Abstract
The mammalian centromeric protein CENP-B shares significant sequence similarity with three proteins in fission yeast (Abp1, Cbh1 and Cbh2) that also bind centromeres and have essential function for chromosome segregation and centromeric heterochromatin formation. Each of these proteins displays extensive sequence similarity with pogo-like transposases, which have been previously identified in the genomes of various insects and vertebrates, in the protozoan Entamoeba and in plants. Based on this distribution, it has been proposed that the mammalian and fission yeast centromeric proteins are derived from ‘domesticated’ pogo-like transposons. Here we took advantage of the vast amount of sequence information that has become recently available for a wide range of fungal and animal species to investigate the origin of the mammalian CENP-B and yeast CENP-B-like genes. A highly conserved ortholog of CENP-B was detected in 31 species of mammals, including opossum and platypus, but was absent from all non-mammalian species represented in the databases. Similarly, no ortholog of the fission yeast centromeric proteins was identified in any of the various fungal genomes currently available. In contrast, we discovered a plethora of novel pogo-like transposons in diverse invertebrates and vertebrates and in several filamentous fungi. Phylogenetic analysis revealed that the mammalian and fission yeast CENP-B proteins fall into two distinct monophyletic clades, each of which includes a different set of pogo-like transposons. These results are most parsimoniously explained by independent domestication events of pogo-like transposases into centromeric proteins in the mammalian and fission yeast lineages, a case of ‘convergent domestication’. These findings highlight the propensity of transposases to give rise to new host proteins and the potential of transposons as sources of genetic innovation.
Keywords: exaptation, transposon domestication, convergence, centromeres, CENP-B, Abp1
Introduction
The origin of new genes is key to our understanding of how genomes evolve and how new biological functions emerge. The most intensively studied mechanism underlying the evolution of new genes involves the duplication or rearrangement of pre-existing genes or exons (Long et al. 2003). Gene duplication may occur at the DNA level, through segmental or whole-genome duplication, or at the RNA level through retroposition, a process by which messenger RNA is reverse-transcribed into a DNA copy that is reintegrated elsewhere in the genome. A distinct, less characterized mechanism for the emergence of new genes is the recycling of sequences and activities previously encoded by transposable elements (TEs), also known as TE ‘domestication’ or ‘exaptation’ (Brosius and Gould 1992; Miller et al. 1999; Volff 2006).
TEs are selfish mobile genetic elements, and their genes encode proteins that normally serve only their propagation. On some occasions, however, TE genes can be co-opted or ‘domesticated’ by the host to assume cellular function, a form of ‘exaptation’ (Brosius and Gould 1992). While several cases of exaptation of TE coding sequences have been documented, very few of these TE-derived proteins have been characterized functionally, and thus, for the most part, their biological functions remain unknown (Brosius 1999; Miller et al. 1999; Smit 1999; Britten 2004; Volff 2006; Feschotte and Pritham 2007). Another open question is whether TE domestication is merely an evolutionary incident, resulting from the sheer abundance and nearly ubiquitous nature of TEs in eukaryotic genomes, or whether certain TEs possess intrinsic properties that enhance exaptation and their recycling into functional components of the genome (Cordaux et al. 2006; Feschotte and Pritham 2007).
One of the earliest documented cases of TE exaptation is the gene encoding human centromere-associated protein B (CENP-B) (for review, Masumoto, Nakano, and Ohzeki 2004). CENP-B encodes a ~599-aa protein which localizes densely at the centromere of all human chromosomes, except the Y chromosome (Earnshaw et al. 1987; Earnshaw, Ratrie, and Stetten 1989; Yoda et al. 1992). The CENP-B protein binds as a homodimer specifically to a 17-bp motif called the CENP-B box located within alpha-satellite centromeric DNA (Masumoto et al. 1989; Yoda et al. 1992; Tanaka et al. 2001). CENP-B and the CENP-B box appear to be highly conserved throughout mammals. The mouse homologous protein is 92% identical to human CENP-B and is associated with centromeric satellites through binding of a DNA motif highly similar to the human CENP-B box (Sullivan and Glass 1991; Kipling et al. 1995). Sequences displaying high sequence identity to the human CENP-B have also been isolated in hamster, sheep and in several primates (Haaf et al. 1995; Burkin at al. 1996; Goldberg et al. 1996; Yoda et al. 1996; Bejarano and Valdivia 1996). More recently, a DNA motif similar in sequence to the CENP-B box was identified within the centromeric satellite repeats of the marsupial Macropus rufogriseus (Bulazel et al. 2006). A fragment containing the motif was bound in vitro by recombinant human CENP-B protein, suggesting that it represents a binding site for a yet uncharacterized marsupial CENP-B homolog (Bulazel et al. 2006). There is no convincing report of the isolation of a CENP-B homolog outside mammals, although some have reported the presence of motifs weakly similar to the CENP-B box in the satellite DNA repeats of Xenopus, insects and plants (Coelho et al. 1996; Lopez and Edstrom 1998; Weide et al. 1998; Heslop-Harrison et al. 1999; Nonomura and Kurata 1999; Lorite et al. 2004; Mravinac, Plohl, and Ugarkovic 2004; Edwards and Murray 2005). Despite the apparent selective constraint acting on CENP-B and the CENP-B box in diverse mammals, its exact function at the centromere remains unclear and even controversial, since a mouse null mutant for Cenp-b exhibits no obvious defects in chromosome segregation and only weak phenotypic abnormalities (Hudson et al. 1998; Kapoor et al. 1998; Perez-Castro et al. 1998; Fowler et al. 2000). Thus, it remains unclear whether CENP-B is involved in chromosome segregation.
It was initially noted that CENP-B displays significant similarity throughout its entire sequence with the transposase encoded by the pogo element of Drosophila melanogaster (Tudor et al. 1992). This relationship was later confirmed through the discovery and analysis of distantly related pogo-like elements in human and Arabidopsis (Robertson 1996; Smit and Riggs 1996; Kapitonov and Jurka 1999; Feschotte and Mouchès 2000). These elements are DNA transposons that form a monophyletic subgroup within the extended Tc1/mariner superfamily (Capy et al. 1998; Plasterk, Izsvák, and Ivics 1999). Since the taxonomic distribution of CENP-B was apparently narrower than those of pogo-like elements and other Tc1/mariner transposons, which are widespread in eukaryotes, it has been hypothesized that CENP-B arose from domestication of a pogo-like transposon (Smit and Riggs 1996; Kipling and Warburton 1997)
Subsequent to the isolation of CENP-B in human and mouse, three centromere-binding proteins were identified in the fission yeast Schizosaccharomyces pombe, ARS-binding protein (Abp1), CENP-B homolog 1 (Cbh1) and CENP-B homolog 2 (Cbh2), that share significant sequence similarity to each other and to mammalian CENP-B (Murakami, Huberman, and Hurwitz 1996b; Lee, Huberman, and Hurwitz 1997; Irelan, Gutkin, and Clarke 2001). Abp1, Cbh1 and Cbh2 have been shown to localize and bind distinct degenerate DNA motifs within the centromeres of S. pombe chromosomes (Halverson et al. 1997; Lee, Huberman, and Hurwitz 1997; Ngan and Clarke 1997; Irelan, Gutkin, and Clarke 2001; Nakagawa et al. 2002). In addition, genetic and biochemical analysis indicates that all three proteins have partially redundant function required for centromeric heterochromatin assembly and chromosome segregation in this organism (Baum and Clarke 2000; Irelan, Gutkin, and Clarke 2001; Nakagawa et al. 2002). It has been proposed that the fission yeast proteins represent functional homologs of the mammalian CENP-B. Studies of these proteins in fission yeast are often used as a model to better understand the function of CENP-B in human centromeric function (e.g. Irelan, Gutkin, and Clarke 2001; Nakagawa et al. 2002; Amor et al. 2004), with the underlying, but undemonstrated, assumption that they are orthologous.
The apparent phenotypic redundancy of the three fission yeast CENP-B homologs has led to the hypothesis that the lack of phenotypic deficiencies in mouse cenp-b mutants might stem from the presence in the mouse genome of genes encoding proteins functionally redundant with CENP-B (Hudson et al. 1998; Kapoor et al. 1998; Irelan, Gutkin, and Clarke 2001; Nakagawa et al. 2002). Although several genes encoding proteins distantly related to CENP-B have been identified in mammals, (Toth et al. 1995; Smit and Riggs 1996; Kipling and Warburton 1997; Zeng et al. 1997; Dou et al. 2004), it is unknown whether any of these proteins act redundantly with CENP-B. Furthermore, the evolutionary relationships of these proteins with mammalian CENP-B or with the fission yeast homologs remain unclear.
