Abstract
Rheumatoid arthritis (RA) is an autoimmune disease associated with the HLA-DR4 and DR1 alleles. The target autoantigen(s) in RA is unknown, but type II collagen (CII) is a candidate, and the DR4- and DR1-restricted immunodominant T cell epitope in this protein corresponds to amino acids 261–273 (CII 261–273). We have defined MHC and T cell receptor contacts in CII 261–273 and provide strong evidence that this peptide corresponds to the peptide binding specificity previously found for RA-associated DR molecules. Moreover, we demonstrate that HLA-DR4 and human CD4 transgenic mice homozygous for the I-Abβ0 mutation are highly susceptible to collagen-induced arthritis and describe the clinical course and histopathological changes in the affected joints.
Rheumatoid arthritis (RA) is an autoimmune disease of unknown cause with a high prevalence (1%) (1). RA is characterized by a chronic inflammation of the synovial joints that become infiltrated with a variety of leukocytes, which together with activated synoviocytes form the pannus of proliferative tissue that overgrows the articular cartilage. This leads to a progressive destruction of the articular cartilage and subsequent erosion of the underlying bone. The ultimate result of the inflammatory process is joint deformity and loss of joint function.
Inherited susceptibility to RA is associated with the DRB1 genes encoding the HLA-DR4 and HLA-DR1 molecules (2). Structural and functional studies of the disease-associated HLA-DR molecules suggest that the molecular basis for the disease association is selective binding of autoantigenic peptides that are presented to CD4+ T cells, which in turn may initiate or perpetuate an immune response leading to disease (3–5). The nature of the target autoantigen(s) in RA is unknown, but type II collagen (CII) is a candidate because it is the predominant protein of articular cartilage and because CII-specific immunity is associated with RA (6–12). Moreover, mice, rats, and primates all develop inflammatory arthritis following immunization with CII, i.e., collagen-induced arthritis (CIA) (13–15), which in many aspects resembles RA.
To generate a new animal model for RA, we have established HLA-DR4 (DRB1*0401, DRA1*0101) and human CD4 transgenic mice and have shown that the transgenes are expressed in a tissue-specific manner and are functional (16). Using these mice and a predictive algorithm for peptide binding to DR4, we have identified an immunodominant T cell epitope in CII corresponding to residues 261–273 (CII 261–273) (17). Here, we report a detailed characterization of MHC and T cell receptor contacts in this epitope and provide strong evidence that it corresponds to the peptide binding specificity previously found for RA-associated DR molecules. Moreover, we demonstrate that HLA-DR4 and human CD4 transgenic mice are highly susceptible to CIA and describe the clinical course and histopathological changes in the affected joints.
MATERIALS AND METHODS
Mice.
HLA-DR4 and human CD4 transgenic mice on the DBA1/J background (16) and DBA1/J mice were bred at the Department of Experimental Medicine at the University of Copenhagen. Transgenic mice were backcrossed twice to MHC class II deficient mice (Abβ0/0) (18) (Taconic, Germantown, NY), and transgenic and non-transgenic offspring homozygous for the Abβ0 mutation were selected for this study by flowcytometric analysis of peripheral blood mononuclear cells stained with anti-HLA-DR, -human CD4, and -I-Aq antibodies (Becton Dickinson and PharMingen).
Affinity Purification of HLA Molecules and Peptide Binding Assays.
HLA-DRA1*0101/-DRB1*0401 (DRB1*0401) molecules were purified by affinity chromatography from lysates of the Epstein—Barr virus-transformed human B cell line, Priess, as recently described (19). Peptides were synthesized by fluorenylmethoxycarbonyl chemistry (Schafer-N, Copenhagen, Denmark). Purity (>95%) was verified by reversed-phase HPLC and integrity by mass spectrometry. Peptide binding assays were performed as recently described in detail elsewhere (19).
T Cell Hybridoma Generation.
HLA-DR4 and human CD4 transgenic mice were immunized with the CII 261–273 peptide emulsified in Complete Freund’s Adjuvant (Sigma) by intradermal injections of 100 μg of peptide at the base of the tail. Ten days later, the draining lymph nodes were removed, and single cell suspensions were prepared and restimulated in vitro by plating out 2 × 106/well in 24-well titer plates together with 100 μg/well of the CII 261–273 peptide and fused 48 h later with the T cell receptor α /β negative variant of the BW5147 cell line (17). Selection was carried out, and the resulting positive wells were expanded and assayed for IL-2 production as described below. The CII 261–273 specific T cell lines were subcloned by limiting dilution.
T Cell Antigen Receptor (TCR) cDNA Amplification and Analysis.
