Abstract
Innate and adaptive cellular immunity is initiated, directed and regulated by a vast array of cell surface receptors. Attempts to harness the cellular immune system in translational settings such as immunotherapy and vaccine development require tools to accurately describe and isolate lymphocytes with specific characteristics. One such tool, flow cytometry, is undergoing a revolution in instrumentation and reagents, providing opportunities for high resolution phenotypic and functional analysis of lymphocytes. Here, we demonstrate how nine-color flow cytometry can be adapted, optimized and applied to investigate the phenotypic complexity and functional heterogeneity of human lymphocyte subsets. We provide examples of studies of adaptive T cell responses against viruses, as well as the assessment of CD1d-restricted NKT cells and NK cells. We discuss the importance of this technology for detailed investigations of lymphocyte subsets in studies of infectious diseases and cancer.
Keywords: Flow cytometry, FACS, T cells, NKT cells, NK cells, CD1d, Fluorochromes
1. Introduction
Innate and adaptive cellular immune responses are vital in the defense against many infections and cancers. NK cells play an important role in the innate response, and their activity is regulated by signals from a vast array of germline encoded activating and inhibitory receptors (Fauci et al., 2005; Hamerman et al., 2005; Lanier, 2005). NKT cells are unconventional T lymphocytes, which operate on the border between innate and adaptive immunity. They are activated through an invariant αβ T cell receptor (TCR) that recognizes CD1d rather than a classical MHC molecule (Godfrey and Kronenberg, 2004; Sandberg and Ljunggren, 2005; Bendelac et al., 2006), and have the capacity to regulate the actions of other cell types, including NK cells (Brutkiewicz, 2006; Moody, 2006). Within days of an infection, the adaptive T cell response targets the pathogen-infected cells in an antigen-specific manner and, in case of a successful response, the infection is cleared and T cell memory is established. Some pathogens, however, such as HIV-1, establish a chronic infection that is only partially controlled by the T cell response. T cell responses involve the differentiation of naïve T cells into several types of effector and memory cells with distinct characteristics (Sallusto et al., 2000; Wherry and Ahmed, 2004; Lefrancois and Marzo, 2006).
Flow cytometry has greatly facilitated advances in our understanding of the immune system (Herzenberg and Herzenberg, 2004). Our knowledge about the vast number of receptors and cytokines involved in regulating cellular immunity, and the changes in expression of these factors upon cellular activation and differentiation, has increased rapidly. However, the means to investigate co-expression of proteins at the single cell level have not kept pace with this development, and the heterogeneity of immune cells has probably been significantly underestimated. Limitations in fluorochrome-conjugated reagents and instrumentation have hampered the broad implementation of multi-color, or polychromatic, flow cytometry in basic research as well as in the clinic. Recent progress in the availability of fluorochromes, reagents, instrumentation and software has, however, made this technology a realistic alternative for many laboratories. The highly complex receptor repertoires of T cells, NKT cells, and NK cells make multi-color approaches particularly valuable for identification and detailed characterization of functionally distinct subsets and differentiation stages of these cells. Here, to provide guidance to the prospective user of multi-color flow cytometry in a translational immunology setting, we illustrate the power of this technology and discuss possibilities and limitations in its implementation to the study of highly defined human lymphocyte subsets.
2. Materials and methods
2.1 Blood samples and cells
Heparinized whole blood samples were obtained after informed consent. PBMC were isolated by Lymphoprep gradient centrifugation (Axis-Shield, Oslo, Norway), and washed twice before analysis by flow cytometry. The human HLA class I-negative erythroleukemia cell line K562 transfected with HLA-E*01033 (K562-E, kindly provided by Dr. K. Söderstrom) was kept in RPMI 1640 medium supplemented with 100 μg/ml l-glutamine, 10% heat-inactivated FBS, 100 U/ml penicillin G, 100 μg/ml streptomycin and 1 mg/ml geniticin. In HLA-E stabilization experiments, K562-E cells were pulsed with 100 μM synthetic HLA-G*0101 signal peptide (GSP) VMAPRTLFL (Invitrogen, Eugene, OR, USA) at 26°C and 5% CO2 for 15 hours to stabilize HLA-E expression.
2.2 Fluorescent reagents
Anti-CD3 CasY was from Dako (Glostrup, Denmark). Anti-CD7 PE, anti-CD45RA PacB, anti-CD62L PE-TR, anti-CD127 biotin, purified anti-NKG2A and anti-Vα24 APC were purchased from Coulter Immunotech (Marseilles, France). Anti-CD4 APC-Cy7, anti-CD8 PerCp, anti-CD14 APC-Cy7, anti-CD16 PacB, anti-CD27 APC-Cy7, anti-CD28 FITC, anti-CD56 PE-Cy5, anti-CD56 PE-Cy7, anti-CD107a FITC, anti-CD161 FITC, and anti-CCR7 PE-Cy7 mAbs were purchased from BD Pharmingen (San Diego, CA, USA). Anti-NKG2C PE was obtained from R&D Systems (Minneapolis, MN, USA). Subjects were evaluated for HLA-A2 expression by staining with anti-HLA-A2 (BD Biosciences). CD8 T cells specific for viruses were identified and enumerated using APC-conjugated HLA-A2 tetrameric complexes refolded with the HIV-1 Gag 77-85 peptide epitope SLYNTVATL or the Influenza virus M1 58-66 epitope GILGFVFTL (Coulter Immunotech). CD1d DimerX recombinant fusion protein reagent was purchased from BD Biosciences, loaded with α-galactosyl ceramide (αGalCer ) (Alexis Biochemicals, Lausen, Switzerland), in vitro according to instructions by the manufacturer, and detected using a secondary anti-mouse IgG1 PE. Purified anti-NKG2A was biotinylated using reagents from Pierce (Rockford, IL, USA). Biotinylated anti-NKG2A and anti-CD127 was detected using Qdot® 605 streptavidin conjugate (Invitrogen, Eugene, OR, USA). Intracellular cytokine was detected using anti-IFNγ PE-Cy7 (BD Biosciences), and anti-TNFα Alexa 647 from eBioscience (San Diego, CA, USA). A compensation particle set, consisting of uncoated polystyrene beads, or beads coated with anti-mouse Igκ (BD Biosciences), were stained with mAbs conjugated with each of the fluorochromes used.