The observations summarized above raise two important questions. First, do the fission yeast proteins and mammalian CENP-B descend from a common CENP-B-like ancestor, i.e. are they orthologous, or did they arise independently from domestication events of distinct pogo-like transposases? Second, are there any paralogous genes in mammals that could be functionally redundant with CENP-B, as demonstrated for the S. pombe CENP-B-like proteins? To begin answering these questions, we capitalized on the vast amount of sequence information that recently became available to investigate the origin and evolutionary relationships of the mammalian CENP-B, fission yeast ‘homologs’ and pogo-like transposons. The results point to a remarkable scenario of convergent TE domestication, whereby two different sources of pogo-like transposase were independently recruited into centromere-binding proteins in the mammalian and fission yeast lineages.
Methods
Gene and transposon mining
In order to retrieve all sequences related to mammalian CENP-B and S. pombe Abp1, Cbh1 and Cbh2 proteins, we used these four proteins, as well as representative pogo-like transposases obtained from GenBank and Repbase (Jurka et al. 2005), to perform exhaustive and reiterative similarity searches of all NCBI protein and nucleotide databases using PSI-BLAST and TBLASTN, respectively. All sequences returned that had more than 30% identity to any of the queries were parsed and retained for further analysis. We next sought to distinguish which of these proteins are encoded by pogo-like transposons and which are encoded as stationary, ‘domesticated’ genes by the host. pogo-like elements, like most DNA (class 2) transposons, are characterized by terminal inverted repeats (TIRs) and conserved target site duplications (TSD) (TA for pogo-like elements). We thus inspected the flanking of each sequences retrieved from the databases for TIRs and TSDs. This procedure allowed us to identify numerous novel pogo-like transposons, which were clustered into families in which individual copies shared extensive nucleotide similarity (>80% over their entire length). To assess syntenic relationships among mammalian pogo-derived genes, we used the pairwise “chained” alignments available at the UCSC Genome Browser Database (http://genome.ucsc.edu/). Orthology was also confirmed by observing the synteny conservation of genes flanking CENP-B and other pogo-derived genes. Accession numbers of newly described pogo-derived genes and pogo-like transposons are reported in table 1 and table 2, respectively. Accession numbers of CENP-B orthologs used in supplementary figures 1 and 2 are: Ateles geoffroyi GI:145279281; Aotus nancymaae GI:74096596; Bos taurus GI:112152000; Callithrix jacchus GI:74217361; Canis familiaris GI:63121068; Cavia porcellus GI:78823234; Chlorocebus aethiops GI:78097410; Colobus guereza GI:82491700; Cricetulus griseus, GI:836955; Dasypus novemcinctus GI:64743060; Equus caballus GI:124093125; Erinaceus europaeus GI:87976610; Felis catus GI:94069308; Homo sapiens GI:148138332; Lemur catta GI:83745227; Macaca mulatta GI:86636346; Monodelphis domestica GI:84821469; Muntiacus reevesi GI:151933349; Mus musculus GI:69973226; Myotis lucifugus GI:105819785; Ornithorhynchus anatinus GI:91357513; Oryctolagus cuniculus GI:63919666; Otolemur garnettii GI:106166469; Ovis aries GI:1016291; Pan troglodytes GI:89627452; Papio anubis GI:78097412; Pongo pygmaeus GI:83745199; Rattus norvegicus GI:32740592; Saimiri boliviensis boliviensis GI:60418061; Spermophilus tridecemlineatus GI:107608120; Tupaia belangeri GI:107987561.
Table 1.
Characteristics of pogo-derived genes used for phylogenetic reconstruction in figure 2
| Species | Gene Name | ID a | PRT length | Clade b |
|---|---|---|---|---|
| Anolis carolinensis | Anolis TIGD4 | 125758256 | 455 | CR |
| Aspergillus oryzae | A.oryzae 678 | 83774678 | 476 | AR |
| Candida glabrata | C.glabrata Pdc2 | 50292447 | 794 | AR |
| Homo sapiens | Human CENPB | P07199 | 599 | CR |
| Homo sapiens | Human JRK | AAH43351 | 568 | JR |
| Homo sapiens | Human JRKL | AAI09203 | 524 | JR |
| Homo sapiens | Human TIGD1 | AAH35143 | 591 | JR |
| Homo sapiens | Human TIGD2 | NP_663761 | 525 | JR |
| Homo sapiens | Human TIGD3 | AAH74862 | 471 | CR |
| Homo sapiens | Human TIGD4 | NP_663772 | 512 | CR |
| Homo sapiens | Human TIGD5 | NP_116251 | 593 | JR |
| Homo sapiens | Human TIGD6 | NP_112215 | 521 | CR |
| Homo sapiens | Human TIGD7 | NP_149985 | 549 | JR |
| Homo sapiens | Human POGK | NP_060012 | 609 | TC2 |
| Homo sapiens | Human POGZ | NP_055915 | 1410 | TC2 |
| Monodelphis domestica | Opossum CENPB | XP_001380458 | 588 | CR |
| Monodelphis domestica | Opossum JRK | XP_001381773 | 570 | JR |
| Monodelphis domestica | Opossum JRKL | XP_001367318 | 525 | JR |
| Monodelphis domestica | Opossum TIGD2 | XP_001375789 | 528 | JR |
| Monodelphis domestica | Opossum TIGD6 | XP_001378790 | 523 | CR |
| Monodelphis domestica | Opossum TIGD7 | 84825501 | 553 | JR |
| Mus musculus | Mouse TIGD3 | NP_941036 | 470 | CR |
| Saccharomyces cerevisiae | Saccharomyces Pdc2 | 577808 | 925 | AR |
| Schizosaccharomyces pombe | S.pombe Abp1 | 19860259 | 522 | AR |
| Schizosaccharomyces pombe | S.pombe Cbh1 | 12643496 | 514 | AR |
| Schizosaccharomyces pombe | S.pombe Cbh2 | 26391975 | 514 | AR |
| Xenopus tropicalis | Xenopus TIGD5 | scaffold_737:167,882-169,480 | 535 | JR |
Protein ID for genes, except Opossum TIGD7 (genomic clone) and Xenopus TIGD5 (scaffold)
Major clades reported in figure 2
Table 2.
Characteristics of pogo-like transposons used for phylogenetic reconstruction in figure 2
| Species | TE Name | ID a | TE nt b | TIRs nt b | TPase length | Clade c |
|---|---|---|---|---|---|---|
| Aedes aegypti | Aedes pogo1 | 78134152 | 2511 | 26 | 522 | CR |
| Anolis carolinensis | Anolis pogo1 | Consensus | na | na | 597 | JR |
| Anolis carolinensis | Anolis pogo2 | Consensus | 3396 | 23 | 585 | JR |
| Anolis carolinensis | Anolis pogo3 | 125764245 | 2432 | 24 | 569 | JR |
| Anolis carolinensis | Anolis TIGGU | Consensus | 2463 | 18 | 570 | JR |
| Anopheles gambiae | Anopheles pogo1 | 31249463 | na | na | 559 | JR |
| Aplysia californica | Aplysia pogo1 | 112506762 | na | na | 559 | CR |
| Aplysia californica | Aplysia pogo2 | 112495283 | na | na | 520 | CR |
| Arabidopsis thaliana | Arabidopsis Lemi1 | 20197576 | 2114 | 24 | 457 | AR |
| Aspergillus fumigatus | A.fumigatus mariner 7 | Repbase | 1825 | 23 | 518 | AR |
| Aspergillus nidulans | A.nidulans mariner 3 | Repbase | 1848 | 13 | 525 | AR |
| Aspergillus niger | A.niger Tan1 | 1805250 | 2324 | 45 | 555 | Fot1 |
| Bombyx mori | Bombyx pogo1 | 46726051 | 2148 | 27 | 501 | CR |
| Candida albicans | C.albicans Cirt1 | 10799004 | 1804 | 40 | 526 | Fot1 |
| Coccidioides immitis | Coccidioides pogo1 | 119181148 | 1358 | 22 | 528 | AR |
| Culex pipiens | Culex pogo1 | 145648644 | na | na | 531 | JR |
| Drosophila ananassae | D.ananassae pogo1 | 109914388 | 2069 | 22 | 507 | JR |
| Drosophila melanogaster | D.melanogaster pogo1 | Repbase | 2121 | 21 | 499 | CR |
| Drosophila mojavensis | D.mojavensis pogo1 | 91924523 | 2234 | 23 | 507 | CR |
| Fusarium oxysporum | Fusarium Fot1 | Repbase | 1928 | 42 | 542 | Fot1 |
| Homo sapiens | Human Tigger1 | Repbase | 2418 | 23 | 454 | JR |
| Homo Sapiens | Human Tigger2 | Repbase | 2718 | 24 | 627 | JR |
| Eutheria d | Tigger3 “Golem” | Repbase | 3029 | 23 | 614 | JR |
| Eutheria d | Tigger4 “Zombi” | Repbase | 2806 | 25 | 452 | JR |
| Neosartorya fischeri | Neosartorya pogo1 | 83742782 | 1826 | 23 | 519 | AR |
| Neosartorya fischeri | Neosartorya pogo2 | 83742785 | 1841 | 24 | 516 | AR |
| Nosema bombycis | Nosema pogo1 | 91178026 | na | na | 487 | JR |
| Phytophthora ramorum | P.ramorum pogo1 | 113919901 | 1999 | 18 | 549 | AR |
| Phytophthora sojae | P.sojae pogo1 | 113924094 | 1786 | 22 | 488 | AR |
| Puccinia graminis | Puccinia pogo1 | 123974350 | 2798 | 24 | 599 | AR |
| Schmidtea mediterranea | Schmidtea pogo2 | Consensus | 2263 | 24 | 578 | JR |
| Schmidtea mediterranea | Schmidtea pogo6 | Consensus | 1981 | 27 | 509 | CR |
| Schmidtea mediterranea | Schmidtea pogo9 | Consensus | 2233 | 24 | 561 | JR |
| Schmidtea mediterranea | Schmidtea pogo11 | Consensus | 1936 | 24 | 489 | CR |
| Takifugu rubripes | Takifugu TIGGU2 | Repbase | 2413 | 23 | 486 | JR |
| Takifugu rubripes | Takifugu TC2FR5 | Repbase | 2209 | 17 | 443 | TC2 |
| Tribolium castaneum | Tribolium pogo1 | 73485479 | na | na | 498 | JR |
| Trichinella spiralis | Trichinella pogo1 | 110005903 | 2073 | 24 | 502 | CR |
Nucleotide ID for TEs. “Consensus” means that more than one transposon has been used to build a consensus sequence
