Abstract
Changes in methylation of CpG sites at the interleukin (IL)-4 and interferon (IFN)-γ promoters are associated with T helper (Th) 2 polarization in vitro. No previous studies have examined whether air pollution or allergen exposure alters methylation of these two genes in vivo. We hypothesized that diesel exhaust particles (DEP) would induce hypermethylation of the IFN-γ promoter and hypomethylation of IL-4 in CD4+ T cells among mice sensitized to the fungus allergen Aspergillus fumigatus.We also hypothesized that DEP-induced methylation changes would affect immunoglobulin (Ig) E regulation. BALB/c mice were exposed to a 3-week course of inhaled DEP exposure while undergoing intranasal sensitization to A. fumigatus. Purified DNA from splenic CD4+ cells underwent bisulfite treatment, PCR amplification, and pyrosequencing. Sera IgE levels were compared with methylation levels at several CpG sites in the IL-4 and IFN-γ promoter. Total IgE production was increased following intranasal sensitization A. fumigatus. IgE production was augmented further following combined exposure to A. fumigatus and DEP exposure. Inhaled DEP exposure and intranasal A. fumigatus induced hypermethylation at CpG−45, CpG−53, CpG−205 sites of the IFN-γ promoter and hypomethylation at CpG−408 of the IL-4 promoter. Altered methylation of promoters of both genes was correlated significantly with changes in IgE levels. This study is the first to demonstrate that inhaled environmental exposures influence methylation of Th genes in vivo, supporting a new paradigm in asthma pathogenesis.
Keywords: environmental exposure, respiratory sensitization, cytokines, inhalation toxicology, epigenetic modification
The incidence of asthma is higher among low-income minority children who are more likely to reside near traffic-related air pollution and industrial sites (Brauer et al., 2002; Gehring et al., 2002; Venn et al., 2001). Most likely, the development of asthma in urban areas is influenced by multiple factors, including genes that predispose toward the development of an allergic phenotype and regulators of their molecular signals, and environmental exposures characteristic of the inner city, such as diesel exhaust particles (DEP), other air pollutants, and indoor allergens (Miller, 1998).
The role of T helper (Th) 2 interleukin (IL)-4, IL-5, IL-13 cytokines in promoting allergic sensitization and asthma, and of Th1 cytokine interferon (IFN)-γ in protecting against allergic sensitization and asthma, has been an important area of research for over two decades (Mosmann et al., 1986). The molecular mechanisms responsible for the early and persistent skewing of the cytokine milieu toward a more proallergic phenotype have become a much more recent area of interest. CpG methylation is an example of an epigenetic modification that can affect chromatin remodeling, Th gene locus accessibility, and therefore changes in gene expression that occur in the absence of alterations in DNA sequences. This covalent addition of a methyl group to cytosines in CpG dinucleotides is believed to begin the process by which the Th cells lose their plasticity and differentiate productively toward the Th1 versus the proallergic Th2 pattern of cytokine gene expression. CpG methylation also may confer heritable stabilization of gene expression patterns and maintain Th cell differentiation (Ansel et al., 2003). In some cases, epigenetic alterations may be transmissible beyond a single generation (Anway et al., 2005; Thompson et al., 2001; Vickaryous and Whitelaw, 2005).
It has become evident from several in vitro studies that demethylation of sites at the proximal promoter and conserved intronic regulatory element (CIRE) in the first intron of the IL-4 gene, and hypermethylation of sites in the IFN-γ promoter, result in efficient IL-4 production (Agarwal and Rao, 1998; Jones and Chen, 2006; Lee et al., 2002; Tykocinski et al., 2005; Yano et al., 2003). Th1 differentiation is associated with methylation of a highly conserved DNaseI-hypersensitive region at the 3′ end of the IL-4 gene (Lee et al., 2002; Yano et al., 2003). The environmental triggers of such altered methylation in Th cytokines have not been identified.