2.3 Flow cytometry
For surface staining, freshly purified PBMC were incubated with a panel of fluorochrome-conjugated mAbs, and HLA-A2-tetramers or CD1d DimerX in PBS with 2% FCS for 30 minutes on ice as indicated (Table 1). Samples were next washed three times and fixed in PBS with 1% formaldehyde. Data were acquired immediately after staining using the CyAn™ ADP instrument (Dako, Denmark) equipped with a 20 mW 488 nm laser, a 25 mW 635 nm laser, and a 25 mW 405 nm laser to detect a total of 9 fluorescence parameters plus two scatter parameters (Table 1). Data were analyzed using FlowJo™ software (Tree Star, Ashland, OR).
Table 1.
Instrument configuration and reagent panels for the study of lymphocyte subsetsa
| Detector | Laser (nm) | Filter (nm) | Fluorochrome | CD8 T cell panel | NKT cell panel | NK cell panel |
|---|---|---|---|---|---|---|
| FL1 | 488 | 530/40 | FITC | CD28 | CD57 | CD107a |
| FL2 | 488 | 575/25 | PE | CD7 | CD1d DimerX | NKG2C |
| FL3 | 488 | 613/20 | PE-TR/QD605 | CD62L | CD127 | NKG2A |
| FL4 | 488 | 680/30 | PerCp/PE-Cy5 | CD8 | CD161 | CD56 |
| FL5 | 488 | 750LP | PE-Cy7 | CCR7 | CD56 | IFNγ |
| FL6 | 405 | 450/50 | PacB | CD45RA | CD16 | CD16 |
| FL7 | 405 | 550/30 | CasY | CD3 | CD3 | CD3 |
| FL8 | 635 | 665/20 | APC/Alexa 647 | HLA-A2 tetramer | Vα24 | TNFα |
| FL9 | 635 | 750LP | APC-Cy7 | CD27 | CD4 | CD14 |
Reagents were from BD Biosciences (San Diego, CA, USA),Coulter Immunotech (Marseilles, France), Dako (Glostrup, Denmark) eBioscience (San Diego, CA, USA), Invitrogen (Eugene, OR, USA),One Lambda (Canoga Park, CA),Pierce (Rockford, IL, USA), R&D Systems (Minneapolis, MN, USA).
2.4 Functional NK cell assay
4×105 PBMC were mixed with K562-E cells at a ratio of 10:1 in round-bottom 96-well plates in a final volume of 200 μl, and co-incubated at 37°C and 5% CO2 for 6 hours. Anti-CD107a and the corresponding IgG1 isotype antibody were added at the beginning of the assay.
Monensin (GolgiSTOP, 1:150 dilution, Becton Dickinson) and Brefeldin A (GolgiPLUG, 1:250 dilution, Becton Dickinson) were added after one hour of co-culture (Alter et al., 2004). Following incubation, surface antigen staining was performed. Next, cells were washed three times, fixed and permeabilized with Cytofix/Cytoperm kit (Becton Dickinson). Finally, cells were stained for intracellular cytokine for 30 minutes, and washed three times. To obtain optimal result, cells were kept at 4°C during the entire staining procedure and until flow cytometric acquisition.
3. Results
3.1. Instrument configuration and reagent panel design for nine-color flow cytometry
Flow cytometry technology is currently in transition from instrumentation limited to four fluorescence parameters to multi-color instruments which will help grasp the heterogeneity of lymphocyte populations (De Rosa et al., 2003). Here we have used the CyAn™ ADP instrument equipped with 488 nm, 635 nm, and 405 nm lasers to detect nine fluorescent parameters plus two scatter parameters (Table 1). The 488 nm laser was used to excite fluorescein isotiocyanate (FITC), peridinin chlorophyll protein (PerCp), phycoerythrin (PE), and the PE-based tandem conjugates PE-Cy7 and PE-Texas Red (PE-TR). The 635 nm laser was used to excite allophycocyanin (APC) and its Cy7 tandem conjugate, whereas the violet 405 nm laser was used to excite Pacific Blue (PacB) and Cascade Yellow (CasY). Significant spectral overlap between, in particular, the 488 nm-excited fluorochromes makes considerable compensation necessary. Single-stained anti-mouse Ig-coated beads were used to generate the compensation matrix in the FlowJo software (Table 2). In particular, PE-TR required considerable compensation, and for some applications directly PE-TR-conjugated mAbs were replaced with biotinylated mAbs and the appropriate streptavidin-conjugated quantum dot (QD), QD605. The narrow emission peaks of QD semiconductor nanocrystals radically reduce the need to compensate for spectral overlap (Bruchez et al., 1998; Chattopadhyay et al., 2006) (Table 2). Another important consideration is the intensity of emission by a given fluorochrome. Matching target cell surface molecules with the appropriate means of detection is necessary to obtain good data (Maecker et al., 2004). For example, because CasY excited by the 405 nm laser was the least bright among the fluorochromes tested it was used to detect CD3 which is a highly expressed and readily detectable protein. CasY is not suitable for weakly expressed surface markers. In addition, the design of antibody panels should take into account the gating strategies used in the subsequent analysis. For example, if negative gating is applied, such as exclusion of monocytes by gating on CD14- cells, a fluorochrome that requires significant compensation can be used with that particular mAb to facilitate the analysis. Here, three mAb panels were developed for the assessment of three lymphocyte subsets in freshly isolated human PBMC; virus-specific CD8 T cells, CD1d-restricted NKT cells, and NK cells (Table 1).
Table 2.