TCR Vα and Vβ gene usage were determined by PCR amplification of cDNA from each hybridoma using Vα- and Vβ-specific primers, a Cα-specific primer, and a Cβ primer specific for Cβ1 and Cβ2 (20, 21). The PCR products were subcloned into the phagemid TA vector (Invitrogen) and sequenced on a 373A DNA Sequencer (Applied Biosystems). The resulting DNA sequence was translated into amino acids, and the CDR3 regions were determined as previously described (22).
T Cell Stimulation Assay.
The T cell hybridomas were incubated with the DRB1*0401-positive Epstein—Barr virus-transformed human B cell line, Priess, as antigen-presenting cells, and varying concentrations of peptide. Antigen-presenting cells (5 × 104 cells/well) were irradiated with 2100 rad before use and preincubated with dilutions of the relevant peptide for 2 h at 37°C, before addition of the hybridoma cells (2 × 104 cells/well) and incubation at 37°C for 24 h. Subsequently, 100 μl of the supernatant from each well was aspirated for IL-2 determination using an IL-2 specific sandwich ELISA based on an Eu3+-labeled streptavidin detection system as previously described (17). The resulting fluorescence was measured in a time-resolved fluorometer (Wallac, Finland).
Induction and Evaluation of Arthritis.
Mice were immunized at 7–12 weeks of age intradermally at the base of the tail with 100 μg of bovine CII emulsified in Complete Freund’s Adjuvant and given a boost injection with 50 μg of CII emulsified in Incomplete Freund’s Adjuvant (Sigma) at day 40. The mice were bled at days 35 and 55 after immunization, and the amount of CII-reactive antibodies in the sera was detected by quantitative ELISA (23). Clinical scoring of arthritis was performed as described (24). The animal experiments were approved by the author’s institutional review board.
Immunhistopathology.
Paws from representative mice were taken at day 60 and were analyzed with histopathological and immunohistochemical techniques. One of the paws from each mouse was fixed in formaldehyde, decalcified, and stained with hematoxylin-eosin on paraffin sections. Another paw from each mouse was decalcified in EDTA, snap-frozen, and cryosectioned as described previously (25). Cryosections were subjected to immunohistochemical analyses using techniques as described (25) and using antibodies against CD4 (H129.19), CD8 (53.6.7), CD11b (M1/70), and HLA-DR (L243).
RESULTS
Identification of HLA-DRB1*0401 Binding Residues within CII 261–273.
We have recently identified an immunodominant T cell epitope in CII corresponding to residues 261–273 (CII 261–273) (17). As a first step in defining the structural requirements for the formation of HLA-DRB1*0401:CII 261–273 complexes, a series of CII 261–273 monosubstituted Ala analog peptides were synthesized, and their relative binding affinities to DRB1*0401 molecules were determined and compared with that of the native CII 261–273 peptide (Table 1). The measured IC50 value of the natural CII-261–273 sequence was 180 nM, consistent with our previous results (17). Substitution of Phe-263 by Ala abrogated peptide binding to the DRB1*0401 molecules, which is consistent with data obtained with a shorter analog of the parent sequence truncated after Phe-263 from the N terminus (17). Several other potential MHC contacts were identified within CII 261–273; Ala substitutions of Lys-264, Glu-266, and Pro-269 resulted in peptides that were bound with approximately 5–10 times lower affinities than the parent peptide, whereas Ala substitutions of Gly-265, Gly-271, and Glu-272 resulted in analogs with approximately 5–10 times higher affinity than the parent peptide. Ala substitutions of Gln-267, Gly-268, Lys-270, and Pro-273 only marginally changed the apparent binding affinity. We have recently hypothesized that Phe-263 is occupying the hydrophobic P1 pocket and Glu-266 the positively charged P4 pocket in the HLA-DRB1*0401 peptide binding site (17). This assumption was further substantiated by a new series of selected monosubstituted analog peptides, which were synthesized and tested for their binding affinity (Table 1). Thus, Phe-263 could be substituted with another amino acid with an aromatic side chain, Tyr, with only a very modest loss in detectable binding affinity, whereas a polar amino acid, Asn, in this position was incompatible with peptide binding. Both results are in accordance with the peptide binding requirements for the P1 pocket (5, 26, 27). Substitution of the negatively charged Glu-266 with the negatively charged Asp resulted in a 10-fold loss of binding affinity, whereas replacement with the positively charged Lys and Arg resulted in peptides that were bound with approximately two orders of magnitude lower affinity. These results are in accordance with the peptide binding requirements for the P4 pocket (5, 27).
Table 1.