Length in nucleotides. “na” (non-available) highlights transposons where TIRs and total length cannot be unambiguously determined
Major clades reported in figure 2
The used consensus sequences of these transposons represent putative active pogo-like TEs active before the radiation of eutherians
Evolutionary and phylogenetic analyses
Pairwise alignments were constructed using ClustalX (Chenna et al. 2003) and MAFFT (Katoh et al. 2005), and manually refined using Bioedit v7.0.5.3 (Hall 1999) and GeneDoc v2.6.002 (http://www.psc.edu/biomed/genedoc). K-estimator (Comeron 1999) was used to tabulate the number of non-synonymous sites and substitutions as well as frequencies for each of three classes of synonymous sites and substitutions (2-S, 2-V, and 4-fold). Using these data, we calculated dN/dS after the method of Pamilo and Bianchi (Pamilo and Bianchi 1993). For the maximum likelihood method, we utilized the free ratio branch model of the codeml program in the PAML suite (Yang 1997). We confined this analysis to full length sequences comprising unambiguous ORFs, using the following input tree, aided by Treeview: ((((CENP-B_hs, CENP-B_Papio_anubis), (CENP-B_Ateles_geoffroyi,CENP-B_Aotus_nancymaae, CENP-B_Callithrix_jacchus)), CENP-B_Lemur_catta), ((CENP-B_mus,CENP-B_rat), CENP-B_hamster), CENP-B_opo). To formally test whether the observed ω on each individual branch was significantly < 1, we ran a series of models in which all branches were free, except 1, which was constrained to ω=1. Likelihood ratio tests were then compared to a chi-squared distribution, with 1 degree of freedom. The neighbor-joining tree in supplementary figure 1 has been obtained using MEGA 3.1 (Kumar, Tamura, and Nei 2004).
Phylogenetic trees of the pogo family were inferred with MrBayes (Ronquist and Huelsenbeck 2003), applying a mixed amino acid model with a discrete gamma-distribution with four rate categories and random starting trees. Two independent runs with four Markov chains each were operating for one million generations with a sampling frequency set to 100. When the standard deviation of split frequencies was <0.01, we considered the two runs converged. The temperature difference between the ‘cold’ chain and the ‘heated’ chains was set to 0.1 to improve the chain swap. For the consensus tree the “burnin” parameter was set to 25% of the samples. We also tested a slightly different approach to infer the phylogenetic tree of the pogo family. First, we determined the most likely amino acid model of evolution on the final multialignment of 65 protein sequences using ProtTest1.3 (Abascal, Zardoya, and Posada 2005). ProtTest implements the Akaike Information Criterion and the Bayesian Information Criterion to establish the likelihood of up to ten different models, and produces a maximum-likelihood phylogenetic tree according to the best model. Then, we executed MrBayes using as initial tree the maximum-likelihood tree instead of random trees for both runs, and fixing the model to the best fitting one according to ProtTest. We observed no significant difference between the trees obtained with the two methods.
Results
CENP-B is widespread and highly conserved in mammals, but undetectable in other metazoans
To identify sequences related to CENP-B, we performed reiterative PSI-BLAST and TBLASTN searches of all NCBI protein and translated nucleotide databases using the human CENP-B protein as the initial query (see Methods). These searches yielded several hundred significant hits that fell into two distinct categories. The first category of hits exhibited amino acid identity >70% over >100 amino acids and all were of mammalian origin. The second category of hits had less than 35% amino acid identity and originated from various organisms. The restricted taxonomic distribution of the high-similarity hits, coupled to the absence of hits with intermediate levels of sequence identity, intuitively suggested that the first category of sequences represented CENP-B orthologs. This assumption was subsequently corroborated by phylogenetic analyses (see suppl. fig. 1 and below) and by syntenic relationship across the genomes of human, mouse, rat, dog and opossum (see Methods). Together these data revealed the presence of a CENP-B ortholog in 31 mammalian species (fig. 1). Reciprocal and reiterative blast searches with each query against the databases revealed only one sequence with high sequence identity (74–100% identical over 100 amino acids) to CENP-B in each mammalian species represented in the database, which includes 27 species with genome sequencing projects completed or nearing completion. For 10 of these mammalian species, high coverage, assembled genome data are currently available. Thus, CENP-B appears to be present as a single-copy gene in all mammalian species examined, including the nonplacental species (opossum and platypus). However, because some species have only partial genome coverage, we cannot exclude the possibility that they possess one or several recently generated CENP-B paralog(s).
Figure 1.
Distribution of CENP-B and related pogo-derived genes. Solid figures indicate apparent extant genes, as evidenced by the presence of uninterrupted ORFs and dN/dS <0.5 and significantly <1. Unshaded figures indicate putative genes exhibiting dN/dS <0.5 and significantly <1, which contain one or more apparent frameshift and/or nonsense mutations. The Greek letter ψ indicates probable pseudogenes, which, in addition to harboring apparent frameshift and nonsense mutations, exhibit dN/dS >0.5 and not significantly <1. Assembled, high coverage genomes (NHGRI, NIH, http://www.genome.gov/10002154) are in bold print. A dash indicates the absence of a detected ortholog in an assembled, high coverage genome.
Out of the 31 mammalian CENP-B orthologs detected, 28 genes harbor a single uninterrupted ORF, while 3 genes exhibit at least one frameshift mutation that could not be attributed to obvious sequencing errors based on the available sequence data (indicated by an unshaded symbol in fig. 1). To confirm that the 28 apparently intact orthologs are extant, functional genes, we sought evidence of selective constraint, in the form of ω (dN/dS), by both a maximum likelihood method (Yang 1997) and a counting method using K-estimator (Comeron 1999) (see Methods). Both analyses produced evidence of strong purifying selection acting in all mammalian lineages, with ω estimates no greater than 0.21. All ω estimates were significantly lower than 1 (p values < 0.00001, Likelihood Ratio Test for PAML, and p values < 0.002, z-test, for counting method) and the tree-wide ω estimate was 0.078. Similar ω estimates were obtained for the 3 ambiguous genes noted above, suggesting that the apparent frameshift mutations in these ORFs are either sequencing artifacts or that these genes underwent independent and recent pseudogenization events. However, given the omnipresence of CENP-B and the strong selective constraint operating on the encoded protein in all the mammalian lineages examined, the hypothesis that CENP-B would be lost independently in these three species seems improbable.
In summary, CENP-B appears to be present as a functional, single-copy gene in most (if not all) mammalian species examined (fig. 1). The gene has followed a similar pattern of evolution in all lineages, characterized by strong purifying selection. The mark of selection is particularly intense on the N-terminal DNA-binding domain (Tanaka et al. 2001), which is nearly identical in the 17 mammalian species where sequence coverage is available for this region (see alignment in suppl. fig. 2). We could not detect any potential CENP-B ortholog in any non-mammalian vertebrates, including three tetrapod species for which draft genome assemblies are available (chicken, the squamate Anolis carolinensis, and the amphibian Xenopus tropicalis). Likewise, we could not detect a direct homolog of CENP-B in any of the numerous and diverse invertebrates with draft genome assemblies (including one echinoderm, 3 ascidians, 12 Drosophila, 3 mosquitoes, one beetle, one lepidopteran, 3 nematodes, 2 flatworms and 1 cnidarian). Given the extreme level of sequence conservation of CENP-B in mammals, it seems inconceivable that homology-based searches would systematically fail to identify a possible ortholog in every one of these animals. Therefore, CENP-B must be taxonomically restricted to mammals. These data indicate that CENP-B originated prior to the split of monotremes, marsupials and placentals and, most likely, in the last common ancestor of these lineages.