Compensation matrix for a flow cytometry panel with nine fluorescence parametersa
| Fluorochrome | Detector | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| FL1 | FL2 | FL3 | FL4 | FL5 | FL6 | FL7 | FL8 | FL9 | |
| FITC | 29 | 7.9 | 4.3 | 0.2 | 0 | 1.6 | 0 | 0 | |
| PE | 0.8b | 30.6 | 18.2 | 2.7 | 0 | 1.4 | 0 | 0 | |
| PE-TR/QD605 | 0.2/0 | 13/3.5c | 82/1 | 17/0 | 0/0 | 0/0.4 | 1/0.1 | 0/0 | |
| PE-Cy5/PerCP | 0/0 | 0.6/0 | 0.2/0 | 17.5/13.5 | 0/0 | 0/0 | 71/23.2 | 10.1/2.0 | |
| PE-Cy7 | 0 | 1.4 | 0.4 | 0.6 | 0 | 0 | 0 | 7.7 | |
| PacB | 0 | 0 | 0 | 0 | 0 | 9.0 | 0 | 0 | |
| CasY | 0 | 0 | 0 | 0 | 0 | 2.8 | 0 | 0 | |
| Alexa 647/APC | 0/0 | 0/0 | 0/0 | 0.4/0.6 | 0/0 | 0/0 | 0/0 | 13.4/9.3 | |
| APC-Cy7 | 0 | 0 | 0 | 0.3 | 2.1 | 0 | 0 | 10.0 | |
Compensation matrix generated in the FlowJo software.
A compensation particle set, consisting of uncoated polystyrene beads, or beads coated with anti-mouse Igκ (BD Biosciences), were stained with mAbs conjugated with each of the fluorochromes used.
Grey fields highlight where exchanging fluorochromes has the most significant effect on compensation needs.
3.2 Detailed characterization of maturational status of virus-specific CD8 T cell populations
The adaptive T cell response to antigens goes through stages of priming, effector response and, in the event of a successful response, establishment of immunological memory. The mere identification of epitope-specific CD8 T cells detected by fluorescently labeled HLA-tetramers (Altman et al., 1996), however, requires the use of mAbs to CD3, CD8 and the HLA-tetramer, and occupies three fluorescence channels. It is thus clear that a high number of fluorescence parameters are needed to dissect the complexity of T cell populations.
Here, to demonstrate the utility of this technology in studies of anti-viral immunity, we use nine-color flow cytometry to dissect the maturational status of CD8 T cells specific for HIV-1 Gag or Influenza virus, by the expression patterns of the cell surface maturation markers CD7, CD27, CD28, CD45RA, CD62L, and CCR7 which all help in the identification of maturation stages in HLA-tetramer-defined CD8 cells (Table 1 and Fig. 1) (Hamann et al., 1997; Sallusto et al., 1999; Appay et al., 2002; Aandahl et al., 2003; Aandahl et al., 2004). CD3+CD8+ single cells staining positive for HLA-A2/HIV-1 Gag 77-85 APC-conjugated tetramer were identified in PBMC from a subject with chronic, untreated HIV-1 infection (Fig. 1, left panels). Bi-exponential transformation of logarithmic scales is used in the display and analysis of data to allow proper visualization of all events, including such events that would otherwise be on the axis (Tung et al., 2004; Herzenberg et al., 2006). Also of note is that proper identification of CD7 high and low cells was straightforward when the PE-conjugated mAb was used, but was difficult with FITC-conjugate which gave less distinct staining (data not shown). Hence, the PE-conjugated anti-CD7 mAb was used. Gag-specific CD8 T cells were largely CD7low and CD28-, although a small subset of cells was CD28+. Both subsets, when analyzed for the expression of CCR7 and CD45RA, were found to display a similar profile with a majority of cells being CD45RA-CCR7-. The CD7lowCD28-cells did, however, also contain a minor subset of CD45RAbright cells. These distinct phenotypic subsets were next analyzed for their expression of CD27 and CD62L. The CD7lowCD28-CCR7-CD45RAdim/CD45RA- cells were predominantly CD27+ and CD62L-, with minor populations of CD27- and CD62L+ cells. The CD7lowCD28+CCR7-CD45RAdim/CD45RA- cells were uniformly CD27+ with a minor population of CD62L+ cells. This clearly illustrates the high level of phenotypic complexity in the CD8 T cells responding to a single HIV-1 epitope.
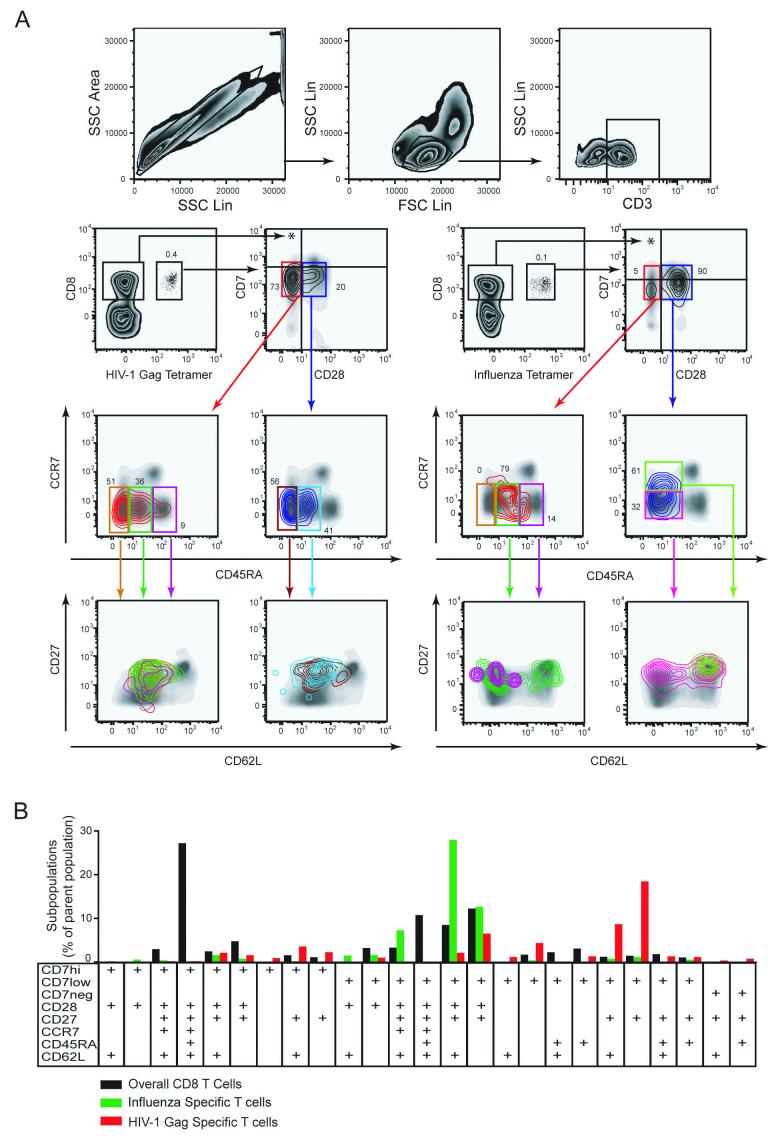
Fig. 1.
