Abstract
UK-1 is a bis(benzoxazole) natural product displaying activity against a wide range of human cancer cell lines. A simplified analog of UK-1, 4-carbomethoxy-2-(2’-hydroxyphenyl)benzoxazole, was previously found to be almost as active as UK-1 against cancer cell lines, and similar to the natural product, formed complexes with variety of metal ions such as Mg2+ and Zn2+. A series 4-substituted-2-(2’-hydroxyphenyl)benzoxazole analogs of this “minimal pharmacophore” of UK-1 were prepared. The anti-cancer activity of these analogs was examined in breast and lung cancer cell lines. Spectrophotometric titrations in methanol were carried out in order to assess the ability of UK-1 and these analogs to coordinate with Mg2+ and Cu2+ ions. Although none of the new analogs were more cytotoxic than 4-carbomethoxy-2-(2’-hydroxyphenyl)benzoxazole, some analogs were identified that display similar cytotoxicity to this simplified UK-1 analog with improved water solubility. UK-1 and all of these new analogs bind Cu2+ ions better than Mg2+ ions, and the nature of the 4-substituent is important for the Mg2+ ion binding ability of these 2-(2’-hydroxyphenyl)benzoxazoles. Previous studies of a limited number of UK-1 analogs demonstrated a correlation between Mg2+ ion binding ability and cytotoxicity; however, within this series of 4-substituted-2-(2’-hydroxyphenyl)benzoxazoles the variations in cytotoxicity do not correlate with either Mg2+ or Cu2+ ion binding ability. These results, together with recent ESI-MS studies of Cu2+-mediated DNA binding by UK-1 and analogs, indicate that UK-1 and analogs may exert their cytotoxic effects by interaction with Cu2+ or other transition metal ions, rather than Mg2+, and that metal ion-mediated DNA binding, rather than metal ion binding affinity, is important for the cytotoxic effect of these compounds. The potential role of Cu2+ ions in the cytotoxic action of UK-1 is further supported by observation that UK-1 in the presence of Cu2+ displays enhanced cytotoxicity to MCF-7 and A549 cells when compared to UK-1 alone.
Four key words: Antibacterial, Metal Ion Binding, Cancer Cell Cytotoxicity, DNA Binding
INTRODUCTION
The bis(benzoxazole) natural product UK-1 (Figure 1) is one of a growing number of structurally related secondary metabolites with interesting biological activity [1–3]. UK-1 displays a wide spectrum of potent anticancer activity against leukemia, lymphoma, and certain solid tumor-derived cell lines; however, this anticancer natural product does not show antibacterial or antifungal activity [1,4]. Recently, synthetic studies of analogs of UK-1 have been undertaken [4–8], and simplified analogs, such as the benzoxaole 1 (Figure 1) have been identified that possess similar, selective cancer cell cytotoxicity to that of UK-1 [6–8]. Studies of UK-1 and these synthetic analogs have implicated metal ion complexation as an important factor in the activity of these agents [6–10]. UK-1 binds a variety of di- and trivalent metal ions, and binds to double-stranded DNA better in the presence of metal ions than in their absence [6,9,10]. Active anticancer analogs of UK-1, such as 1, also display this affinity for metal ions and metal-mediated DNA binding ability [6,10]. In contrast, analogs such as 2 neither bind DNA in a metal ion-mediated fashion nor display cancer cell cytotoxicity [5,6]. However, the exact role of metal ion coordination in the mechanism of action of UK-1 and analogs remains to be determined. There is a growing interest in the role of metal ions and their complexes in drug design [11–14], particularly in metal complex–DNA interactions [15–21]. Studies of UK-1 and its analogs may offer important insights, provided by nature, into how metal ion complexation can be harnessed in the design of selective cytotoxins.
Figure 1.