Although DEP exposure has been associated with upregulation of Th2 cytokine and chemokine production in multiple in vitro and in vivo studies (Bommel et al., 2000; Fujieda et al., 1998), only a few studies have examined whether exposure to DEP or other air pollutants can induce or inhibit gene methylation or other epigenetic alterations. In vitro exposure to particulate matter less than 10 μm in aerodynamic diameter (PM)10 induced chromatin remodeling by histone acetylation in human lung type II alveolar-like epithelial cells (Gilmour et al., 2003). Inhaled DEP and black carbon have been shown to cause methylation of the p16INK4a gene in lung tumors in rats (Belinsky et al., 2002). But no previous studies have examined the role of ambient air pollution, including DEP, on methylation patterns in vivo among genes associated with Th differentiation or the asthma phenotype. Finding an association between inhaled diesel exposure and Th regulation would provide a critical link in our understanding of the mechanisms involved in gene–environment interactions in asthma causation.
We hypothesized that chronically inhaled exposure to DEP would induce hypermethylation of the IFN-γ gene promoter, and demethylation of the proximal promoter and the CIRE of IL-4 gene in CD4+ T cells among mice sensitized to the allergens from the fungus Aspergillus fumigatus. We also hypothesized that DEP-induced methylation changes would affect immunoglobulin (Ig) E regulation, a biomarker associated with allergic sensitization and asthma. A finding that ambient air pollution in the setting of allergen exposure can modulate airway disease through epigenetic modification would suggest a new paradigm for asthma etiology. Ultimately, this work may have important implications for asthma prevention.
MATERIALS AND METHODS
Overview of study design and sensitization to A. fumigatus
The study consists of four groups of five female BALB/c mice: (1) aerosol vehicle (saline) alone and negative control air; (2) aerosol vehicle alone and DEP; (3) A. fumigatus only and negative control air; and (4) A. fumigatus and DEP. All protocols were approved by the IACUC of the Health Sciences Division of Columbia University and New York University.
Sensitization to A. fumigatus was induced according to previously described protocols that reliably elicited a strong proallergic IgE response associated with airway hyperreactivity and eosinophilia (Grunig et al., 1997; Padilla et al., 2005; Zimmermann et al., 2003). Briefly, 11-week-old BALB/c mice were exposed intranasally to 62.5 μg of aerosolized A. fumigatus protein extract (Hollister Stier, Spokane, WA) (measured endotoxin dose < 0.16 EU/ml; Endotoxin Testing Service, Cambrex Bio Science Walkersville, Inc, MD) in 50 μl PBS every 4 days for a total of six doses.
In vivo exposure to DEP
DEP exposure occurred 5 h per day for 3 weeks concurrent with the A. fumigatus treatment. Diesel exhaust was produced by a 5500-watt single-cylinder diesel engine generator at New York University (Yanmar, Model YDG 5500E) that contains a 406 cc displacement air-cooled engine. The engine is operated using Number 2 Diesel Certification Fuel (Phillips Chemical Company) and 40 weight motor oil (Rotella T, Shell). Exhaust was diluted approximately 150:1 and routed to a 1-m3 flow-through exposure chamber. The exhaust composition from the single-cylinder engine is consistent with “typical” diesel exhaust reported in the literature (McDonald et al., 2004) and a larger on-road engine, a 2000 Model Cummins 5.9L ISB engine operated on a variable duty cycle on a dynamometer using the same fuel and lube-oil. The average particle concentration was 1.28 mg/m3. This system reliably mimics outdoor exposure (McDonald et al., 2004). The control animals were exposed to HEPA (high efficient particle) filtered ambient air in parallel and for the same duration of time as the diesel exposure.
IgE production
Sera were collected by retro-orbital bleed prior to the inhaled administration of A. fumigatus and 4 days after the sixth dose. Levels of IgE were measured by enzyme-linked immunosorbent assay using complementary capture and detection antibody pairs (Pharmingen, San Diego, CA). IgE levels were calculated based on a standard curve using recombinant IgE.
Bronchoalveolar lavage and histological analysis of the lungs
Bronchoalveolar lavage (BAL) was performed with normal saline and collected lavage fluid centrifuged for 10 min at 1500 rpm at 4°C. Slides were prepared, air-dried, fixed in methanol, and stained (Wright-Giemsa; Scientific Products). For differential cell counts, 10 fields of cells were enumerated for each sample and identified as macrophages, lymphocytes, neutrophils, or eosinophils. After lavage, dissected lungs from each mouse were fixed in 10% formalin, mounted in paraffin, sectioned, and stained with hematoxylin and eosin. The slides were analyzed by a blinded reader using a Nikon EclipseE800 microscope (Leica Microsystems GmbH, Wetzlar, Germany).