High definition phenotypic analysis of virus-specific CD8 T cells. A) T cells were identified by gating on single cells, using SSC-Area versus SSC-Height (upper left panel), followed by gating on lymphocytes (upper middle panel), and CD3+ cells (upper right panel). The lower left panels display identification and subset analysis of CD8 T cells specific for the HIV-1 Gag 77-85 epitope. The lower right panels display identification and subset analysis of CD8 T cells specific for Influenza virus M1 58-66 epitope. Tetramer-defined cells are displayed as a contour plot overlay on a background density plot that represents all CD3+ lymphocytes included as a reference. In subset analysis the contour plot color follows the gate color upstream in the gating scheme. B) Subset distribution analysis of T cells specific for Influenza virus (green) and HIV-1 Gag (red) in comparison with the overall CD8 T cells from the healthy donor in A). Bars represent occurrence of a particular subset as percentage of the parent populations. Anti-CD3 mAb was conjugated with CasY, HLA-A2 tetramer with APC, and anti-CD8 mAb with PerCp. Subsets were defined by expression of CD7 (PE), CD28 (FITC), CD27 (APC-Cy7), CCR7 (PE-Cy7), CD45RA (PacB), and CD62L (PE-TR) as indicated in the figure. Only the 25 subsets which represented at least 1% of total CD8 T cells or at least 0.2% of tetramer-defined T cells were included in the analysis.
To compare the phenotype of HIV-specific CD8 T cells with that of cells specific for a different type of viral infection, CD3+CD8+ single cells staining positive for HLA-A2/Influenza M1 58-66 APC-conjugated tetramer were identified in PBMC from a healthy blood donor (Fig. 1, right panels). These cells displayed a radically different phenotypic profile with a vast majority being CD28+ with a mixed high and low expression of CD7. Detailed subset analysis revealed that the Influenza M1-specific CD8 T cells were very different from the HIV-1 Gag-specific cells. The CD28+ cells were predominantly CD45RAdimCCR7+ and CD27+CD62L+, and the small population of CD28-CD7low cells was largely CD45RAdimCCR7- with a mixed expression of CD62L and CD27.
To better illustrate the complex subset distribution of the two virus-specific CD8 T cell populations in this example, these data can be plotted as a bar chart with healthy donor overall CD8 T cells included as a reference (Fig. 1B). The number of cells appearing in each discrete sub-population is expressed as a percentage of the parent population, such as the HIV-1 gag-tetramer defined CD8 T cells. This analysis helps to clarify this complicated data set, and clearly illustrates, in this example, a divergent maturation marker expression pattern of CD8 T cells specific for different viruses. These data demonstrate that multi-color flow cytometry and graphic visualization strategies together becomes a very useful tool to help evaluate the heterogeneity of CD8 T cell populations, and identify discrete phenotypic subsets that can be subject to further analysis.
3.3 Phenotypic investigation of rare subsets of CD1d-restricted NKT cells
The rare invariant CD1d-restricted NKT cells are regulatory lymphocytes on the border between the innate and adaptive arms of the immune system. They recognize both pathogen-derived and endogenous lipid antigens presented by CD1d, and they display strong reactivity to the model antigen αGalCer (Godfrey and Kronenberg, 2004; Sandberg and Ljunggren, 2005; Bendelac et al., 2006). NKT cells may be of clinical significance in diverse disease settings such as autoimmunity, tumor immunity and infectious diseases (van der Vliet et al., 2004). Proper identification of these rare cells require staining for the TCR α-chain segment Vα24, in conjunction with Vβ11 or an αGalCer-loaded CD1d tetramer or CD1d DimerX reagent (Matsuda et al., 2000; Schumann et al., 2003), on a population pre-gated on CD3+ cells. NKT cells are thus a good example of a cell type for which detailed characterization absolutely requires a significant number of fluorescence parameters.