Experimentally determined IC50 values for the binding of monosubstituted CII261-273 analogs to HLA-DRB1*0401
| Peptide | Amino acid sequence and putative position in the binding pocket
|
IC50 (nM) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P-1 | P-2 | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 | P11 | ||
| CII261-273 | A | G | F | K | G | E | Q | G | P | K | G | E | P | 180 |
| CII262A | A | A | F | K | G | E | Q | G | P | K | G | E | P | 52 |
| CII263A | A | G | A | K | G | E | Q | G | P | K | G | E | P | >35,000 |
| CII264A | A | G | F | A | G | E | Q | G | P | K | G | E | P | 2022 |
| CII265A | A | G | F | K | A | E | Q | G | P | K | G | E | P | 21 |
| CII266A | A | G | F | K | G | A | Q | G | P | K | G | E | P | 1249 |
| CII267A | A | G | F | K | G | E | A | G | P | K | G | E | P | 115 |
| CII268A | A | G | F | K | G | E | Q | A | P | K | G | E | P | 65 |
| CII269A | A | G | F | K | G | E | Q | G | A | K | G | E | P | 781 |
| CII270A | A | G | F | K | G | E | Q | G | P | A | G | E | P | 77 |
| CII271A | A | G | F | K | G | E | Q | G | P | K | A | E | P | 40 |
| CII272A | A | G | F | K | G | E | Q | G | P | K | G | A | P | 24 |
| CII273A | A | G | F | K | G | E | Q | G | P | K | G | E | A | 429 |
| CII263N | A | G | N | K | G | E | Q | G | P | K | G | E | P | >35,000 |
| CII263S | A | G | S | K | G | E | Q | G | P | K | G | E | P | >35,000 |
| CII263Y | A | G | Y | K | G | E | Q | G | P | K | G | E | P | 541 |
| CII266D | A | G | F | K | G | D | Q | G | P | K | G | E | P | 2222 |
| CII266K | A | G | F | K | G | K | Q | G | P | K | G | E | P | >35,000 |
| CII266R | A | G | F | K | G | R | Q | G | P | K | G | E | P | >35,000 |
T Cell Receptor Recognition of HLA-DRB1*0401/CII Complex.
To begin to analyze the structural requirements of T cell receptor recognition of the DRB1*0401:CII 261–273 complex, four DRB1*0401-restricted and CII 261–273-specific T cell hybridomas (3838, C4, D3, and E11) generated from HLA-DR4 and human CD4 transgenic mice immunized with the CII 261–273 peptide were used in this study. The primary structures of the four CII 261–273-specific T cell receptors were determined, and T cell receptor contacts in the CII 261–273 sequence were mapped. The four hybridomas each used a different Vα gene segment: Vα11 (3838), Vα2 (C4), Vα4 (D3), and Vα5T (E11) and also had different CDR3 sequences (data not shown). In contrast, three of the hybridomas (3838, C4, and D3) used the same Vβ14 gene segment, whereas the E11 hybridoma used the Vβ 1/10 segment. All four β chains used different CDR3 sequences (data not shown).
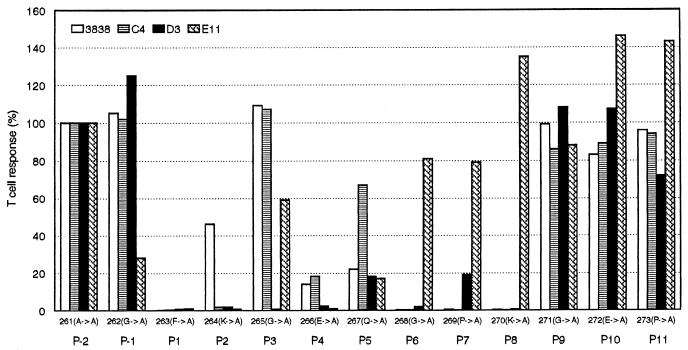
To determine the TCR contact residues in the CII 261–273 peptide, the four hybridomas were tested for their ability to recognize the complete set of monosubstituted Ala analogs of CII 261–273 presented by a HLA-DRB1*0401-positive Epstein—Barr virus-transformed human B cell line. To compare the individual T cell hybridoma responses to the Ala analogs, the IL-2 ELISA results were expressed as percentage of the respective T cell response to the parent peptide (Fig. 1). As expected on the basis of the MHC binding data, Ala substitutions of Phe-263, Lys-264, and Gln-266 led to either disrupted or reduced T cell stimulation. The individual T cell hybridomas had different T cell receptor contact points, and no single contact point was shared by all four hybridomas. Three of the hybridomas (3838, D3, C4) recognized similar contact points, whereas one (E11) had different structural requirements for stimulation. Thus, individual Ala replacements of Gly-268, Pro-269, or Lys-270 all led to disrupted stimulation of hybridomas 3838, D3, and C4. These three hybridomas differed from each other with respect to sensitivity to Ala substitutions of Lys-264, Gly-265, and Gln-267. Thus, Ala replacement of Gly-265 only led to disrupted stimulation of hybridoma D3, whereas Ala substitution of Gln-267 affected both hybridomas 3838 and D3 but not C4. Ala substitution of the presumed MHC anchor, Lys-264, reduced the stimulation of hybridoma 3838 by 50% but disrupted stimulation of C4 and D3. This result indicates that Lys-264 also provides T cell contact for at least the C4 and D3 hybridomas, because the reduction in binding affinity of the corresponding Ala-substituted analog does not fully explain the abrogated T cell stimulation. Three T cell contact points were identified for hybridoma E11: Gly-262, which was specific for this hybridoma, Lys-264, which was shared with hybridomas C4 and D3, and Gln-267 which was also recognized by hybridomas 3838 and D3. Three of the identified T cell receptor contact points, Gly-262, Gly-265, and Gly-268, involved a Gly that has no side chain for T cell receptor interaction, which therefore is likely to be represented by the MHC:peptide complex in these positions.