CD4+ cell isolation/genomic DNA purification
Splenic cell suspensions from harvested organs were prepared by gently pressing tissues through a nylon mesh cell strainer. CD4+ cells were isolated using MACs sorting kits (CD4 (L3T4) microbeads; Miltenyi Biotec, Auburn, CA). Genomic DNA was purified from CD4+ cells using Wizard SV Genomic DNA Purification System from Promega (Madison WI) as per manufacturer's instruction.
Pyrosequencing/determination levels of gene methylation
Purified DNA underwent bisulfite treatment (i.e., to convert unmethylated cytosines into uracils and keep unchanged methylated cytosines) and PCR amplification as recommended by Biotage/EpigenDx (Worcester, MA, http://www.pyrosequencing.com). Briefly, the MethylDetector Bisulfite Modification Kit (Active motif, Carlsbad, CA) was used for bisulfite treatment according to the manufacturer's instructions. Differential methylation was measured at all six CpG sites in the proximal promoter of the IFN-γ gene: CpG−205, CpG−190, CpG−170, CpG−53, CpG−45, CpG−34 (relative to the transcription starting site) (Jones and Chen, 2006; Kersh et al., 2006). CpG−408, CpG−393, CpG−339, and CpG−314 in the IL-4 proximal promoter region, and CpG+101, CpG+113, CpG+185 CpG+225, and CpG+229 in the first intron (all positions relative to the first start codon of the IL-4 gene) were selected for testing based on previous reports of hypomethylation in Th2 cells and hypermethylation in Th1 and naïve T cells (Tykocinski et al., 2005). PCR amplification of converted and purified DNA was conducted with the primers listed in Table 1. PCR reactions were performed using Eppendorf PCR Mastermix (Westbury, NY) with cycling parameters consisted of heating at 95°C for 5 min, and 45 cycles of 95°C for 30 s, 52°C for 30 s, 72°C for 30 s. Samples subsequently were sent to EpigenDex for commercial pyrosequencing. Briefly, the methods involves sequencing-by-synthesis in real time and provides a rapid and accurate quantification of consecutive CpG methylation sites with built-in quality controls (Dupont et al., 2004; Tost and Gut, 2007). The analysis is based on an indirect bioluminometric assay of the pyrophosphate that is released from each deoxynucleotide upon DNA-chain elongation. The extent of methylation at each CpG site was automatically calculated and is sensitive enough to detect 2−5% changes in methylation levels (http://www.pyrosequencing.com).
TABLE 1.
Primers Used for PCR Amplification and Pyrosequencing Experiments
| Primer name | Gene | Type of primer | Primer sequence 5′-3′ | 5′ Modification |
|---|---|---|---|---|
| ADS156FP | IFN-γ promoter | PCR forward | TGGTGTGAAGTAAAAGTGTTTTTAGA | |
| ADS156RPB | IFN-γ promoter | PCR reverse | TACACCTCTCTAACTTCCAATTTT | Biotin |
| ADS156FS1 | IFN-γ promoter | Pyrosequencing | GAATGGTATAGGTGGGTA | |
| ADS156FS2 | IFN-γ promoter | Pyrosequencing | AAAACAAATTTGTGAAAATA | |
| ADS018FPB | IL-4 promoter | PCR forward | GGAAGTAGTTAGGTTTAGGTGTGT | Biotin |
| ADS018RP | IL-4 promoter | PCR reverse | CCCCCTTTTTTTTTAAATCTACAA | |
| ADS018RS | IL-4 promoter | Pyrosequencing | CAACATAAAAAATTACACCA | |
| ADS017FP | IL-4 CIRE | PCR forward | GGATGYGATAAAAATTATTTGAGAG | |
| ADS017RPB | IL-4 CIRE | PCR reverse | TAATCCTACCTCAACCACCTA | Biotin |
| ADS017FS1 | IL-4 CIRE | Pyrosequencing | AAAAATTATTTGAGAGAGAT |
Statistical analysis
Data analysis started with calculation of descriptive statistics and examination of the distribution of each variable and bivariate association between variables. The outcome variables include the methylation levels of IL-4 and IFN-γ genes. One mouse failed to generate an IgE response following administration of A. fumigatus and was classified as an outlier and dropped from all future analyses. All tests were two-sided with significance level of 0.05. Mann–Whitney tests were used to compare means between groups. Correlations using rank order nonparametric testing were conducted to compare methylation with IgE levels.