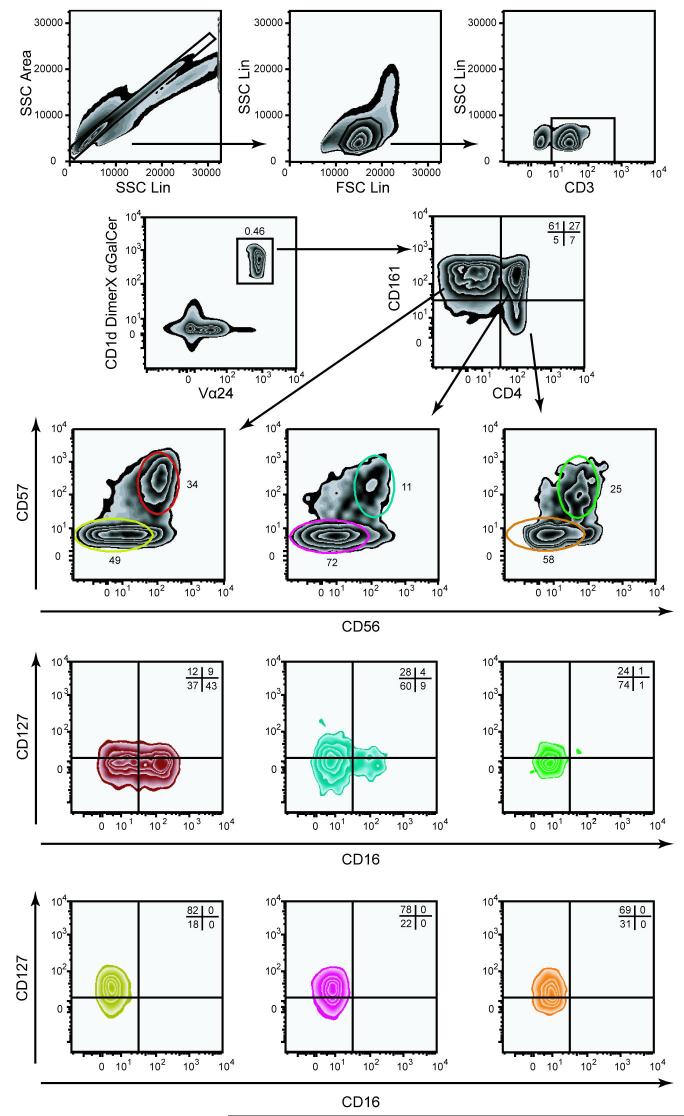
To illustrate the usefulness of nine-color flow cytometry in the study of rare lymphocytes, we provide an example of the use of this technology to investigate the expression of CD16, CD56, CD57, CD127, and CD161 on the CD4+ and CD4- NKT cell subsets (Fig. 2 and Table 1) (Gumperz et al., 2002). CD16 (FcγRIIIA) and CD56 are the classical NK cell markers and CD127 is the receptor for the homeostatic regulatory cytokine IL-7. CD57 is associated with replicative senescence in an end-stage effector phenotype in CD8 T cells (Brenchley et al., 2003). CD161 is a marker of NKT cell maturation (Baev et al., 2004; Sandberg et al., 2004; Berzins et al., 2005). CD3+ single cells staining positive for both TCR Vα24 and CD1d DimerX loaded with αGalCer were identified in PBMC from a healthy blood donor (Fig. 2). This population of CD1d-restricted cells segregated into two main populations of CD4+CD161+ cells and CD4-CD161+ cells, with a minor subset of CD4+CD161- cells. The three subsets were further scrutinized for expression of the classical NK markers CD56 and CD57. All three NKT cell subsets defined by CD4 and CD161 expression patterns contained CD56+CD57+ double-positive cells, whereas essentially no CD56 or CD57 single-positive cells were found. Thus far, utilization of seven fluorescence parameters had allowed us to identify six subsets of CD3+Vα24+ CD1d -restricted NKT cells; CD4+CD161+CD56+CD57+ cells, CD4+CD161+CD56-CD57- cells, CD4-CD161+CD56+CD57+ cells, CD4-CD161+CD56-CD57- cells, CD4+CD161-CD56+CD57+ cells, and CD4+CD161-CD56-CD57- cells. With the final two fluorescence parameters at our disposal, we next analyzed the expression of CD16 and CD127 on these subsets. In the cells from this healthy subject used to generate this data example, CD16 expression was chiefly found in about half of the CD4-CD161+CD56+CD57+ NKT cells, whereas CD127 was highly expressed in all CD56-CD57- subsets but less so in the CD56+CD57+ subsets. These data clearly illustrates the power of polychromatic flow cytometry in generating high resolution phenotypic data that may in turn lead to hypotheses for testing in the laboratory.
Fig. 2.
Identification and characterization of rare invariant CD1d-restricted NKT cells by nine-color flow cytometry. Top panels show the gating to identify CD3+ single lymphocytes, using SSC-Area versus SSC-Height (upper left panel), followed by gating on lymphocytes (upper middle panel), and CD3+ cells (upper right panel). NKT cells were identified among CD3+ cells as staining double positive for αGalCer-loaded CD1d DimerX and anti-Vα24, and further subdivided as CD4-CD161+, CD4+CD161+, and CD4+CD161-. In subset analysis the zebra plot color follows the gate color upstream in the gating scheme. CD1d DimerX was detected with anti-mouse IgG1 PE, biotinylated anti-CD127 was detected with streptavidin-conjugated QD605. Other reagents include: Anti-CD57 FITC, anti-CD161 PE-Cy5, anti-CD56 PE-Cy7, anti-CD16 PacB, anti-CD3 CasY, anti-Vα24 APC, anti-CD4 APC-Cy7.
3.4 Assessment of polyfunctionality in highly defined NK cell subsets
NK cells are an important component of the innate cellular immune response (Fauci et al., 2005; Hamerman et al., 2005; Malmberg and Ljunggren, 2006). A primary effector mechanism of these cells is killing of target cells via the directed release of granules containing perforin and granzymes (Bossi and Griffiths, 2005). Furthermore, NK cells mediate their effects via the production of different cytokines such as IFNγ and TNFα (Biron et al., 1999). Monitoring of NK cell effector functions was for a long time limited to the usage of chromium release assays measuring direct cytotoxicity. However, flow cytometric assessment of NK cell degranulation by the detection of cell surface CD107a was recently described as a surrogate for measurements of cytolysis (Alter et al., 2004). The combination of CD107a staining and intracellular staining for cytokine should allow the simultaneous assessment of several distinct functions on the single cell level. This type of investigation is likely to be important for studies of many diseases where NK cells play a role.