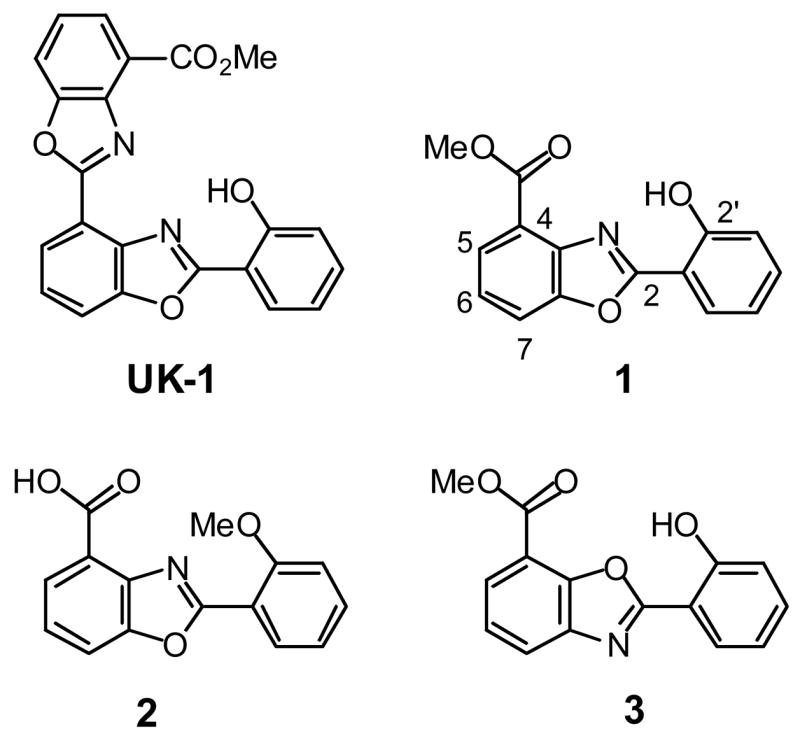
Structure of UK-1 and 2-(2’-hydroxylphenyl)benzoxazole analogs.
The current work focuses on the synthesis and biological evaluation of new benzoxazole analogs of 1. These analogs were modified at the C4 position, retaining the 2-(2’-hydrophenyl)benzoxazole functionality of UK-1 and 1 that is thought to be important for chelating metal ions. The cytotoxicity and metal ion binding ability of these benzoxazoles were determined. The results indicate that metal ion binding per se is not a predictor of cytoxicity in this series. However, the ability of these compounds to undergo metal ion-mediated DNA binding may be related to their cytotoxicity, particularly in the case of Cu2+ or other transition metals.
RESULTS AND DISCUSSION
Analog Design and Synthesis
It was previously determined that compound 1 was the minimum pharmacophore structure that retained the selective cytotoxicity of UK-1 [6]. The key structural elements that affect metal binding and cytotoxicity were addressed by comparing 1 with its 7-substituted benzoxazole analog 3 (Figure 1). Compound 3 differs with 1 solely in the disposition of the benzoxazole N and O atoms, but it is less cytotoxic than 1 [10]. In addition, the complexation of Mg2+ by 1 and 3, as determined by 1H NMR and ESI-mass spectra were quite distinct [10]. There was also minimal DNA-metal mediated binding of 3 compared to 1 observed by ESI-MS [10]. This data highlighted the importance of the arrangement between the phenolic hydroxyl group, the benzoxazole nitrogen atom and the ester for metal coordination, DNA binding, and cytotoxicity. A similar observation was made by Wang and co-workers by comparing a different series of UK-1 analogs and their inhibition of topoisomerase II [7]. Their research suggested that the analogs required a structural motif comprised of an isosceles triangular arrangement between the oxygen atom of carbonyl group, the heterocyclic nitrogen atoms, and the phenolic hydroxyl for enzyme inhibition.
Keeping these considerations in mind, a new series of compounds that explore the influence on metal ion binding and cytotoxicity of different 4-substitutents of 1 while retaining the benzoxazole core and 2-(2’-hydroxypenyl) substituent were prepared. In particular, the effect of homologation of the 4-methyl ester and its replacement with an amide were addressed. Given the evidence that metal ion complexation is important for cytotoxicity in these UK-1 derivatives, additional potential metal ion coordination sites were also introduced in some of these amide and ester analogs.