RESULTS
As shown in Figure 1, total IgE production was increased following sensitization to A. fumigatus. IgE production was augmented further following combined exposure to A. fumigatus and DEP. In addition, the mean eosinophil percentage on BAL was greater following sensitization to A. fumigatus and increased even further following combined exposure to A. fumigatus and DEP. Histopathological analysis revealed a greater degree of goblet cell hyperplasia and eosinophilic and mononuclear cell inflammatory infiltrate around the airways and blood vessels of mice sensitized to A. fumigatus alone or in combination with DEP compared with saline or DEP-treated mice.
FIG. 1.
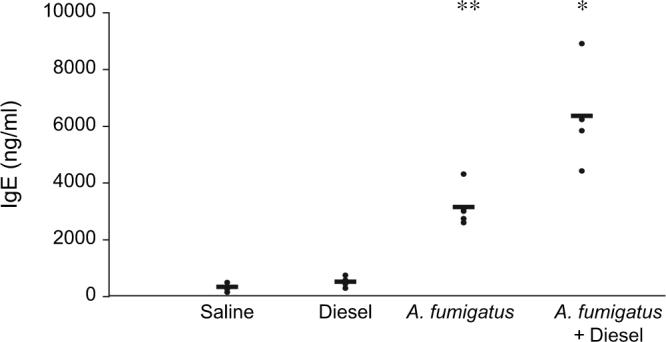
IgE production following exposure to Aspergillus fumigatus and DEP. *p < 0.05 two-tailed t test, compared with saline, DEP or A. fumigatus alone. **p < 0.05 two-tailed t test, compared with saline, or diesel.
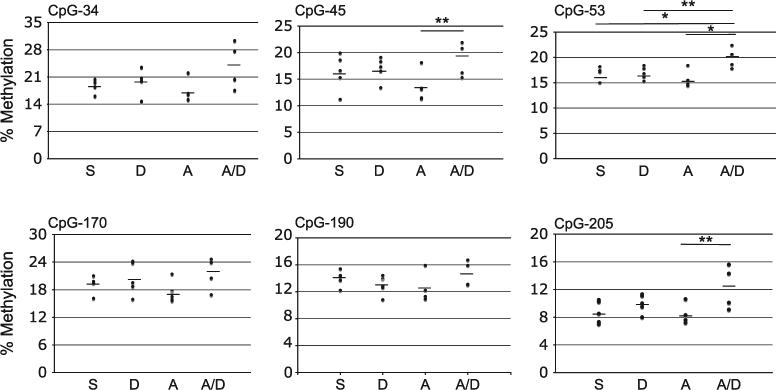
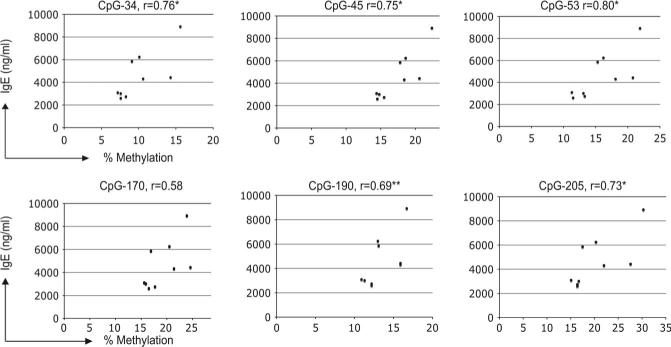
For the CpG−53 site of the IFN-γ promoter, greater methylation was detected following concomitant A. fumigatus and DEP exposure compared with A. fumigatus alone. For CpG−45 and CpG−205, the increase in methylation following concomitant A. fumigatus and DEP exposure reached borderline significance (Fig. 2). Also at CpG−53, greater methylation may have been detected following concomitant A. fumigatus and DEP exposure compared with DEP alone. DEP, in the absence of concomitant exposure to A. fumigatus, did not alter methylation patterns in the IFN-γ promoter (Fig. 2). These results suggest that at select CpG sites, combined exposure to allergen and DEP may induce methylation of the IFN-γ promoter. In addition, at CpG−205, CpG−53, CpG−45, and CpG−34, the extent of methylation correlated significantly with IgE production. A similar trend at CpG−190 was noted as well (Fig. 3). These findings suggest that the altered methylation may exert functional downstream effects associated with a heightened allergic immune response.