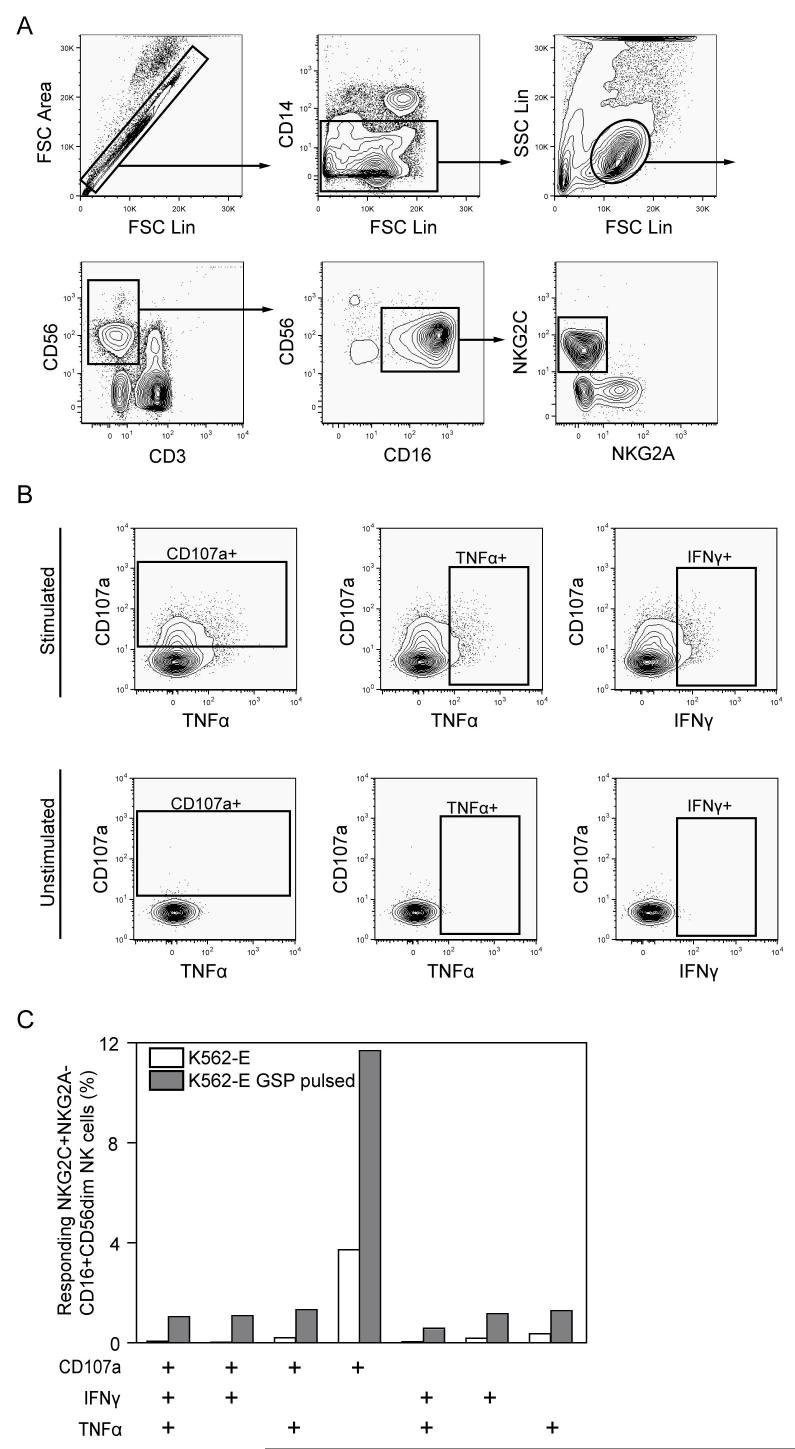
To exemplify how nine-color flow cytometry can be used to investigate the functional response profile of NK cells, we investigated the NK cell response against HLA-E-expressing target cells. NK cells can recognize HLA-E, in complex with peptides derived from signal sequences of certain other HLA molecules, via the inhibitory receptor CD94/NKG2A and the activating receptor CD94/NKG2C (Houchins et al., 1997; Braud et al., 1998). Cell surface expression of HLA-E was stabilized on K562 cells transfected with HLA-E by pulsing with a HLA-E binding signal peptide from HLA-G. These cells were co-incubated with PBMC from a healthy blood donor, and the NKG2C+NKG2A- NK cell subset responsive to HLA-E was identified within the CD16+CD56dimCD3-CD14- NK cells (Fig. 3A), using six fluorescence parameters (Table 1). We assessed surface CD107a, as well as intracellular IFNγ and TNFα expression in the NKG2C+NKG2A- NK cell subset (Fig. 3B). The magnitude of the NK cell response increased after specific NKG2C triggering and triple-functional NK cells co-expressing CD107a, IFNγ and TNFα were detected (Fig. 3C). However, the most common NK cell response profile was degranulation without concurrent cytokine production. Altogether, the combination of CD3, CD14, and CD56 to identify NK cells, with markers pinpointing NK cell subsets expressing distinct receptor profiles (CD16, NKG2A, and NKG2C), and a multi-functional readout system (CD107a, IFNγ, and TNFα), opens up the possibility to perform detailed functional evaluation of multiple highly specialized human NK cell subsets. This methodology will likely prove particularly rewarding when working with limited patient material, and for investigations of NK cell function and phenotype in tissues that have low percentages of NK cells.
Fig. 3.
Multi-functional response of NK cell subsets visualized using nine-color flow cytometry. A) Gating scheme used to identify NKG2C+NKG2A- CD16+CD56dimCD3-CD14- NK cells. FSC parameters were used to discriminate single cell events, followed by exclusion of monocytes based on CD14 expression and gating on lymphocytes. NK cells were defined by gating on CD3-CD56+ cells. B) The functional response of NK cells as determined by combined staining for CD107a, IFNγ and TNFα. Unstimulated control cells are shown in the bottom row. C) A Boolean gate platform was used to analyze all possible combinations of CD107a, IFNγ, and TNFα expression . One representative experiment is shown. Biotinylated anti-NKG2A was detected using QD605 streptavidin conjugate. Other reagents: Anti-CD107a FITC, anti-NKG2C PE, anti-CD56 PE-Cy5, anti-IFNγ PE-Cy7, anti-CD16 PacB, anti-CD3 CasY, anti-TNFα Alexa 647, anti-CD14 APC-Cy7.