Figure 1.
Stimulatory effect of monosubstituted CII 261–273 alanine analogs on four different HLA-DRB1*0401 and CII 261–273 specific T cell hybridomas. Putative positions in the HLA-DRB1*0401 binding pocket (P-2 to P11) of the amino acids in the CII261–273 peptide are indicated.
Induction of CIA in the DR4 and Human CD4 Transgenic Mice.
To facilitate the functional analysis of the role of HLA-DR4 in the pathogenesis of RA, HLA-DR4 and human CD4 transgenic mice were backcrossed twice to MHC class II-deficient mice (Abβ0/0) (18). In the resulting offspring mice, the expression of the transgenes was similar to that previously described for HLA-DR4 and human CD4 transgenic mice on the DBA1/J background, and the peripheral T cell compartment, deficient in the Abβ0/0 mice, was reconstituted as recently reported (28) (data not shown).
Two experimental groups of DR4 and human CD4 transgenic/I-Abβ0/0 mice, nontransgenic Abβ0/0 mice and DBA/1 mice, were tested for susceptibility to CIA (Table 2). After the first CII immunization, arthritis developed in 25% of the DR4 and human CD4 transgenic/I-Abβ0/0 mice and in 56% of the DBA/1 mice in the first experimental group and in 14% of the DR4 and human CD4 transgenic/I-Abβ0/0 mice and in 18% of the DBA/1 mice in the second experimental group. After the second CII immunization given at day 40, arthritis developed in 88% of the DR4 and human CD4 transgenic/I-Abβ0/0 mice and 89% DBA/1 mice in group 1 and 57% of the DR4 and human CD4 transgenic/I-Abβ0/0 mice and 82% of the DBA/1 mice in group 2, whereas the nontransgenic Abβ0/0 littermates were resistant to disease (Table 2, data not shown).
Table 2.
Frequency, onset, and severity of arthritis
| Frequency of disease | Mean day of onset† | Mean max score‡ | |
|---|---|---|---|
| Group 1 | |||
| DBA/1 | 8/9 | 38 (25–51) | 9.9 (2, 1) |
| DR4/huCD4 tg | 7/8 | 47 (39–51) | 11.3 (0, 4) |
| Non tg | 0/13 | — | — |
| Group 2 | |||
| DBA/1 | 9/11 | 45 (21–51) | 7.4 (2, 0) |
| DR4/huCD4 tg | 8/14 | 46 (27–54) | 9.7 (2, 5) |
| Non tg | 0/15 | — | — |
Includes all mice with clinical arthritis. Range of day of onset is given in parentheses.
Mice sacrificed for photography or histopathology during the experiment are not included. The standard deviation is given in parentheses. Tg, transgenic.