FIG. 2.
Aspergillus fumigatus and DEP exposure are associated with increased methylation of IFN-γ promoter at select CpG sites in vivo. S, intranasal saline; D, diesel exhaust particles; A, A. fumigatus; A/D, A. fumigatus + diesel exhaust particles. *p < 0.05 two-tailed t test; **p < 0.05 one-tailed t test.
FIG. 3.
Level of IFN-γ methylation correlates with IgE production. Methylation level at each CpG site was plotted versus IgE level among mice that underwent sensitization to A. fumigatus in the absence or presence of DEP exposure. *p < 0.05 two tailed, Rank order correlation; **p < 0.05 one tailed, Rank order correlation.
Furthermore, hypomethylation in the IL-4 promoter was detected following concomitant exposure to A. fumigatus and DEP, when compared with exposure to either saline, DEP or A. fumigatus alone at CpG−408. Altered methylation was not detected at any of the other sites tested. The extent of hypomethylation was associated significantly with a reduction in IgE when mice that received A. fumigatus (in presence or absence of DEP) were compared (Fig. 4).
FIG. 4.
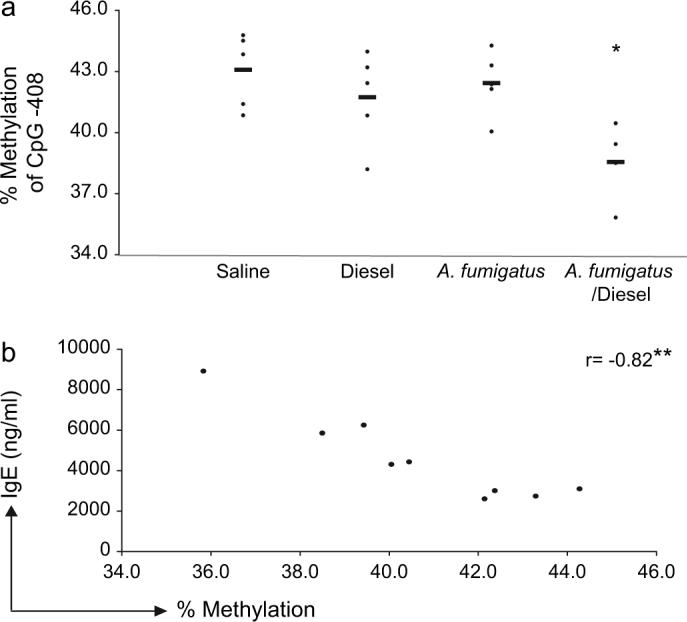
In vivo Aspergillus fumigatus and DEP exposure are associated with hypomethylation of IL-4 promoter at CpG−408 and decreased IgE production. (a) Decreased methylation occurs following A. fumigatus and DEP exposure when compared with either saline or A. fumigatus alone. (b) Level of methylation is associated inversely with IgE production. Methylation level at CpG−408 was plotted versus IgE among mice that underwent A. fumigatus sensitization in the absence or presence of DEP exposure. *p < 0.05 two-tailed compared with saline or A. fumigatus. **p < 0.05 two tailed, Rank order correlation.