4. Concluding remarks
Our knowledge about the vast array of receptors and cytokines involved in regulating cellular immunity, and the changes in expression of these factors upon cellular activation and differentiation, has increased rapidly over the last decade. The means to investigate co-expression of proteins at the single cell level have not kept pace with this development, and the heterogeneity of immune cells has probably been underestimated when receptors are studied one, two or three at a time. Limitations in fluorochrome-conjugated reagents, instrumentation and software have long slowed implementation of the polychromatic flow cytometry needed to address this complexity. There has, however, been considerable progress in these areas in recent years. So while these are still relatively expensive and technically challenging experiments to do, polychromatic flow cytometry is now becoming practically feasible for many translational immunology laboratories.
In studies of the human immune system in infectious diseases and also in cancer, the small number of cells that can be retrieved from patients is almost always a limiting factor. This is true for blood samples but even more so for tissue biopsies from, for example, the liver or the intestines. Therefore, it is desirable to increase the number of parameters that can be assessed in a limited number of cells by adding more fluorescence parameters to the experimental setup. This incremental increase in the amount of data that can be retrieved is, however, not as important as the new information that can be gathered about highly refined subsets of cells based on their co-expression of many phenotypical or functional parameters. This type of qualitative assessment at the single cell level promises to prove very useful in clinical settings. Infectious disease research has been pioneering the translational and clinical applications of polychromatic flow cytometry, and this technology should be applicable to the diagnosis of, for example, hematological malignancies. In basic studies of immune cell function it could be very useful to interrogate several cell types in a single sample, where for example activation of one cell type may influence the actions of another. Advanced flow cytometry will be very useful in this type of applications. Unquestionable, polychromatic flow cytometry will be among the most important tools in deciphering the full functional heterogeneity of cellular immunity, and will most likely also improve the diagnosis and treatment of many severe diseases.
Acknowledgements
This work was supported by grants from the Swedish International Development Agency, the Swedish Research Council, the Swedish Foundation for Strategic Research, the Harald Jeansson Foundation, the Clas Groschinsky Foundation, the National Institute of Health (NIH AI52731), and Karolinska Institutet. Author contributions: V.D.G. co-wrote manuscript and generated data; N.K.B. co-wrote manuscript and generated data; K.J.M., M.M., C.K., J.M. took part in technology and method development; H.G.L. co-wrote manuscript; J.K.S. directed technology and method development and wrote manuscript. The authors declare no competing financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aandahl EM, Quigley MF, Moretto WJ, Moll M, Gonzalez VD, Sonnerborg A, Lindback S, Hecht FM, Deeks SG, Rosenberg MG, Nixon DF, Sandberg JK. Expansion of CD7(low) and CD7(negative) CD8 T-cell effector subsets in HIV-1 infection: correlation with antigenic load and reversion by antiretroviral treatment. Blood. 2004;104:3672–8. doi: 10.1182/blood-2004-07-2540. [DOI] [PubMed] [Google Scholar]
- Aandahl EM, Sandberg JK, Beckerman KP, Tasken K, Moretto WJ, Nixon DF. CD7 Is a Differentiation Marker That Identifies Multiple CD8 T Cell Effector Subsets. J. Immunol. 2003;170:2349–55. doi: 10.4049/jimmunol.170.5.2349. [DOI] [PubMed] [Google Scholar]
- Alter G, Malenfant JM, Altfeld M. CD107a as a functional marker for the identification of natural killer cell activity. J. Immunol. Methods. 2004;294:15–22. doi: 10.1016/j.jim.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Altman JD, Moss PAH, Goulder PJR, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–6. [PubMed] [Google Scholar]
- Appay V, Dunbar PR, Callan M, Klenerman P, Gillespie GM, Papagno L, Ogg GS, King A, Lechner F, Spina CA, Little S, Havlir DV, Richman DD, Gruener N, Pape G, Waters A, Easterbrook P, Salio M, Cerundolo V, McMichael AJ, Rowland-Jones SL. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat. Med. 2002;8:379–85. doi: 10.1038/nm0402-379. [DOI] [PubMed] [Google Scholar]
- Baev DV, Peng XH, Song L, Barnhart JR, Crooks GM, Weinberg KI, Metelitsa LS. Distinct homeostatic requirements of CD4+ and CD4- subsets of Valpha24-invariant natural killer T cells in humans. Blood. 2004;104:4150–6. doi: 10.1182/blood-2004-04-1629. [DOI] [PubMed] [Google Scholar]
- Bendelac A, Savage PB, Teyton L. The Biology of NKT Cells. Annu. Rev. Immunol. 2006 doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- Berzins SP, Cochrane AD, Pellicci DG, Smyth MJ, Godfrey DI. Limited correlation between human thymus and blood NKT cell content revealed by an ontogeny study of paired tissue samples. Eur. J. Immunol. 2005;35:1399–407. doi: 10.1002/eji.200425958. [DOI] [PubMed] [Google Scholar]
- Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu. Rev. Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- Bossi G, Griffiths GM. CTL secretory lysosomes: biogenesis and secretion of a harmful organelle. Semin. Immunol. 2005;17:87–94. doi: 10.1016/j.smim.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Braud VM, Allan DS, O’Callaghan CA, Soderstrom K, D’Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, Lanier LL, McMichael AJ. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–9. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- Brenchley JM, Karandikar NJ, Betts MR, Ambrozak DR, Hill BJ, Crotty LE, Casazza JP, Kuruppu J, Migueles SA, Connors M, Roederer M, Douek DC, Koup RA. Expression of CD57 defines replicative senescence and antigen-induced apoptotic death of CD8+ T cells. Blood. 2003;101:2711–20. doi: 10.1182/blood-2002-07-2103. [DOI] [PubMed] [Google Scholar]