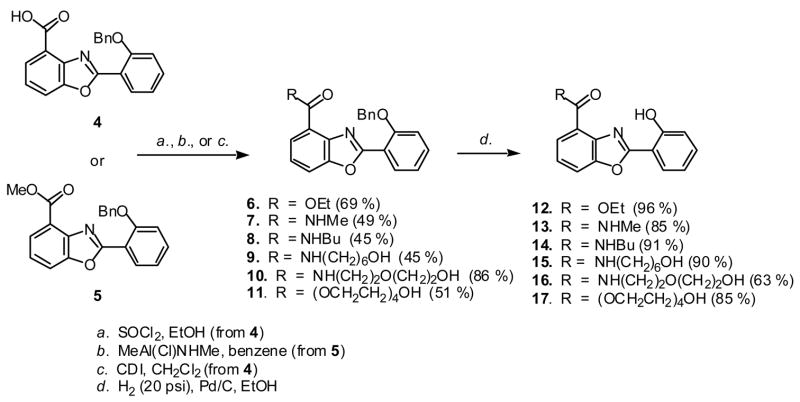
The synthesis of these analogs was accomplished through straightforward adaptation of the previously reported synthesis of UK-1 and 1 [5,6]. The key intermediate, benzyl-protected carboxylic acid 4 was esterified (6,11) or coupled with a primary amine (8–10) (Scheme 1). The methylamide 7 was prepared from the methyl ester 5 (Scheme 1). The resulting esters and amides were subjected to hydrogenolysis to remove the benzyl phenol protecting group to afford the 2-(2’-hydroxyphenyl)benzoxazoles 12-17 in 63–91 % yield.
Scheme 1.
Synthesis of 2-(2’-hydroxyphenyl)benzoxazole analogs of UK-1.
In addition to providing further sites for metal ion coordination, the incorporation of polar groups at the C4 position in some of the compounds of this series also favorably impacted their solubility in aqueous buffers. As previously reported, UK-1 [1] and compound 1 [6] have low aqueous solubilities. Compounds 15–17 have increased water solubility, relative to 1. Compound 17 with a tetraethyleneglycol side chain was the most soluble in this series, and the only one which remained in solution in aqueous buffers at concentrations up to 100 μM.
Cytotoxicity
The cytotoxicity of the new 2-(2’-hydroxyphenyl)benzoxazole analogs 12–17 was tested using the AlamarBlue assay in the breast cancer cell line MCF-7 and the lung cancer cell line A549. For comparison, UK-1, the isomeric 2-(2’-hydroxphenyl)benzoxazole 3 and the benzoxazole carboxylic acid 2 (Figure 1) were also tested. As expected based on previous reports [6], the anticancer activity of compound 1 is somewhat less than that of UK-1, whereas the benzoxazole carboxylic acid 2 is not cytotoxic at the highest concentration tested (50 μM). Benzoxazole 3, which is regioisomeric to 1, is less cytotoxic than 1, particularly against the A549 cell line. Similar results had been observed previously for 3 in the PC-3 and HT-29 cell lines, in which compound it is several fold less cytotoxic than 1 [10].
Although none of the new benzoxazoles reported here are superior to UK-1 or to 1 in their cytotoxicity to these cancer cell lines, there are subtle variations in activity that can be understood in structural terms. Replacing the methyl ester group of 1 with a methyl amide, as in 13, leads to comparable cytotoxicity. However, as the size of the amide group increases from the N-butyl analog 14 to the N-(6-hydroxyhexyl) analog 15 to the N-(hydroxyethoxyethyl) analog 16, the cytotoxicity decreases. While it is also true that the ethyl ester 12 is less active than the methyl ester 1, the tetraethyleneglycol ester 17 has a similar cytotoxic profile to 1, retaining activity against both cell lines.