DISCUSSION
Epigenetic regulation appears to be an important modifier of disease susceptibility. Environmental exposures are believed to induce these epigenetic changes. Previous studies have shown that altered methylation affects in vitro Th differentiation, raising the possibility that the risk for developing allergic sensitization or asthma also may be affected. We show for the first time that concomitant exposure to inhaled DEP and allergen can induce hypermethylation at select CpG sites in the IFN-γ promoter, and induce hypomethylation at one proximal CpG site in the IL-4 promoter in vivo. In addition, these patterns were associated with changes in IgE production, suggesting that altered methylation exerts molecular effects downstream.
In general, the methylation levels of CD4+ splenocytes at select CpG sites in the IFN-γ promoter averaged higher than observed in previous murine studies of CD4+ thymocytes (Jones and Chen, 2006), especially following combined inhaled exposure to DEP and allergen. However, the levels appeared lower than measured from human CD4+CD45RA+ naïve T cells following in vitro Th2 polarizing conditions, or isolated from adult atopics (White et al., 2002, 2006), suggesting fundamental differences exist across species and cell types.
A growing body of in vitro data has begun to reveal how these methylation patterns may modulate downstream molecular signaling pathways. For example, CpG−53 and CpG−190 are conserved in rat, dog, chimpanzee, and human (Jones and Chen, 2006; White et al., 2002). CpG−53 is located in the proximal AP1-binding site of the IFN-γ promoter and its methylation resulted in a change in factor binding (Young et al., 1994). A subsequent study demonstrated that methylation of CpG−53 in the IFN-γ promoter significantly inhibited CREB and ATF2/c-Jun transcription factor binding to the CpG-containing AP1 site, augmenting proallergic Th2 polarization. Methylation of CpG−53 alone was sufficient to inhibit the IFN-γ promoter-driven reporter gene expression in a Th1 cell line (Jones and Chen, 2006). CpG−190 also interacted with AP-1–CREB DNA binding complexes (White et al., 2002; Ye et al., 1994).
In the present study, DEP and A. fumigatus sensitization induced demethylation in only one of the studied CpG sites of the IL-4 promoter (i.e., CpG−408), and not in any of the sites in the conserved element in the first intron (CIRE). However, demethylation along the IL-4 gene tends to progress in sequential order during Th2 differentiation from 5′ to 3′ (Ansel et al., 2006). Specifically, demethylation during Th2 differentiation has been shown to begin at CpG−408 (Tykocinski et al., 2005). Interestingly, CpG−408 has been identified as a putative transcription factor AP-2 site, although not associated with IL-4 gene transcription to date (Comb and Goodman, 1990).
We recognize several limitations to our approach, including the possibility that DEP and/or A. fumigatus exposure also could act via more distant elements in the IFN-γ or IL-4 genes or other genes. Additional airway studies (i.e., airway hyperreactivity) would have provided more information on the effects at local target tissues. BALB/c mice have been shown to be predisposed toward Th2 allergic responses in some experiments (Whitehead et al., 2003), raising the possibility that the selection of mouse strain may have introduced biased. Finally, it may be difficult to distinguish the independent effects of DEP on CpG methylation from those associated with a general enhancement of Th2-mediated immune responses. Nonetheless, DEP repeatedly augmented the molecular changes observed following sensitization to A. fumigatus alone (Figs. 2, 4), suggesting a direct effect of DEP exposure on T cells.
In sum, chronically inhaled exposure to DEP-induced hypermethylation at several CpG sites of the IFN-γ gene promoter, and hypomethylation of CpG−408 in the proximal promoter of IL-4 gene in CD4+ T cells among mice sensitized to A. fumigatus. Hypermethylation of the IFN-γ gene promoter was associated with greater IgE production, and hypomethylation of the CpG−408 site in the proximal promoter of IL-4 gene was associated with reduced IgE production. This study is the first to demonstrate that inhaled environmental exposures implicated in asthma can alter methylation of Th genes in vivo. These results suggest that combined ambient allergen exposure and air pollution can modulate airway disease through epigenetic modification, supporting a new paradigm in asthma pathogenesis.
ACKNOWLEDGMENTS
We thank Gabriele Grunig, Divya Shah, Huaijie Zhu, and Lin Lin for their assistance with the manuscript.
FUNDING
National Institutes of Health (1R21ES013063) to R.L.M., (P01ES09600) to R.L.M. and F.P., (EPA RD-83214101) to R.L.M. and F.P., (R01ES015495) to L.C.C., and (ES00260) to L.C.C.
