acad Sci USA. 1995;92:12505–12509. doi: 10.1073/pnas.92.26.12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.