Metal Ion Binding
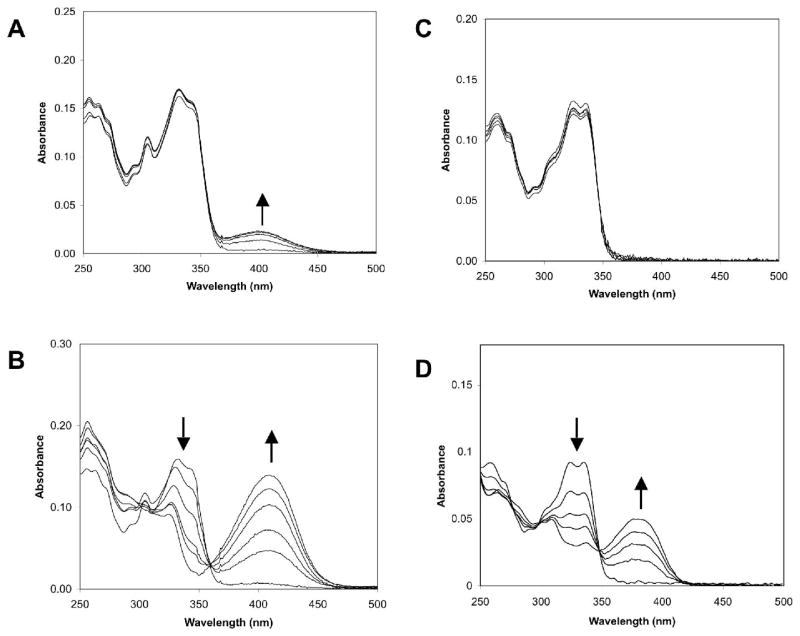
Because metal binding has been correlated with cytotoxicity in other benzoxazole compounds [4,6,9,10], the affinity of the new benzoxazole analogs 12–17 for various metal ions was studied. Solutions of these compounds in methanol change from colorless to yellow upon the addition of divalent cations such as Mg2+, Zn2+, Ni2+, and Cu2+, indicating complexation between the analogs and the metal ions. The most pronounced change occurs in the presence of copper ions. Therefore, the metal complex formation with both the physiologically abundant magnesium ions as well copper ions for all of the analogs was monitored by UV/VIS spectral changes upon addition of increasing concentrations of the metal ions salts in methanol. A marked hyperchromycity in the 350–450 nm region was observed upon addition of the metal ions, concomitant with a hypochromicity in the 315–350 region. A clear isosbestic point was present at approximately 350 nm, with slight variation of the wavelength of maximum absorbance depending on the analog studied. Representative data for compounds 1 and 3 for magnesium and copper binding are shown in Figure 2.
Figure 2.
Absorbance spectra of compounds 1 (A, B) and 3 (C, D) in the presence of increasing concentrations of Mg2+ (A, C) or Cu2+ (B, D). Arrows denote the changes in absorbance with increasing metal ion concentrations.
The change in absorbance of the peak in the 410 nm region was plotted against metal ion concentration to obtain binding isotherms, from which the Mg2+ and Cu2+ association constants of all of the analogs were determined and are summarized in Table 2. UK-1, 1, and all of the other 4-substituted benzoxazole analogs 12–17 bind copper with high affinity as 1:1 complexes. The binding curves for these compounds displayed saturation at all concentrations of added Cu2+, so only lower limits to the association constants could be determined. In contrast to the 4-substituted benzoxazoles, the 7-substituted benzoxazole 3 is a relatively poor ligand for Cu2+. Compound 3 is the only substituted 2-(2’-hydroxyphenyl)-benzoxazole that does not appear to bind Mg2+ by these spectrophotometric titrations; although, a weak, alternative Mg2+ binding mode for this compound has been proposed based on 1H NMR and ESI-MS studies [10]. The methyl phenyl ether 2 does not form complexes with either Mg2+ or Cu2+. In contrast to the Cu2+ binding ability of the 4-substituted benzoxazoles UK-1, 1, and 12–17, which is high regardless of changes in the 4-substituent, the Mg2+ binding ability of these compounds is dependent on the nature of the 4-substituent. The analogs with amide substituents (e.g., 12–14) do not bind Mg2+ as well as those analogs with ester substituents. As a result, these amides, particularly 13, are more selective ligands for Cu2+ versus Mg2+. The critical role played by the benzoxazole 4-substituent in metal ion recognition is also demonstrated by a comparison of these substituted benzoxazoles with 2-(2’-hydroxypehenyl)benzoxazole (HPBO). Under these conditions, HPBO does not bind Mg2+ (data not shown). HPBO does complex Cu2+ [22], however; the association constant for Cu2+ under these conditions (4±1 x 105 M−1) is very similar to that of the 7-substituted benzoxazole 3 (3.8 x 104 M−1) and significantly lower than any of the 4-substituted benzoxazoles (Table 2).