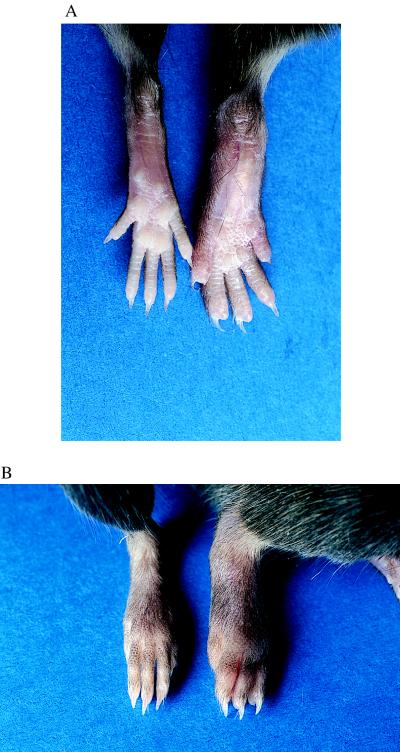
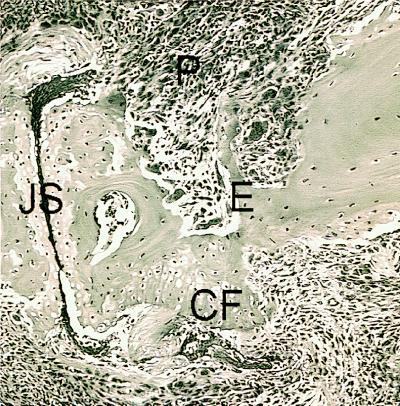
In both DR4 and human CD4 transgenic/Abβ0/0 mice and DBA/1 mice, the peripheral joints in the front and rear paws were severely affected (Table 2, Fig. 2), showing erythema and swelling, which decreased after 3–4 weeks, leaving the affected paws deformed. Day of onset and severity of disease were similar for DR4 and human CD4 transgenic/Abβ0/0 mice and DBA/1 mice (Table 2). Histopathology of joints from arthritic paws from DR4 and human CD4 transgenic/I-Abβ0/0 mice showed severely deformed joints and areas of both active inflammation and new bone and cartilagenous formation (Fig. 3). Immunohistopathologic analysis showed the presence of infiltrating CD4+ T cells and a pronounced pannus formation with activated macrophages and neutrophils as determined by their strong CD11b expression. Expression of HLA-DR was seen on some of the synovial macrophages and on Langerhans cells in the epidermis (data not shown).
Figure 2.
Collagen-induced arthritis in DR4 and human CD4 transgenic/Abβ0/0 mice. Plantar (A) and dorsal (B) views of rear paws of a DR4 and human CD4 transgenic/Abβ0/0 mouse (right) and a transgene negative Abβ0/0 littermate (left). Peripheral joints of the transgenic mouse are severely affected showing erythema and swelling, which decreased after 3–4 weeks leaving the affected paws deformed.
Figure 3.
Histopathology of an arthritic joint from a DR4 and human CD4 transgenic/I-Abβ0/0 mouse immunized with CII. Hematoxylin-eosin staining of a distal interphalangeal joint obtained 60 days after CII immunization. Both inflammatory pannus tissue (P) eroding the bone (E) and cartilage surface and new cartilage formation (CF) are seen. The joint space (JS) is filled with fibrin.
The DR4 and human CD4 transgenic/Abβ0/0 mice and the DBA/1 mice developed high levels of antibodies to bovine CII, crossreactive to mouse CII, whereas no response was seen in the I-Abβ0/0 mice (Table 3). The DR4 and human CD4 transgenic/I-Abβ0/0 mice tended to make a stronger response than the DBA/1 with a higher fraction crossreacting to mouse CII especially at late stages (day 55), but these differences were not significant. Furthermore, the response was dominated by IgG2a antibodies, indicating a Th1 type of response. Again, the DR4 and human CD4 transgenic/I-Abβ0/0 mice made a relatively stronger IgG2a than IgG1 response.
Table 3.
Antibody specific for bovine CII (bCII) and murine CII (mCII) at days 35 and 55 after bovine CII immunization
| Strain | Mean IgG (μg/ml)† | ||
|---|---|---|---|
| bCII day 55 | mCII
|
||
| day 35 | day 55 | ||
| DBA/1 | 698 (430–966) | 111 (46–176) | 620 (87–1153) |
| DR4/huCD4 tg | 1023 (415–1631) | 68 (8–120) | 1188 (308–2068) |
| Non tg | 0 | ND | ND |
| Strain | Mean IgG1 (μg/ml) mCII
|
Mean IgG2a (μg/ml) mCII
|
||
|---|---|---|---|---|
| day 35 | day 55 | day 35 | day 55 | |
| DBA/1 | 26 (1–51) | 101 (6–196) | 22 (9–35) | 128 (15–241) |
| DR4/huCD4 tg | 30 (2–56) | 233 (74–392) | 29 (2–56) | 496 (158–834) |
| Non tg | ND | ND | ND | ND |
Only results from experimental group 1 are shown but are analogous to those obtained in experimental group 2. No antibody response was observed in unimmunized mice. The 95% probability limits of the mean is given in parentheses. ND, not determined. Tg, transgenic.
In summary, the arthritis development and immune response against CII in DR4 and human CD4 transgenic/I-Abβ0/0 mice were similar to CIA as seen in H-2q mice with the exception that autoantibody response to CII tended to be stronger in the DR4 and human CD4 transgenic/I-Abβ0/0 mice.