- Bruchez M, Jr., Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013–6. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- Brutkiewicz RR. CD1d ligands: the good, the bad, and the ugly. J. Immunol. 2006;177:769–75. doi: 10.4049/jimmunol.177.2.769. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay PK, Price DA, Harper TF, Betts MR, Yu J, Gostick E, Perfetto SP, Goepfert P, Koup RA, De Rosa SC, Bruchez MP, Roederer M. Quantum dot semiconductor nanocrystals for immunophenotyping by polychromatic flow cytometry. Nat. Med. 2006;12:972–7. doi: 10.1038/nm1371. [DOI] [PubMed] [Google Scholar]
- De Rosa SC, Brenchley JM, Roederer M. Beyond six colors: a new era in flow cytometry. Nat. Med. 2003;9:112–7. doi: 10.1038/nm0103-112. [DOI] [PubMed] [Google Scholar]
- Fauci AS, Mavilio D, Kottilil S. NK cells in HIV infection: paradigm for protection or targets for ambush. Nat Rev Immunol. 2005;5:835–43. doi: 10.1038/nri1711. [DOI] [PubMed] [Google Scholar]
- Godfrey DI, Kronenberg M. Going both ways: immune regulation via CD1d-dependent NKT cells. J. Clin. Invest. 2004;114:1379–88. doi: 10.1172/JCI23594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumperz JE, Miyake S, Yamamura T, Brenner MB. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by CD1d tetramer staining. J. Exp. Med. 2002;195:625–36. doi: 10.1084/jem.20011786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann D, Baars PA, Rep MH, Hooibrink B, Kerkhof-Garde SR, Klein MR, van Lier RA. Phenotypic and functional separation of memory and effector human CD8+ T cells. J. Exp. Med. 1997;186:1407–18. doi: 10.1084/jem.186.9.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamerman JA, Ogasawara K, Lanier LL. NK cells in innate immunity. Curr. Opin. Immunol. 2005;17:29–35. doi: 10.1016/j.coi.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Herzenberg LA, Herzenberg LA. Genetics, FACS, immunology, and redox: a tale of two lives intertwined. Annu. Rev. Immunol. 2004;22:1–31. doi: 10.1146/annurev.immunol.22.012703.104727. [DOI] [PubMed] [Google Scholar]
- Herzenberg LA, Tung J, Moore WA, Herzenberg LA, Parks DR. Interpreting flow cytometry data: a guide for the perplexed. Nat Immunol. 2006;7:681–5. doi: 10.1038/ni0706-681. [DOI] [PubMed] [Google Scholar]
- Houchins JP, Lanier LL, Niemi EC, Phillips JH, Ryan JC. Natural killer cell cytolytic activity is inhibited by NKG2-A and activated by NKG2-C. J. Immunol. 1997;158:3603–9. [PubMed] [Google Scholar]
- Lanier LL. NK cell recognition. Annu. Rev. Immunol. 2005;23:225–74. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- Lefrancois L, Marzo AL. The descent of memory T-cell subsets. Nat Rev Immunol. 2006;6:618–23. doi: 10.1038/nri1866. [DOI] [PubMed] [Google Scholar]
- Maecker HT, Frey T, Nomura LE, Trotter J. Selecting fluorochrome conjugates for maximum sensitivity. Cytometry A. 2004;62:169–73. doi: 10.1002/cyto.a.20092. [DOI] [PubMed] [Google Scholar]
- Malmberg KJ, Ljunggren HG. Escape from immune- and nonimmune-mediated tumor surveillance. Semin. Cancer Biol. 2006;16:16–31. doi: 10.1016/j.semcancer.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Matsuda JL, Naidenko OV, Gapin L, Nakayama T, Taniguchi M, Wang CR, Koezuka Y, Kronenberg M. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J. Exp. Med. 2000;192:741–54. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody DB. The surprising diversity of lipid antigens for CD1-restricted T cells. Adv. Immunol. 2006;89:87–139. doi: 10.1016/S0065-2776(05)89003-0. [DOI] [PubMed] [Google Scholar]
- Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–12. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- Sallusto F, Mackay CR, Lanzavecchia A. The role of chemokine receptors in primary, effector, and memory immune responses. Annu. Rev. Immunol. 2000;18:593–620. doi: 10.1146/annurev.immunol.18.1.593. [DOI] [PubMed] [Google Scholar]
- Sandberg JK, Ljunggren HG. Development and function of CD1d-restricted NKT cells: influence of sphingolipids, SAP and sex. Trends Immunol. 2005;26:347–9. doi: 10.1016/j.it.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Sandberg JK, Stoddart CA, Brilot F, Jordan KA, Nixon DF. Development of innate CD4+ alpha-chain variable gene segment 24 (Valpha24) natural killer T cells in the early human fetal thymus is regulated by IL-7. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7058–63. doi: 10.1073/pnas.0305986101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann J, Voyle RB, Wei BY, MacDonald HR. Cutting edge: influence of the TCR V beta domain on the avidity of CD1d:alpha-galactosylceramide binding by invariant V alpha 14 NKT cells. J. Immunol. 2003;170:5815–9. doi: 10.4049/jimmunol.170.12.5815. [DOI] [PubMed] [Google Scholar]
- Tung JW, Parks DR, Moore WA, Herzenberg LA, Herzenberg LA. New approaches to fluorescence compensation and visualization of FACS data. Clin. Immunol. 2004;110:277–83. doi: 10.1016/j.clim.2003.11.016. [DOI] [PubMed] [Google Scholar]
- van der Vliet HJ, Molling JW, von Blomberg BM, Nishi N, Kolgen W, van den Eertwegh AJ, Pinedo HM, Giaccone G, Scheper RJ. The immunoregulatory role of CD1d-restricted natural killer T cells in disease. Clin. Immunol. 2004;112:8–23. doi: 10.1016/j.clim.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Wherry EJ, Ahmed R. Memory CD8 T-cell differentiation during viral infection. J. Virol. 2004;78:5535–45. doi: 10.1128/JVI.78.11.5535-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]