Table 2.
Association constants for Mg+2 and Cu+2 binding in MeOH (M-1)
| Compound | Mg2+ | Cu2+ |
|---|---|---|
| UK-1 | 5±1 x 105 | > 5 x 107 |
| 1 | 7±3 x 105 | > 4 x 107 |
| 2 | No binding | No binding |
| 3 | No binding | 3.8±0.4 x 104 |
| 12 | 1.5±0.4 x 105 | > 3 x 107 |
| 13 | 6±2 x 104 | > 3 x 107 |
| 14 | 1.1±0.2 x 104 | > 6 x 107 |
| 15 | 2.4±0.4 x 104 | > 2 x 107 |
| 16 | 3±1 x 105 | > 7 x 107 |
| 17 | 8±3 x 105 | > 7 x 107 |
The Role of Metal Ion Coordination
The phenol ether 2 does not bind Mg2+ or Cu2+ and is not cytotoxic to either of the two cancer cell lines examined here. However, within the series of 2-(2’-hydroxypehnyl)benzoxazoles, there is no correlation between either Mg2+ or Cu2+ ion binding affinity and cytotoxicity. Although 3 is the weakest ligand for Mg2+ and Cu2+ ions in the 2-(2’-hydroxyphenyl)benzoxazole series, it is cytotoxic, although somewhat less so than 1. The tetraethylene glycol ester 17 binds both Mg2+ and Cu2+ ions better than UK-1 or 1, yet it is not more cytotoxic than these compounds.
While metal ion binding affinity per se does not predict cytotoxicity within this series of 2-(2’-hydroxyphenyl)benzoxazoles, the ability of these compounds to bind to DNA in the presence of metal ions is correlated with cytotoxicity. The cytotoxic analogs 1, 12–14, and 17 all demonstrate enhanced DNA binding by ESI-MS in the presence of Cu2+ or Ni2+; whereas, the relatively non-cytotoxic analogs 15 and 16 do not [23]. Even the 7-substituted benzoxazole 3 forms ligand-metal ion-DNA complexes, although these complexes are formed in much lower abundance when compared to those formed in the presence of 1 [10]. Thus, despite its greatly diminished metal ion binding ability, 3 retains some ability to undergo metal ion-mediated DNA binding and is also moderately cytotoxic, compared to 1.
These studies cast doubt on the role of magnesium binding in the cytotoxicity of these 2-(2’-hydroyphenyl)benzoxazoles. In methanol, 7-substituted benzoxazole 3 does not bind Mg2+ ions, yet this compound is moderately cytotoxic. Despite the pronounced effect of 4-position substituents on Mg2+ ion binding affinity in the 2-(2’-hydroxyphenyl)benzoxazole series, there are only relatively subtle variations in cytotoxicity. Our recent ESI-MS investigation of the metal ion binding of these compounds failed to demonstrate formation of Mg2+ complexes in aqueous buffers, although complexes were observed in methanol [23]. These and previous ESI-MS studies [10] failed to demonstrate formation of ligand-Mg2+-DNA complexes with UK-1 or these analogs, although ligand-Cu2+-DNA complexes were readily observed. ESI-MS also confirmed the formation of 1:1 and in some cases 2:1 complexes between UK-1 or these benzoxazoles and Cu2+ in aqueous solution.
Increased Cytotoxicity of UK-1 in the presence of Copper
The cytotoxicity of UK-1 was directly compared with that of UK-1 in the presence of added CuCl2 in both MCF-7 and A549 cells (Table 3). In both cell lines, the addition of 10 μM CuCl2 did not affect the growth rate (data not shown). The combination of CuCl2 and UK-1 displays enhanced cytotoxicity relative to UK-1 alone. This enhanced cytotoxicity is most pronounced after 24 h incubation the case of MCF-7 cells. In contrast, in the case of A549 cells, the enhanced cytotoxicity of UK-1 plus CuCl2 relative to UK-1 alone is more pronounced after 72 h exposure than 24 h exposure.