DISCUSSION
Inherited susceptibility to RA is in most ethnic groups associated with the genes encoding the HLA-DR4 and HLA-DR1 molecules (2). Structural and functional studies of these molecules provide strong circumstantial evidence that selective binding of peptides is the molecular basis for the HLA-DR association (3–5, 17). Thus, the crystal structures of the HLA-DR4 and HLA-DR1 molecules have revealed that amino acids 67–74 of the β chain, which are shared between RA-associated HLA-DR alleles, are located in the antigen binding cleft of the molecule where they contribute to the formation of the P4 pocket (3, 4). Functional studies have shown that the peptide binding specificity of HLA-DR4 molecules correlates with RA association and depends largely on a single amino acid corresponding to position 71 of the β chain, which is found in the P4 pocket. Thus, a negative charge is accepted and a positive charge is not accepted by the RA-associated DR4 subtypes in position 71 of the DR β chain, and the opposite is the case for the non-associated 0402 molecule (5). However, it is currently unknown which autoantigenic peptides are presented by the RA-associated HLA-DR molecules to disease-mediating CD4+ T cells, but it is likely that they are derived from cartilage proteins.
Thus, a specific immune response toward the major cartilage collagen, CII, clearly exists in many RA patients. IgG antibodies of the IgG type specific for CII have been observed in peripheral blood (6), in the synovial fluid (7), bound to cartilage (8), and secreted by blood and synovial B lymphocytes (9). The B lymphocyte response to CII is also higher in DR4-positive RA patients than in DR4 negative (10, 11). Moreover, CII-specific T cells have been shown to persist in the synovial membrane of active rheumatoid joints (12).
We have recently identified a DRB1*0401-restricted immunodominant T cell epitope in human and bovine CII corresponding to residues 261–273 (CII 261–273) (17). On the basis of a model generated from the crystal structure of HLA-DR1 complexed with an influenza hemagglutinin peptide, we have suggested that Phe-263 is occupying the large nonpolar P1 pocket and Glu-266 the positively charged P4 pocket in the HLA-DRB1*0401 peptide binding site (17). Our new studies with monosubstituted CII 261–273-derived peptide analogs confirm this hypothesis, which is also supported by a recent model of the same complex based on the crystal structure of HLA-DR4 complexed with another CII-derived peptide (4). Further, according to the latter model Gly-268 (P6), Pro-269 (P7), and Gly-271 (P9) are at least partially buried in pockets, whereas Ala-261 (P-2), Gly-262 (P-1), Lys-264 (P2), Gly-265 (P3), Gln-265 (P5), Lys-270 (P8), Glu-272 (P10), and Pro-273 (P11) are substantially exposed to solvent with the side chains of Gln-265 (P5) and Lys-270 (P8) extending directly into solution, which also, in general, is in accordance with our MHC binding and T cell recognition results. Based on the model proposed by Dessen et al. (4), peptide residues from P-2 to P8 are expected to be covered by the footprint of a TCR based on the orientation and contact area seen in the MHC class I-peptide interface (29). In accordance, we have identified seven different T cell receptor contacts in a sequence core extending from position Gly-262 (P-1) through Lys-270 (P8). Our analysis does not include P-2 because Ala is the native amino acid in this position. Together, these data provide a detailed understanding of the DRB1*0401-restricted T cell recognition of the CII261–273 peptide.
We also demonstrate in this study that HLA-DR4 and human CD4 transgenic mice homozygous for the MHC class II “knock out” mutation (I-Abβ0/0) are highly susceptible to CIA that clinically and histopathologically share significant similarities with those found in mice with CIA-susceptible H-2 haplotypes, including composition of infiltrating cells, erosion/remodeling, and severity (30). Interestingly, HLA-DR4 and human CD4 transgenic mice on the CIA-susceptible DBA/1 H-2q background, expressing I-A but no I-E molecules, develop less arthritis than both the native DBA/1 mice and the HLA-DR4 and human CD4 transgenic mice expressing no endogenous MHC class II molecules (31). The reason for this dichotomy in susceptibility between these two strains of HLA-DR4 and human CD4 transgenic mice is unclear but could reflect undefined epistatic interactions that are different in the two genetic backgrounds, including a protective effect of the HLA-DR4 and/or human CD4 transgenes on the DBA/1 background, which could be mediated through a changed cytokine balance or T cell repertoire.
HLA-DR1 (DRB1*0101) transgenic mice has recently been reported also to be susceptible to CIA (32), and the clinical course and histological joint changes seem to be similar to those observed in our DR4 and human CD4 transgenic mice. Moreover, the DR1-restricted immunodominant CII peptide apparently corresponds to the DR4-restricted epitope (32). Neither of these findings is unexpected owing to the structural homology of the peptide binding pockets between these two RA-associated HLA-DR molecules. Interestingly, the DR1 transgenes confer CIA susceptibility on the B10.M/Sn (H-2f) arthritis-resistant background expressing I-A but no I-E molecules. The I-Af molecules also restrict a T cell response toward CII (32), which may therefore have contributed to, or possibly partially protected from, the arthritis development. The simultaneous expression of the endogenous I-Af molecule might therefore complicate a general interpretation of the specific role of DR1 in shaping of the T cell repertoire and MHC class II-restricted immune responses and consequently also complicate a broader understanding of its role in arthritis causing autoimmune responses. This concern is underscored by a recent study demonstrating that thymic deletion mediated by disease-protective MHC class II molecules seems to be a major factor in the development of autoimmune diseases (33). Nevertheless, both DR4 and DR1 transgenic mice will most likely be valuable in the understanding of the pathogenesis of HLA-DR-associated inflammatory arthritis and development of novel therapeutic strategies.