Other pogo-derived genes in metazoans
The second category of BLAST hits to CENP-B in mammalian genomes appears to be the result of sequence similarity to distantly related transposase pseudogenes (Tigger elements, see next section)(Smit and Riggs 1996) or to several additional ‘host’ genes with distant homology to CENP-B. Besides CENP-B, nine orthologous clusters of pogo-derived genes can be recognized in mammals: Tigger-derived genes 1–7 (TIGD1-7) (Robertson 2002; Dou et al. 2004), JERKY (JRK) (Toth et al. 1995) and Jerky-like (JRKL) (Zeng et al. 1997) (see table 1). Only the mouse jerky gene has been functionally characterized. Disruption of this gene in mice causes a phenotype that is characterized by recurrent limbic seizures reminiscent of some forms of human inherited epilepsy (Toth et al. 1995). The mouse JRK protein has both DNA and RNA-binding activity and specific neuronal localization (Liu et al. 2003). Like CENP-B, JRK and other pogo-derived genes seem to be restricted to mammals. Based on the current data, the only two exceptions appear to be TIGD4 and TIGD5. We identified an ortholog of TIGD4 in Anolis carolinensis and an ortholog of TIGD5 in Xenopus tropicalis and chicken (though it may be a pseudogene in the latter species, data not shown) (table 1). Thus, TIGD4 and TIGD5 are the only two mammalian pogo-derived genes found in other vertebrates.
Overall, each of these additional pogo-derived proteins has relatively weak similarity to CENP-B (18–28% identity). TIGD3, TIGD4 and TIGD6 seem most closely related to CENP-B (see below) and, like CENP-B, they are widely distributed and highly conserved in mammals (fig. 1). Within a given mammalian genome, TIGD6 is consistently the closest relative to CENP-B, with an average pairwise amino acid identity of 28%. Phylogenetic distance analysis based on synonymous sites (Ks tree, not shown) with an application of an average neutral substitution rate of ~2.2−9 in mammals (Kumar and Subramanian 2002) is indicative of a coalescence time congruent with a much earlier divergence of CENPB, TIGD3, TIGD4 and TIGD6 than is suggested by their taxonomic distribution (i.e. mammalian-wide or possibly amniote-wide in the case of TIGD4). These observations are in line with the hypothesis that CENP-B and TIGD3, 4 and 6 are not paralogous genes that arose by ‘classic’ duplication of an ancestral gene. It is more probable that each of these genes originated by independent domestication of a different source of pogo-like transposase. These events must have taken place during an evolutionary timeframe ranging from the emergence of amniotes (~360 Mya) to the split of monotremes and marsupials (~230 Mya) (Hedges and Kumar 2003; van Rheede et al. 2006). This scenario is further supported by the phylogenetic placement and relationships of the encoded proteins to each other and to various pogo-like transposases (see below). Thus, it appears that pogo-like transposons were a recurrent source of new protein-coding genes in mammals.
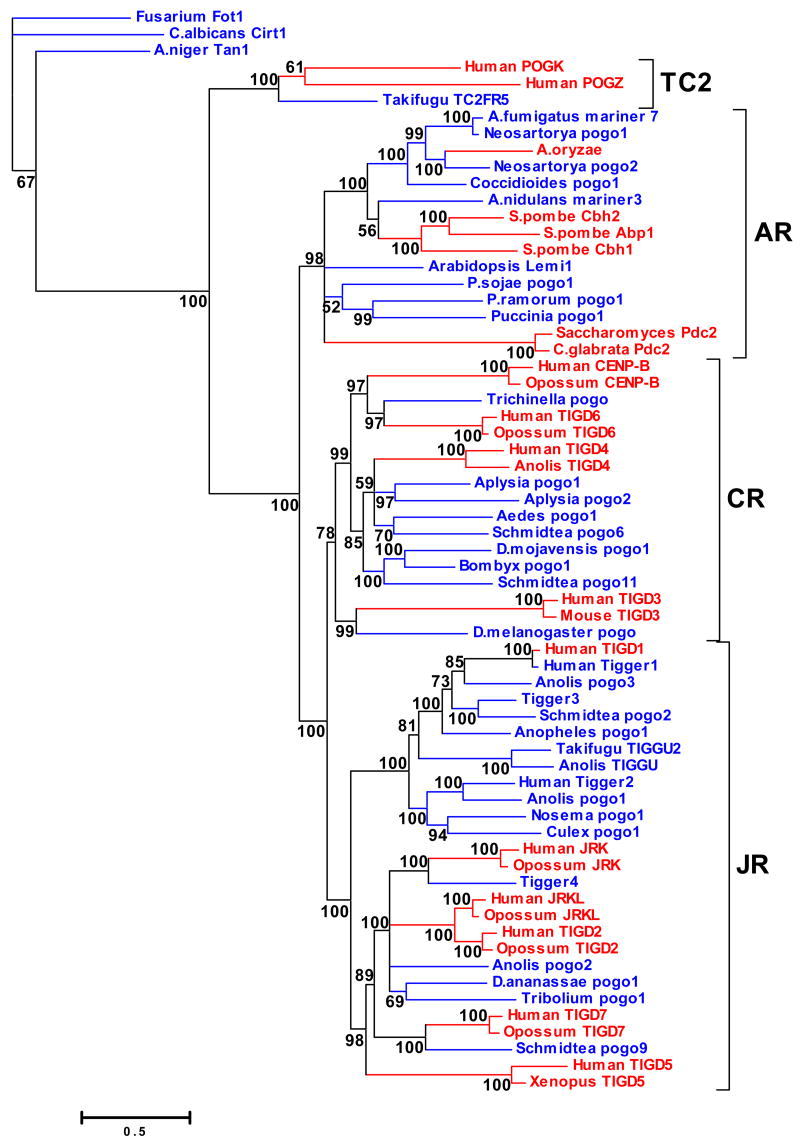
A plethora of newly identified pogo-like transposons in various metazoans
While we could not detect an ortholog of CENP-B in any of the non-mammalian metazoan species represented in the databases, we could readily identify multiple pogo-like transposons in many of these species (table 2). Some of these transposons have been previously described, such as the founding pogo element in D. melanogaster (Tudor et al. 1992) and the Tigger elements of H. sapiens (Smit and Riggs 1996; Robertson 1996), but most represent newly identified families from the lizard Anolis carolinensis, the nematode Trichinella spiralis, the sea hare Aplysia californica, the flatworm Schmidtea mediterrranea, the starlet sea anemone Nematostella vectensis and several insects (table 2). Most of these elements have TIRs very similar to those of known pogo-like transposons and are flanked by canonical 5′-TA-3′ TSD (fig. 3). Each family is represented by multiple copies dispersed throughout the respective host genome, suggesting relatively recent transpositional activity. Although the transposase sequences from different families or from different species may sometimes be highly divergent (16–52% identity, but in most cases less than 30% identity), phylogenetic analysis unequivocally places these elements within the pogo subgroup of the Tc1/mariner superfamily (fig. 2). These data underscore the high diversity and evolutionary persistence of pogo-like transposons in metazoans, and particularly in invertebrates. Moreover, the widespread distribution of pogo-like transposons in metazoans stands in contrast with the taxonomic restriction of CENP-B to mammals. These findings illustrate that highly divergent pogo-like sequences can be readily retrieved from a wide diversity of animals using conventional homology-based searches. With one exception (TIGD4 in A. carolinensis), these transposases represent the most closely related sequences to the mammalian CENP-B in each of the non-mammalian species represented in the databases. Hence, we believe that our inability to detect CENP-B in non-mammalian species is not due to a lack of sensitivity of the BLAST algorithms, but rather reflects the absence of CENP-B orthologs in these species.
Figure 3.
Terminal inverted repeats of pogo-like transposons. Multialignments of TIR sequences of transposons belonging to the three clades of pogo-like elements identified in this study: CENP-B-related (CR), Abp1-related (AR) and JERKY-related (JR). Nucleotides conserved in at least 50% of sequences are shaded.
Figure 2.
Phylogenetic tree of pogo-like transposases and pogo-derived genes. Protein sequences were aligned using the MAFFT package and the alignment refined manually with Bioedit to produce a final multialignment of about 310 residues. The tree shown here was inferred using the Bayesian algorithms implemented in MrBayes as described in Methods. Numbers in the nodes show posterior probabilities. pogo-derived genes and pogo-like transposons are highlighted in red and blue, respectively. Detailed information about each sequence are reported in table 1 and table 2. CR: CENP-B-related clade; AR:Abp1-related clade; JR: JERKY-related clade: TC2: TC2-related clade. Fot1, Cirt1 and Tan1 belong to the Fot1 clade.