REFERENCES
- Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- Ansel K, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and IL-4 locus accessibility. Annu. Rev. Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat. Immunol. 2003;7:616–623. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belinsky SA, Snow SS, Nikula KJ, Finch GL, Tellez CS, Palmisano WA. Aberrant CpG island methylation of the p16(INK4a) and estrogen receptor genes in rat lung tumors induced by particulate carcinogens. Carcinogenesis. 2002;23:335–339. doi: 10.1093/carcin/23.2.335. [DOI] [PubMed] [Google Scholar]
- Bommel H, Li-Weber M, Serfling E, Duschl A. The environmental pollutant pyrene induces the production of IL-4. J. Allergy Clin. Immunol. 2000;105:796–802. doi: 10.1067/mai.2000.105124. [DOI] [PubMed] [Google Scholar]
- Brauer M, Hoek G, Vliet PV, Meliefste K, Fisher PH, Wijga A, Koopman LP, Neijens HJ, Geritsen J, Kerkhof M, et al. Air pollution from traffic and the development of respiratory infections and asthmatic and allergic symptoms in children. Am. J. Respir. Crit. Care Med. 2002;166:1092–1098. doi: 10.1164/rccm.200108-007OC. [DOI] [PubMed] [Google Scholar]
- Comb M, Goodman HM. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990;18:3975–3982. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont JM, Tost J, Jammes H, Gut IG. De novo quantitative bisulfite sequencing using the pyrosequencing technology. Anal. Biochem. 2004;333:119–127. doi: 10.1016/j.ab.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Fujieda S, Diaz-Sanchez D, Saxon A. Combined nasal challenge with diesel exhaust particles and allergen induces in vivo IgE isotype switching. Am. J. Respir. Cell. Mol. Biol. 1998;19:507–512. doi: 10.1165/ajrcmb.19.3.3143. [DOI] [PubMed] [Google Scholar]
- Gehring U, Cyrys J, Sedlimeir G, Brunekreef B, Bellander T, Fischer P, Bauer CP, Reinhardt D, Wichmann HE, Heinrich J. Traffic-related air pollution and respiratory health during the first 2 years of life. Eur. Respir. J. 2002;19:690–698. doi: 10.1183/09031936.02.01182001. [DOI] [PubMed] [Google Scholar]
- Gilmour PS, Rahman I, Donaldson K, MacNee W. Histone acetylation regulates epithelial IL-8 release mediated by oxidative stress from environmental particles. Am. J. Physiol. Lung Cell. Mol. Physiol. 2003;284:L533–L540. doi: 10.1152/ajplung.00277.2002. [DOI] [PubMed] [Google Scholar]
- Grunig G, Corry DB, Leach MW, Seymour BW, Kurup VP, Rennick DM. Interleukin-10 is a natural suppressor of cytokine production and inflammation in a murine model of allergic bronchopulmonary aspergillosis. J. Exp. Med. 1997;185:1089–1099. doi: 10.1084/jem.185.6.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B, Chen J. Inhibition of IFN-g transcription by site-specific methylation during T helper cell development. EMBO J. 2006;25:2443–2452. doi: 10.1038/sj.emboj.7601148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersh EN, Fitzpatrick DR, Murali-Krishna K, Shires J, Speck SH, Boss JM, Ahmed R. Rapid demethylation of the IFN-g gene occurs in memory but not naive CD8 T cells. J. Immunol. 2006;176:4083–4093. doi: 10.4049/jimmunol.176.7.4083. [DOI] [PubMed] [Google Scholar]
- Lee DU, Agarwal S, Rao A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity. 2002;16:649–660. doi: 10.1016/s1074-7613(02)00314-x. [DOI] [PubMed] [Google Scholar]
- McDonald JD, Barr EB, White RK. Design, characterization, and evaluation of a small-scale diesel exhaust exposure system. Aerosol Sci. Technol. 2004;38:62–78. [Google Scholar]