It has recently been proposed that HLA-DQ and not HLA-DR alleles mediate susceptibility to RA (34). This hypothesis is based on the well known linkage disequlibrium between the RA-associated DR4 alleles and the DQ7 and DQ8 alleles combined with the recent observations that HLA-DQ8 transgenic mice are susceptible to CIA (35) and that the immune response in these mice to peptides from the third hypervariable region of HLA-DRB1 molecules correlates with predisposition to RA (36). According to this hypothesis, the DRB1 locus is in general conferring protection to RA, whereas the RA-associated DRB1 alleles are not responsible for the primary disease association but merely permissive for the susceptibility conferred by the HLA-DQ alleles with which they are in linkage disequlibrium (34). If DQ8 indeed was the HLA class II molecule responsible for susceptibility to RA, whereas the RA-associated DRB1 molecules were merely playing a permissive role, one would expect that the vast majority of RA patients carrying the RA-associated DR4 molecules would be DQ8 positive. To test this hypothesis, we have performed a critical review of the literature on the HLA association in RA with special emphasis on studies in which both an HLA-DR and -DQ association has been investigated (37). Our analyses provide strong evidence against the hypothesis that HLA-DQ molecules play a major role in the general susceptibility to RA and demonstrate the strongest association in RA is with DRB1 genes rather than DQB1 genes.
Acknowledgments
This paper is dedicated the memory of the late professor Jørn Hess Thaysen. We thank Margareta Svejme for help with histopathologic analysis. This study was supported by grants from the Alfred Benzon Foundation of Denmark, The Danish Medical Research Council, The Danish Arthritis Foundation, The Swedish Medical Research Council, and The Swedish Rheumatism Foundation.
ABBREVIATIONS
- RA
rheumatoid arthritis
- CIA
collagen-induced arthritis
- CII
collagen type II
- TCR
T cell antigen receptor
References
- 1.Feldmann M, Brennan F M, Maini R N. Cell. 1996;85:307–310. doi: 10.1016/s0092-8674(00)81109-5. [DOI] [PubMed] [Google Scholar]
- 2.Gregersen P K, Silver J, Winchester R J. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 3.Stern L J, Brown J H, Jardetsky T S, Gorga J C, Urban R G, Strominger J L, Wiley D C. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 4.Dessen A, Lawrence C M, Cupo S, Zaller D M, Wiley D C. Immunity. 1997;7:473–481. doi: 10.1016/s1074-7613(00)80369-6. [DOI] [PubMed] [Google Scholar]
- 5.Hammer J, Gallazzi F, Bono E, Karr R W, Guenot J, Valsasnini P, Nagy Z A, Sinigaglia F. J Exp Med. 1995;181:1847–1855. doi: 10.1084/jem.181.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cook A D, Rowley M J, Mackay I R, Gough A, Emery P. Arthritis Rheum. 1996;39:1720–1727. doi: 10.1002/art.1780391015. [DOI] [PubMed] [Google Scholar]
- 7.Clague R B, Moore L J. Arthritis Rheum. 1984;27:1370–1377. doi: 10.1002/art.1780271207. [DOI] [PubMed] [Google Scholar]
- 8.Jasin H E. Arthritis Rheum. 1985;28:241–248. doi: 10.1002/art.1780280302. [DOI] [PubMed] [Google Scholar]
- 9.Tarkowski A, Klareskog L, Carlsten H, Herberts P, Koopman W J. Arthritis Rheum. 1989;32:1087–1092. doi: 10.1002/anr.1780320906. [DOI] [PubMed] [Google Scholar]
- 10.Rowley M J, Tait B, Doran T, Emery P, Mackay I R. Ann Rheum Dis. 1990;49:578–581. doi: 10.1136/ard.49.8.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ronnelid J, Lysholm J, Engstrom-Laurent A, Klareskog L, Heyman B. Arthritis Rheum. 1994;37:1023–1029. doi: 10.1002/art.1780370707. [DOI] [PubMed] [Google Scholar]
- 12.Londei M, Savill C M, Verhoef A, Brennan F, Z A L, Duance V, Maini R N, Feldman M. Proc Natl Acad Sci USA. 1988;86:636–640. doi: 10.1073/pnas.86.2.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Courtenay J S, Dallman M J, Dayan A D, Martin A, Mosedale B. Nature. 1980;283:666–668. doi: 10.1038/283666a0. [DOI] [PubMed] [Google Scholar]