Pogo transposons and their derived genes in fungi
We employed the same BLAST-based strategy to identify sequences related to the fission yeast Abp1, Cbh1 and Cbh2 proteins and to pogo-like transposases in all fungal genomes available. These searches revealed several fungi sequences related to the S. pombe genes. With two exceptions (discussed hereafter), each of these sequences exhibits typical features of transposons, such as multiple interspersed copies, TIRs and TSDs (table 2). Inspection of the TIR sequences and phylogenetic analyses of the predicted transposases indicate that these transposons belong to the pogo subgroup (fig. 2). To our knowledge, these elements are the first characterized fungal pogo-like transposons sensu stricto. Indeed, these elements are clearly distinct from Fot1-like transposons, another subgroup of the Tc1/mariner superfamily that is abundant in filamentous fungi (Daboussi and Capy 2003). In fact, some genomes harbor both Fot1-like and pogo-like elements (e.g. Aspergillus), akin to the single-celled eukaryotes Entamoeba (Pritham, Feschotte, and Wessler 2005). Thus, pogo-like and Fot1-like transposons diverged prior to the divergence of fungi and Entamoeba. The newly discovered fungal pogo-like transposons are the only relatives of the S. pombe CENP-B-like genes in most of the fungi for which we have genome sequence data. Conversely, S. pombe lacks detectable pogo-like transposons and, in fact, this species does not appear to contain any DNA transposons (Wood et al. 2002; Feschotte and Pritham 2007).
Neither Saccharomyces cerevisiae nor other Saccharomycetales harbor detectable pogo-like transposons. Instead, each of these yeasts possesses a single gene, first described as Pdc2 in S. cerevisiae (Mojzita and Hohmann 2006), which is distantly related to the fission yeast ‘CENP-B homologs’ (Smit and Riggs 1996). Orthologs of Pdc2 could be identified in 23 fungi species, all of which are Saccharomycetales. Several lines of evidence suggest that Pdc2 is not orthologous to the fission yeast CENP-B-like genes. First, Pdc2 functions as a transcription factor in the pyruvate decarboxylase pathway (Mojzita and Hohmann 2006) and there is no evidence that the protein has centromere-binding activity or that it plays a role in chromosome segregation. Second, the entire Pdc2 protein is about 1.5 times longer and aligns poorly with the fission yeast proteins (25% identity over 526 amino acid in the most conserved region). Pdc2 and the closely related protein in other Saccharomycetales contain a fast-evolving C-terminal extension with essentially no homology to the fission yeast proteins (data not shown). Third, Pdc2 and the S. pombe proteins fall into two separate phylogenetic clades of pogo-derived proteins intermingled with different pogo-like transposases (see fig. 2 and further description below). Thus, the Pdc2 orthologous gene cluster appears to be derived from yet another source of pogo-like transposase than the S. pombe CENP-B-like proteins.
Two Aspergillus species possess, in addition to pogo-like transposons, a single-copy orthologous gene predicted to encode a protein related to pogo-like transposases (table 1). These two ORFs could not be associated with obvious transposon features. The protein sequences do not group with the fission yeast proteins or with the Pdc2 proteins in phylogenetic reconstructions (see below, fig. 2), suggesting that these two Aspergillus genes are unlikely to be orthologous to the fission yeast CENP-B-like genes. Most likely, they represent previously undescribed stationary pogo-derived genes with an independent origin.
Together these data indicate that the CENP-B ‘homologs’ of S. pombe are restricted to this species, or possibly to the Schizosaccharomycetales. The three fission yeast proteins share 39–47% amino acid identity over their entire length and have redundant functions in chromosome segregation, although each protein appear to bind distinct DNA sites at S. pombe centromeres (Lee, Huberman, and Hurwitz 1997; Ngan and Clarke 1997; Baum and Clarke 2000; Irelan, Gutkin, and Clarke 2001; Nakagawa et al. 2002). These data are consistent with a scenario whereby Abp1, Cbh1 and Cbh2 arose from a single domestication event of a pogo-like transposase in the lineage of S. pombe followed by ‘standard’ gene duplication and subfunctionalization (Prince and Pickett 2002).
Taxonomic distribution suggests independent origins of centromere-binding proteins in mammals and fission yeast
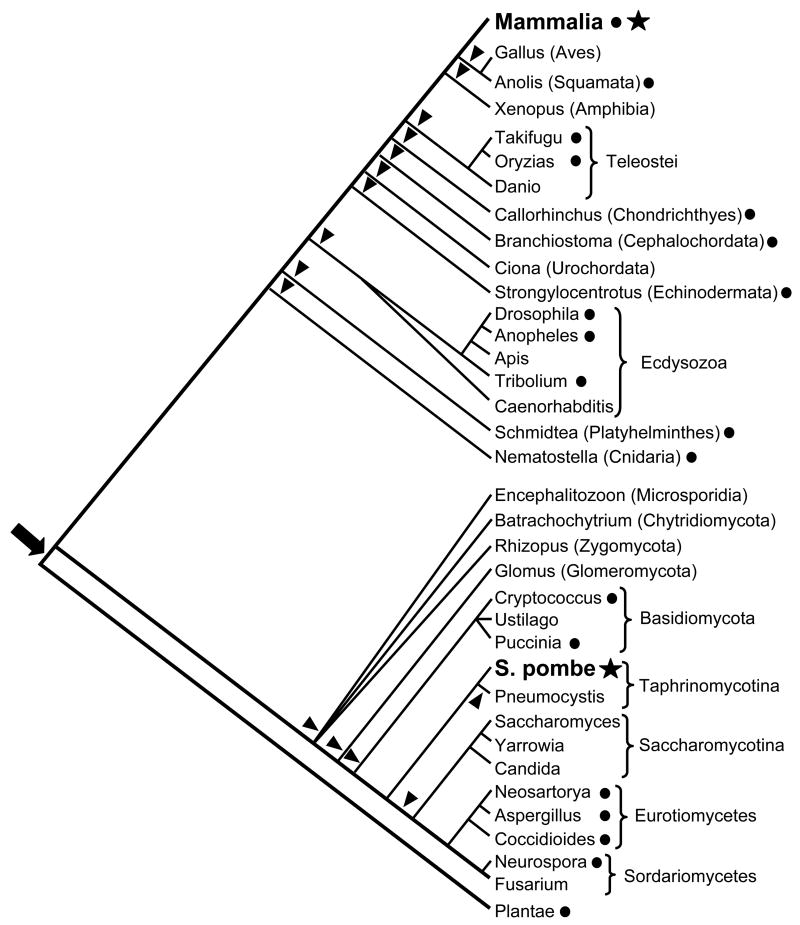
The above data raise the question whether the fission yeast and mammalian centromere-binding proteins descend from a common ancestral gene domesticated prior to the divergence of metazoa and fungi (i.e. they are orthologous) or originated independently from different transposase sources. In light of the taxonomic distribution described above, and taking into account only the taxa wherein one or more whole-genome sequences are available, the first scenario would require a minimum of 15 independent losses of the CENP-B gene during animal and fungi evolution (10 losses in animals and 5 in fungi, see fig. 4). Although gene loss is a common process in eukaryotic evolution (Aravind et al. 2000; Krylov et al. 2003), this scenario is difficult to reconcile with the critical function of the fission yeast genes for chromosome segregation and cell cycle progression (Halverson et al. 1997; Irelan, Gutkin, and Clarke 2001; Nakagawa et al. 2002; Locovei et al. 2006). Additionally, it seems unlikely that a sequence domesticated prior to divergence of fungal and animal lineages would be so highly constrained in a single animal lineage (i.e. mammals) while independently becoming dispensable in most other animal lineages. Thus, a scenario that invokes two separate domestication events of pogo-like transposases in the lineages of fission yeast and mammals is indeed more parsimonious (fig. 4).
Figure 4.
Convergent domestication of pogo-like transposases in fission yeast and mammals. A star indicates the presence of pogo-derived centromere-binding protein. A circle indicates the presence of pogo-like transposons. Under an orthology model, arrowheads indicate inferred gene loss events following a single domestication event indicated by the arrow. Representative taxa for which whole-genome sequencing is complete are shown. The cladogram has been inferred using the Tree of Life Web Project (Maddison and Schulz 1996–2006) and the fungi phylogeny described in James et al. (2006).
Phylogenetic analyses point to independent origins of centromere-binding proteins in mammals and fission yeast
Two distinct predictions as to the topology of a phylogenetic tree of mammalian and fission yeast centromere-binding proteins and pogo-like transposases arise as a result of these observations.