- Miller RL. The need for asthma research on gene-environment interactions. Am. J. Public Health. 1998;89:819–822. doi: 10.2105/ajph.89.6.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- Padilla J, Daley E, Chow A, Robinson K, Parthasarathi K, McKenzie AN, Tschernig T, Kurup VP, Donaldson DD, Grunig G. L-13 regulates the immune response to inhaled antigens. J. Immunol. 2005;174:8097–8105. doi: 10.4049/jimmunol.174.12.8097. [DOI] [PubMed] [Google Scholar]
- Thompson SL, Konfortova G, Gregory R, Reik W, Dean W, Feil R. Environmental effects on genomic imprinting in mammals. Toxicol. Lett. 2001;120:143–150. doi: 10.1016/s0378-4274(01)00292-2. [DOI] [PubMed] [Google Scholar]
- Tost J, Gut I. DNA methylation analysis by pyrosequencing. Nat. Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- Tykocinski LO, Hajkova P, Chang HD, Stamm T, Sozeri O, Lohning M, Hu-Li J, Niesner U, Kreher S, Friedrich B, et al. A critical control element for interleukin-4 memory expression in T helper lymphocytes. J. Biol. Chem. 2005;280:28177–28185. doi: 10.1074/jbc.M502038200. [DOI] [PubMed] [Google Scholar]
- Venn AJ, Lewis SA, Cooper M, Hubbard R, Britton J. Living near a main road and the risk of wheezing illness in children. Am. J. Respir. Crit. Care Med. 2001;164:2217–2180. doi: 10.1164/ajrccm.164.12.2106126. [DOI] [PubMed] [Google Scholar]
- Vickaryous N, Whitelaw E. The role of early embryonic environment on epigenotype and phenotype. Reprod. Fertil. Dev. 2005;17:335–340. doi: 10.1071/rd04133. [DOI] [PubMed] [Google Scholar]
- White GP, Hollams EM, Yerkovich ST, Bosco A, Holt BJ, Bassami MR, Kusel M, Sly PD, Holt PG. CpG methylation patterns in the IFNg promoter in naive T cells: Variations during Th1 and Th2 differentiation and between atopics and non-atopics. Pediatr. Allergy Immunol. 2006;17:557–564. doi: 10.1111/j.1399-3038.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- White GP, Watt PM, Holt BJ, Holt PG. Differential patterns of methylation of the IFN-g promoter at CpG and non-CpG sites underlie differences in IFN-g gene expression between human neonatal and adult CD45RO-T cells. J. Immunol. 2002;168:2820–2827. doi: 10.4049/jimmunol.168.6.2820. [DOI] [PubMed] [Google Scholar]
- Whitehead GS, Walker JK, Berman KG, Foster WM, Schwartz DA. Allergen-induced airway disease is mouse strain dependent. Am. J. Physiol. Lung Cell. Mol. Physiol. 2003;285:L32–L42. doi: 10.1152/ajplung.00390.2002. [DOI] [PubMed] [Google Scholar]
- Yano S, Ghosh P, Kusaba H, Buchholz M, Longo DL. Effect of promoter methylation on the regulation of IFNg gene during in vitro differentiation of human peripheral blood T cells into a Th2 population. J. Immunol. 2003;171:2510–2516. doi: 10.4049/jimmunol.171.5.2510. [DOI] [PubMed] [Google Scholar]
- Ye J, Ghosh P, Cippitelli M, Subleski J, Hardy KJ, Ortaldo JR, Young H. Characterization of a silencer regulatory element in the human interferon-gamma promoter. J. Biol. Chem. 1994;269:25728–25734. [PubMed] [Google Scholar]
- Young H, Ghosh P, Ye J, Lederer J, Lichtman A, Gerard J, Penix L, Wilson C, Melvin A, McGurn M, et al. Differentiation of the T helper phenotypes by analysis of the methylation state of the IFN-gamma gene. J. Immunol. 1994;153:3603–3610. [PubMed] [Google Scholar]
- Zimmermann N, King N, Laporte J, Yang M, Mishra A, Pope S, Muntel E, Witte D, Pegg A, Foster P, et al. Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis. J. Clin. Invest. 2003;111:1863–1874. doi: 10.1172/JCI17912. [DOI] [PMC free article] [PubMed] [Google Scholar]