- 14.Trentham D E, Townes A S, Kang A H. J Exp Med. 1977;146:857–868. doi: 10.1084/jem.146.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cathcart E S, Hayes K C, Gonnerman W A, Lazzari A A, Franzblau C. Lab Invest. 1986;54:26–31. [PubMed] [Google Scholar]
- 16.Fugger L, Michie S A, Rulifson I, Lock C B, McDevitt G S. Proc Natl Acad Sci USA. 1994;91:6151–6155. doi: 10.1073/pnas.91.13.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fugger L, Rothbard J, McDevitt G S. Eur J Immunol. 1996;26:928–933. doi: 10.1002/eji.1830260431. [DOI] [PubMed] [Google Scholar]
- 18.Grusby, M. J., Johnson, R. S., Papaioannou, V. E. & Glimcher, L. H. (1991) 253, 1417–1420. [DOI] [PubMed]
- 19.Hansen B E, Andersson E C, Madsen L S, Engberg J, Søndergaard L, Svejgaard A, Fugger L. Tissue Antigens. 1998;51:119–128. [PubMed] [Google Scholar]
- 20.Casanova J L, Romero P, Widmann C, Kourilsky P, Maryanski J L. J Exp Med. 1991;174:1371–1383. doi: 10.1084/jem.174.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osman G E, Toda M, Kanagawa O, Hood L E. J Exp Med. 1993;177:387–395. doi: 10.1084/jem.177.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rock E P, Sibbald P R, Davis M M, Chien Y H. J Exp Med. 1994;179:323–328. doi: 10.1084/jem.179.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michaëlsson E, Malmström V, Reis S, Engström Å, Burkhardt H, Holmdahl R. J Exp Med. 1994;180:745–749. doi: 10.1084/jem.180.2.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmdahl R, Carlsen S, Mikulowska A, Vestberg M, Brunsberg U, Hansson A-S, Sundvall M, Jansson L, Pettersson U. In: Human Genome Methods. Adolpho K, editor. New York: CRC Press; 1998. pp. 215–238. [Google Scholar]
- 25.Holmdahl R, Jonsson R, Larsson P, Klareskog L. Lab Invest. 1988;58:53–60. [PubMed] [Google Scholar]
- 26.Marshall K, Wilson K J, Liang J, Woods A, Zaller D, Rothbard J B. J Immunol. 1995;154:5927–5933. [PubMed] [Google Scholar]
- 27.Rammensee H-G, Friede T, Stevanovic S. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 28.Patel S D, Cope A P, Congia M, Chen T T, Kim E, Fugger L, Wherrett D, McDevitt G. Proc Natl Acad Sci USA. 1997;94:8082–8087. doi: 10.1073/pnas.94.15.8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garboczi D N, Ghosh P, Utz U, Fan Q R, Biddison W E, Wiley D C. Nature. 1996;384:134–141. [PubMed] [Google Scholar]
- 30.Holmdahl R, Andersson M, Goldschmidt T J, Gustafsson K, Jansson L, Mo J A. Immunol Rev. 1990;118:193–232. doi: 10.1111/j.1600-065x.1990.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 31.Cope A, Fugger L, Chu W, Sønderstrup-McDevitt G. In: Functional and Medical Implication. Charron D, editor. Paris: EDK; 1997. pp. 634–637. [Google Scholar]
- 32.Rosloniec E F, Brand D D, Myers L K, Whittington K B, Gumanovskaya M, Zaller D M, Woods A, Altmann D M, Stuart J M, Kang A H. J Exp Med. 1997;185:1113–1122. doi: 10.1084/jem.185.6.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt D, Verdaguer J, Averill N, Santamaria P. J Exp Med. 1997;186:1059–1075. doi: 10.1084/jem.186.7.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zanelli E, Gonzales-Gay M A, David C S. Immunol Today. 1995;16:274–278. doi: 10.1016/0167-5699(95)80181-2. [DOI] [PubMed] [Google Scholar]
- 35.Nabozny G H, Baisch J, Cheng S, Cosgrove D, Griffiths M M, Luthra H S, David C S. J Exp Med. 1996;183:27–37. doi: 10.1084/jem.183.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zanelli E, Krco C J, Baisch J M, Cheng S, David C S. Proc Natl Acad Sci USA. 1996;93:1814–1819. doi: 10.1073/pnas.93.5.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fugger L, Svejgaard A. Tissue Antigens. 1997;50:494–500. doi: 10.1111/j.1399-0039.1997.tb02905.x. [DOI] [PubMed] [Google Scholar]