Under the single domestication event hypothesis, one would expect that the mammalian and fission yeast CENP-B proteins will form a monophyletic clade sister to a clade of pogo-like transposases descended from the transposase source that gave rise to CENP-B in the common ancestor of fungi and animals. Alternatively, the vast evolutionary gap between mammals and yeasts might hinder the phylogenetic clustering of the two groups of proteins and obscure their relationship to each other and to a particular clade of transposases. Under the independent domestication hypothesis, the fission yeast proteins and mammalian CENP-B proteins should fall into distinct clades together with fungal and animal transposases from which they respectively derived.
To test these predictions, we selected 65 sequences from a data set of non-redundant transposases and pogo-derived proteins and built a multiple alignment using the MAFFT program (Katoh et al. 2005) (tables 1 and 2). After manually adjusting the alignment and removing regions with extremely low conservation introducing long gaps, we reconstructed the evolutionary tree from our data set using a Bayesian approach (see Methods). The Bayesian tree (fig. 2) shows that the mammalian pogo-derived proteins, including CENP-B, and the fission yeast centromeric proteins Abp1, Cbh1 and Cbh2 fall into two separate clades with robust statistical support.
As predicted under the independent domestication hypothesis, the mammalian pogo-derived proteins cluster with transposases isolated from various animal genomes (insects, nematodes, mollusks and flatworms). However, we note that the pogo-like transposases currently found in vertebrate genomes (Tiggers from mammals and amniotes and TIGGU from pufferfish) are not the most closely related to CENP-B. Tiggers and TIGGU transposases fall into a different clade (denoted ‘JR’ for JERKY-related in fig. 2) together with several newly identified transposases from various invertebrates and from the microsporidian Nosema bombycis and several pogo-derived proteins (JRK, JRKL and TIGD2, 5 and 7 in one subclade and TIGD1 in a second subclade; see fig. 2, tables 1 and 2). This topology suggests that there are two major, anciently diverged clades of pogo-like transposons in metazoans: the CR clade (CENP-B-related) and the JR clade. Most likely, the two clades were already separated prior to the split of invertebrates and vertebrates, but transposons from the CR clade have gone extinct and are presently undetectable in the dataset of vertebrate genomes currently available (mostly mammals). CENP-B, TIGD3, 4 and 6 might be viewed as derivatives of these transposons that have persisted through evolutionary time due to the action of natural selection.
All the fungi sequences group into a strongly supported clade that includes the fission yeast centromeric proteins, the Pdc2 proteins and pogo-like transposases from fungi, plants and the oomycete Phytophthora. We designate this clade ‘AR’ for Abp1-related. Within the AR clade, the fission yeast proteins are significantly closer to transposases found in the ascomycetes class of Eurotiomycetes, which comprises the genera Aspergillus, Neosartorya and Coccidioides. These transposases thus appear to represent the closest extant relatives of the transposases that gave rise to the fission yeast centromeric proteins. This group of transposons has now completely disappeared from the S. pombe genome, together with all other DNA transposons. Finally, the Pdc2 group of proteins falls outside of this group, consistent with its independent origination from yet another source of pogo-like transposase.
Discussion
Our analysis provides evidence that two distinct sources of pogo-like transposases gave rise independently to mammalian and fission yeast proteins with centromere-binding activity. Based on this common activity and overall sequence similarity, these proteins have been previously considered functional homologs (Murakami, Huberman, and Hurwitz 1996b; Halverson et al. 1997; Lee, Huberman, and Hurwitz 1997; Irelan, Gutkin, and Clarke 2001; Nakagawa et al. 2002; Amor et al. 2004; Masumoto, Nakano, and Ohzeki 2004). Consequently, the results drawn from the analysis of fission yeast proteins have often been used to speculate on the function of the mammalian CENP-B and vice-versa. While functional parallels clearly exist between these proteins and in the general centromeric protein–DNA architecture of fission yeast and mammals (Clarke 1998; Amor et al. 2004; Henikoff and Dalal 2005), our results indicate that the sequence relationship between these proteins does not derive from vertical inheritance of an ancestral host protein with centromere-binding activity. Instead, our data indicate that the relationship of these proteins reflects a process of convergent domestication, whereby the same type of transposases (i.e. pogo-like transposases) were exapted independently in the lineage of fission yeast and mammals to give rise to host proteins with centromere-binding activity.
At first, it may seem surprising that the same type of transposase would be recruited twice independently in evolutionarily distant lineages to become centromere-binding proteins. However, we note that the association of TEs and centromeres appears to be a recurrent theme in molecular evolution. Many intimate links have been uncovered between various kinds of TEs and the structure and/or function of centromeres in diverse species (reviewed in Kipling and Warburton 1997; Dawe 2003; Wong and Choo 2004). Furthermore, it is conceivable that pogo-like transposases possess a predisposition to be recruited as centromeric proteins. For example, one can envision that these transposases have an intrinsic ability to interact with centromeric DNA, either directly (all transposases so far characterized have DNA-binding activity) or indirectly via interaction with a host protein. This host factor could be a constitutive component of the kinetochore or a protein transiently associated with centromeric chromatin.
Genetic and biochemical analyses of the three fission yeast centromeric proteins have clearly established their role in the function and organization of the centromeres (Halverson et al. 1997; Lee, Huberman, and Hurwitz 1997; Ngan and Clarke 1997; Baum and Clarke 2000; Irelan, Gutkin, and Clarke 2001; Nakagawa et al. 2002). Individually, each protein is not essential for viability, but double deletion mutants display loss of viability and dramatic morphological changes, including abnormal branching and cell elongation. All three proteins are also required for the recruitment of the major heterochromatin protein Swi6 to the centromeres, although Abp1 seems to make the greatest contribution to this process (Nakagawa et al. 2002). Thus the function of the three proteins seems to be partially redundant, but together essential for cell cycle progression and specification of a chromatin state at the centromeres that is competent for chromosome segregation. Consistent with some level of functional partitioning, the three proteins have different DNA binding affinities in vitro (Murakami, Huberman, and Hurwitz 1996a; Halverson et al. 1997 Lee, Huberman, and Hurwitz 1997) and distinct targets in vivo. Cbh1 binds the outer repeats of the S. pombe centromere and perhaps also non-centromeric regions (Baum and Clarke 2000), while Cbh2 appears to bind predominantly the inner centromeric region (Irelan, Gutkin, and Clarke 2001). Abp1 binds specifically the outer centromeric repeats, where it promotes specific histone modifications that lead to the recruitment of Swi6 and the formation of centromeric heterochromatin (Nakagawa et al. 2002). The notion of subfunctionalization is compatible with the results of our analyses, although we cannot at the moment determine whether the three S. pombe proteins originated from a single domestication event followed by subsequent gene duplication events or by domestication of three closely related transposase genes.
The function of CENP-B at mammalian centromeres is less clear than for the fission yeast proteins (for review, Warburton 2001; Masumoto, Nakano, and Ohzeki 2004). On one hand, it has been demonstrated that CENP-B and its binding sites on alphoid satellite DNA (CENP-B box) are required for de novo centromere formation in human cells and faithful segregation of human artificial chromosomes (Harrington et al. 1997; Ohzeki et al. 2002). On the other hand, the CENP-B box and CENP-B are not found on the Y chromosome and CENP-B is dispensable for the activation of neo-centromeres (Broccoli, Miller, and Miller 1990; Depinet et al. 1997; Warburton 2001). Finally, in CENP-B knockout mouse cells, functional kinetochores are maintained and null mutant mice exhibit only mild growth and reproductive abnormalities in the laboratory (Hudson et al. 1998; Kapoor et al. 1998; Perez-Castro et al. 1998; Fowler et al. 2000). While what is mild in the laboratory is not necessarily invisible to natural selection, these observations and others (Goldberg et al. 1996; Masumoto, Nakano, and Ohzeki 2004) raise the hypothesis that CENP-B might have functionally redundant homologs within mammalian genomes. Our analysis suggests that all mammalian genomes possess indeed other transposase-derived proteins related to CENP-B. However, the overall level of similarity of these proteins to CENP-B is relatively weak and it is lower than among the three proteins of fission yeast. Based on our analysis, the best candidates as functional homologs of CENP-B are TIGD3, 4 and 6, with the latter being the closest relative to CENP-B (fig. 2). Recently, it was found that recombinant human JRK-GFP fusion proteins densely co-localize with CREST antigens (which recognize CENP-A, -B and -C) at distinct chromosomal foci of cultured cells in S and G2 phase (Waldron and Moore 2004). Thus JRK might also interact directly or indirectly with some centromeric components. Given the distant relationship of JRK to CENP-B, but its close proximity to mammalian Tigger4 transposons (see fig. 2), it could be that JRK represents yet another case of independently domesticated pogo-like transposase with a centromeric function. Further experiments are needed to explore the function of mammalian pogo-derived proteins and to assess whether any could act redundantly and/or cooperatively with CENP-B at the centromere.
Supplementary Material
Acknowledgments
We thank Ellen Pritham for critical reading of the manuscript and Esther Betrán for providing logistical and financial support to C.C. We also thank the following Sequencing Centers: Agencourt, Inc., WUSTL School of Medicine, Joint Genome Institute, TIGR, Broad Institute and Baylor College of Medicine for prepublication access to their genome data. This work was supported by start-up funds from UT Arlington and by grant R01GM77582-01 from the National Institute of Health to C.F.
Literature Cited
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Amor DJ, Kalitsis P, Sumer H, Choo KH. Building the centromere: from foundation proteins to 3D organization. Trends Cell Biol. 2004;14:359–368. doi: 10.1016/j.tcb.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Aravind L, Watanabe H, Lipman DJ, Koonin EV. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc Natl Acad Sci U S A. 2000;97:11319–11324. doi: 10.1073/pnas.200346997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum M, Clarke L. Fission yeast homologs of human CENP-B have redundant functions affecting cell growth and chromosome segregation. Mol Cell Biol. 2000;20:2852–2864. doi: 10.1128/mcb.20.8.2852-2864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano LA, Valdivia MM. Molecular cloning of an intronless gene for the hamster centromere antigen CENP-B. Biochim Biophys Acta. 1996;1307:21–25. doi: 10.1016/0167-4781(96)00039-5. [DOI] [PubMed] [Google Scholar]
- Britten RJ. Coding sequences of functioning human genes derived entirely from mobile element sequences. Proc Natl Acad Sci U S A. 2004;101:16825–16830. doi: 10.1073/pnas.0406985101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broccoli D, Miller OJ, Miller DA. Relationship of mouse minor satellite DNA to centromere activity. Cytogenet Cell Genet. 1990;54:182–186. doi: 10.1159/000132989. [DOI] [PubMed] [Google Scholar]
- Brosius J. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene. 1999;238:115–134. doi: 10.1016/s0378-1119(99)00227-9. [DOI] [PubMed] [Google Scholar]
- Brosius J, Gould SJ. On “genomenclature”: a comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA”. Proc Natl Acad Sci U S A. 1992;89:10706–10710. doi: 10.1073/pnas.89.22.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulazel K, Metcalfe C, Ferreri GC, Yu J, Eldridge MD, O’Neill RJ. Cytogenetic and molecular evaluation of centromere-associated DNA sequences from a marsupial (Macropodidae: Macropus rufogriseus) X chromosome. Genetics. 2006;172:1129–1137. doi: 10.1534/genetics.105.047654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkin DJ, Jones C, Burkin HR, McGrew JA, Broad TE. Sheep CENPB and CENPC genes show a high level of sequence similarity and conserved synteny with their human homologs. Cytogenet Cell Genet. 1996;74:86–89. doi: 10.1159/000134388. [DOI] [PubMed] [Google Scholar]
- Capy P, Bazin C, Higuet D, Langin T. Dynamics and evolution of transposable elements. Springer-Verlag; Austin, Texas: 1998. [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L. Centromeres: proteins, protein complexes, and repeated domains at centromeres of simple eukaryotes. Curr Opin Genet Dev. 1998;8:212–218. doi: 10.1016/s0959-437x(98)80143-3. [DOI] [PubMed] [Google Scholar]
- Coelho PA, Nurminsky D, Hartl D, Sunkel CE. Identification of Porto-1, a new repeated sequence that localises close to the centromere of chromosome 2 of Drosophila melanogaster. Chromosoma. 1996;105:211–222. [PubMed] [Google Scholar]
- Comeron JM. K-Estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics. 1999;15:763–764. doi: 10.1093/bioinformatics/15.9.763. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Udit S, Batzer MA, Feschotte C. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc Natl Acad Sci U S A. 2006 doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daboussi MJ, Capy P. Transposable elements in filamentous fungi. Annu Rev Microbiol. 2003;57:275–299. doi: 10.1146/annurev.micro.57.030502.091029. [DOI] [PubMed] [Google Scholar]
- Dawe RK. RNA interference, transposons, and the centromere. The Plant Cell. 2003;15:297–301. doi: 10.1105/tpc.150230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depinet TW, Zackowski JL, Earnshaw WC, Kaffe S, Sekhon GS, Stallard R, Sullivan BA, Vance GH, Van Dyke DL, Willard HF, et al. Characterization of neo-centromeres in marker chromosomes lacking detectable alpha-satellite DNA. Hum Mol Genet. 1997;6:1195–1204. doi: 10.1093/hmg/6.8.1195. [DOI] [PubMed] [Google Scholar]
- Dou T, Gu S, Zhou Z, Ji C, Zeng L, Ye X, Xu J, Ying K, Xie Y, Mao Y. Isolation and characterization of a Jerky and JRK/JH8 like gene, tigger transposable element derived 7, TIGD7. Biochem Genet. 2004;42:279–285. doi: 10.1023/b:bigi.0000034428.95802.35. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Ratrie H, 3rd, Stetten G. Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma. 1989;98:1–12. doi: 10.1007/BF00293329. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Sullivan KF, Machlin PS, Cooke CA, Kaiser DA, Pollard TD, Rothfield BF, Cleveland DW. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J Cell Biol. 1987;104:817–829. doi: 10.1083/jcb.104.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards NS, Murray AW. Identification of xenopus CENP-A and an associated centromeric DNA repeat. Mol Biol Cell. 2005;16:1800–1810. doi: 10.1091/mbc.E04-09-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Mouchès C. Evidence that a family of miniature inverted-repeat transposable elements (MITEs) from the Arabidopsis thaliana genome has arisen from a pogo-like DNA transposon. Mol Biol Evol. 2000;17:730–737. doi: 10.1093/oxfordjournals.molbev.a026351. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annual Review of Genetics. 2007;41 doi: 10.1146/annurev.genet.40.110405.090448. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler KJ, Hudson DF, Salamonsen LA, Edmondson SR, Earle E, Sibson MC, Choo KH. Uterine dysfunction and genetic modifiers in centromere protein B-deficient mice. Genome Res. 2000;10:30–41. [PMC free article] [PubMed] [Google Scholar]
- Goldberg IG, Sawhney H, Pluta AF, Warburton PE, Earnshaw WC. Surprising deficiency of CENP-B binding sites in African green monkey alpha-satellite DNA: implications for CENP-B function at centromeres. Mol Cell Biol. 1996;16:5156–5168. doi: 10.1128/mcb.16.9.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T, Mater AG, Wienberg J, Ward DC. Presence and abundance of CENP-B box sequences in great ape subsets of primate-specific alpha-satellite DNA. J Mol Evol. 1995;41:487–491. doi: 10.1007/BF00160320. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Halverson D, Baum M, Stryker J, Carbon J, Clarke L. A centromere DNA-binding protein from fission yeast affects chromosome segregation and has homology to human CENP-B. J Cell Biol. 1997;136:487–500. doi: 10.1083/jcb.136.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Kumar S. Genomic clocks and evolutionary timescales. Trends Genet. 2003;19:200–206. doi: 10.1016/S0168-9525(03)00053-2. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Dalal Y. Centromeric chromatin: what makes it unique? Curr Opin Genet Dev. 2005;15:177–184. doi: 10.1016/j.gde.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison JS, Murata M, Ogura Y, Schwarzacher T, Motoyoshi F. Polymorphisms and genomic organization of repetitive DNA from centromeric regions of Arabidopsis chromosomes. The Plant Cell. 1999;11:31–42. doi: 10.1105/tpc.11.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson DF, Fowler KJ, Earle E, Saffery R, Kalitsis P, Trowell H, Hill J, Wreford NG, de Kretser DM, Cancilla MR, et al. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J Cell Biol. 1998;141:309–319. doi: 10.1083/jcb.141.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irelan JT, Gutkin GI, Clarke L. Functional redundancies, distinct localizations and interactions among three fission yeast homologs of centromere protein-B. Genetics. 2001;157:1191–1203. doi: 10.1093/genetics/157.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. Molecular paleontology of transposable elements from Arabidopsis thaliana. Genetica. 1999;107:27–37. [PubMed] [Google Scholar]
- Kapoor M, Montes de Oca Luna R, Liu G, Lozano G, Cummings C, Mancini M, Ouspenski I, Brinkley BR, May GS. The cenpB gene is not essential in mice. Chromosoma. 1998;107:570–576. doi: 10.1007/s004120050343. [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipling D, Mitchell AR, Masumoto H, Wilson HE, Nicol L, Cooke HJ. CENP-B binds a novel centromeric sequence in the Asian mouse Mus caroli. Mol Cell Biol. 1995;15:4009–4020. doi: 10.1128/mcb.15.8.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipling D, Warburton PE. Centromeres, CENP-B and Tigger too. Trends Genet. 1997;13:141–145. doi: 10.1016/s0168-9525(97)01098-6. [DOI] [PubMed] [Google Scholar]
- Krylov DM, Wolf YI, Rogozin IB, Koonin EV. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 2003;13:2229–2235. doi: 10.1101/gr.1589103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Subramanian S. Mutation rates in mammalian genomes. Proc Natl Acad Sci U S A. 2002;99:803–808. doi: 10.1073/pnas.022629899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lee JK, Huberman JA, Hurwitz J. Purification and characterization of a CENP-B homologue protein that binds to the centromeric K-type repeat DNA of Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1997;94:8427–8432. doi: 10.1073/pnas.94.16.8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Seto J, Sibille E, Toth M. The RNA binding domain of Jerky consists of tandemly arranged helix-turn-helix/homeodomain-like motifs and binds specific sets of mRNAs. Mol Cell Biol. 2003;23:4083–4093. doi: 10.1128/MCB.23.12.4083-4093.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Betran E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nat Rev Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- Lopez CC, Edstrom JE. Interspersed centromeric element with a CENP-B box-like motif in Chironomus pallidivittatus. Nucleic Acids Res. 1998;26:4168–4172. doi: 10.1093/nar/26.18.4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorite P, Carrillo JA, Tinaut A, Palomeque T. Evolutionary dynamics of satellite DNA in species of the Genus Formica (Hymenoptera, Formicidae) Gene. 2004;332:159–168. doi: 10.1016/j.gene.2004.02.049. [DOI] [PubMed] [Google Scholar]
- Maddison DR, Schulz K-S, editors. The Tree of Life Web Project. 1996–2006 http://tolweb.org.
- Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol. 1989;109:1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Nakano M, Ohzeki J. The role of CENP-B and alpha-satellite DNA: de novo assembly and epigenetic maintenance of human centromeres. Chromosome Res. 2004;12:543–556. doi: 10.1023/B:CHRO.0000036593.72788.99. [DOI] [PubMed] [Google Scholar]
- Miller WJ, McDonald JF, Nouaud D, Anxolabehere D. Molecular domestication--more than a sporadic episode in evolution. Genetica. 1999;107:197–207. [PubMed] [Google Scholar]
- Mojzita D, Hohmann S. Pdc2 coordinates expression of the THI regulon in the yeast Saccharomyces cerevisiae. Mol Genet Genomics. 2006 doi: 10.1007/s00438-006-0130-z. [DOI] [PubMed] [Google Scholar]
- Mravinac B, Plohl M, Ugarkovic D. Conserved patterns in the evolution of Tribolium satellite DNAs. Gene. 2004;332:169–177. doi: 10.1016/j.gene.2004.02.055. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Huberman JA, Hurwitz J. Identification, purification, and molecular cloning of autonomously replicating sequence-binding protein 1 from fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1996a;93:502–507. doi: 10.1073/pnas.93.1.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Huberman JA, Hurwitz J. Identification, purification, and molecular cloning of autonomously replicating sequence-binding protein 1 from fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1996b;93:502–507. doi: 10.1073/pnas.93.1.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H, Lee JK, Hurwitz J, Allshire RC, Nakayama J, Grewal SI, Tanaka K, Murakami Y. Fission yeast CENP-B homologs nucleate centromeric heterochromatin by promoting heterochromatin-specific histone tail modifications. Genes Dev. 2002;16:1766–1778. doi: 10.1101/gad.997702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngan VK, Clarke L. The centromere enhancer mediates centromere activation in Schizosaccharomyces pombe. Mol Cell Biol. 1997;17:3305–3314. doi: 10.1128/mcb.17.6.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura KI, Kurata N. Organization of the 1.9-kb repeat unit RCE1 in the centromeric region of rice chromosomes. Mol Gen Genet. 1999;261:1–10. doi: 10.1007/s004380050935. [DOI] [PubMed] [Google Scholar]
- Ohzeki J, Nakano M, Okada T, Masumoto H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol. 2002;159:765–775. doi: 10.1083/jcb.200207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamilo P, Bianchi NO. Evolution of the Zfx and Zfy genes: rates and interdependence between the genes. Mol Biol Evol. 1993;10:271–281. doi: 10.1093/oxfordjournals.molbev.a040003. [DOI] [PubMed] [Google Scholar]
- Perez-Castro AV, Shamanski FL, Meneses JJ, Lovato TL, Vogel KG, Moyzis RK, Pedersen R. Centromeric protein B null mice are viable with no apparent abnormalities. Dev Biol. 1998;201:135–143. doi: 10.1006/dbio.1998.9005. [DOI] [PubMed] [Google Scholar]
- Plasterk RHA, Izsvák Z, Ivics Z. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 1999;15:326–332. doi: 10.1016/s0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- Prince VE, Pickett FB. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 2002;3:827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- Pritham EJ, Feschotte C, Wessler SR. Unexpected diversity and differential success of DNA transposons in four species of entamoeba protozoans. Mol Biol Evol. 2005;22:1751–1763. doi: 10.1093/molbev/msi169. [DOI] [PubMed] [Google Scholar]
- Robertson HM. Members of the pogo superfamily of DNA-mediated transposons in the human genome. Mol Gen Genet. 1996;252:761–766. doi: 10.1007/BF02173985. [DOI] [PubMed] [Google Scholar]
- Robertson HM. Evolution of DNA transposons. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. American Society for Microbiology Press; Washington, DC: 2002. pp. 1093–1110. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Devel. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Riggs AD. Tiggers and DNA transposon fossils in the human genome. Proc Natl Acad Sci U S A. 1996;93:1443–1448. doi: 10.1073/pnas.93.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KF, Glass CA. CENP-B is a highly conserved mammalian centromere protein with homology to the helix-loop-helix family of proteins. Chromosoma. 1991;100:360–370. doi: 10.1007/BF00337514. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Nureki O, Kurumizaka H, Fukai S, Kawaguchi S, Ikuta M, Iwahara J, Okazaki T, Yokoyama S. Crystal structure of the CENP-B protein-DNA complex: the DNA-binding domains of CENP-B induce kinks in the CENP-B box DNA. Embo J. 2001;20:6612–6618. doi: 10.1093/emboj/20.23.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth M, Grimsby J, Buzsaki G, Donovan GP. Epileptic seizures caused by inactivation of a novel gene, jerky, related to centromere binding protein-B in transgenic mice. Nat Genet. 1995;11:71–75. doi: 10.1038/ng0995-71. [DOI] [PubMed] [Google Scholar]
- Tudor M, Lobocka M, Goodwell M, Pettitt J, O’Hare K. The pogo transposable element family of Drosophila melanogaster. Mol Gen Genet. 1992;232:126–134. doi: 10.1007/BF00299145. [DOI] [PubMed] [Google Scholar]
- van Rheede T, Bastiaans T, Boone DN, Hedges SB, de Jong WW, Madsen O. The platypus is in its place: nuclear genes and indels confirm the sister group relation of monotremes and Therians. Mol Biol Evol. 2006;23:587–597. doi: 10.1093/molbev/msj064. [DOI] [PubMed] [Google Scholar]
- Volff JN. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays. 2006;28:913–922. doi: 10.1002/bies.20452. [DOI] [PubMed] [Google Scholar]
- Waldron R, Moore T. Complex regulation and nuclear localization of JRK protein. Biochem Soc Trans. 2004;32:920–923. doi: 10.1042/BST0320920. [DOI] [PubMed] [Google Scholar]
- Warburton PE. Epigenetic analysis of kinetochore assembly on variant human centromeres. Trends Genet. 2001;17:243–247. doi: 10.1016/s0168-9525(01)02283-1. [DOI] [PubMed] [Google Scholar]
- Weide R, Hontelez J, van Kammen A, Koornneef M, Zabel P. Paracentromeric sequences on tomato chromosome 6 show homology to human satellite III and to the mammalian CENP-B binding box. Mol Gen Genet. 1998;259:190–197. doi: 10.1007/s004380050804. [DOI] [PubMed] [Google Scholar]
- Wong LH, Choo KH. Evolutionary dynamics of transposable elements at the centromere. Trends Genet. 2004;20:611–616. doi: 10.1016/j.tig.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Wood VR, Gwilliam MA, Rajandream M, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML:a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yoda K, Kitagawa K, Matsumoto H, Muro Y, Okazaki T. A human centromere protein, CENP-B, has a DNA binding domain containing four potential alpha helices at the NH2 terminus, which is separable from dimerizing activity. J Cell Biol. 1992;119:585–596. doi: 10.1083/jcb.119.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K, Nakamura T, Masumoto H, Suzuki N, Kitagawa K, Nakano M, Shinjo A, Okazaki T. Centromere protein B of African green monkey cells: gene structure, cellular expression, and centromeric localization. Mol Cell Biol. 1996;16:5169–5177. doi: 10.1128/mcb.16.9.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z, Kyaw H, Gakenheimer KR, Augustus M, Fan P, Zhang X, Su K, Carter KC, Li Y. Cloning, mapping, and tissue distribution of a human homologue of the mouse jerky gene product. Biochem Biophys Res Commun. 1997;236:389–395. doi: 10.1006/bbrc.1997.6935. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.