Table 3.
Cytotoxicity of UK-1 Alone and in the Presence of Cu2+.
| Compound | IC50 (μM) MCF-7Cells | IC50 (μM) A549 Cells | ||
|---|---|---|---|---|
| 24 h | 72 h | 24 h | 72 h | |
| UK-1 | 31 ± 5 | 1.7 ± 0.2 | 17 ± 2 | 1.0 ± 0.1 |
| UK-1 + 10 μM CuCl2 | 4.6 ± 0.6 | 1.2 ± 0.2 | 4.5 ± 0.5 | 0.2 ± 0.04 |
| Mitomycin-C | 5 ± 2 | 1.2 ± 0.2 | 4.5 μM | 0.24 ± 0.03 |
The enhanced cytotoxicity of the UK-1 in the presence of copper supports a role for the copper complex in the mode of action of UK-1. The formation of the copper complex of UK-1 in cells may be limited by the relatively low availability of free Cu2+ ions. Addition of Cu2+ ions increases the effectiveness of UK-1 by increasing the amount of the complex available to the cellular target.
CONCLUSIONS
This study has shown that it is possible to modify the minimalized UK-1 analog 4-carbomethoxy-2-(2’-hydroxyphenyl)benzoxazole (1) at the C4 position with certain groups and still retain activity against cancer cell lines. In particular, changing the methyl ester of 1 to a small amide group, as in 13, retains anticancer activity, but as the length of the amide N-substituent increases, cytotoxicity decreases. This trend of decreasing cytotoxicity with increasing chain length is also seen with the ethyl ester 12; however, the incorporation of additional metal ion coordination sites in the side chain, as in the tetraethyleneglycol ester 17, can recapture this lost activity while also increasing water solubility.
The Mg2+ and Cu2+ ion binding ability of these UK-1 analogs were determined spectrophotometrically in methanol. While the Cu2+ ion binding affinity of these 4-substituted benzoxazole analogs is relatively unaffected by the nature of the 4-position substituents examined in this study, the Mg2+ ion binding affinities vary with the nature of this substituent. Both Mg2+ and Cu2+ ion binding ability are greatly diminished in the regioisomeric 7-substituted benzoxazole 3. All of these 2-(2’-hydroxyphenyl)benzoxazoles, including UK-1, bind to copper with greater affinity than magnesium, but the variations in cytotoxicity for these benzoxazoles do not correlate with either Mg2+ or Cu2+ ion binding ability. Recent ESI-MS studies have shown that UK-1 does not form Mg2+ ion complexes in water, but that UK-1 and analogs can form complexes with Cu2+ and undergo Cu2+-mediated DNA binding. Taken together, these results indicate that UK-1 and analogs may exert their cytotoxic effects by interaction with Cu2+ or other transition metal ions, rather than Mg2+, and that metal ion-mediated DNA binding, rather than metal ion binding affinity, is important for the cytotoxic effect of these compounds. The potential role of Cu2+ ions in the cytotoxic action of UK-1 is further supported by observation that UK-1 in the presence of CuCl2 displays enhanced cytotoxicity to MCF-7 and A549 cells when compared to UK-1 alone.
EXPERIMENTAL
General
Compounds 1, 2, 3, 4, 5 and UK-1 were synthesized as described previously [5,6,10]. AlamarBlue reagent was purchased from Biosource and mitomycin-C (MMC) was purchased from MP Biomedicals. Cell viability assays were carried out in sterile black polystyrene 96-well plates from Nunc. Spectroscopic metal ion binding studies were carried out in HPLC grade MeOH. The MCF-7 and A549 cell lines were kind gifts from Profs. Shawn Bratton and Bob Krug (University of Texas at Austin) respectively, from ATCC Biology Products. The media for the MCF7 cell line was the improved MEM (Richter’s Modification) with L-glutamine, supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (from MediaTech). The A549 cells were cultured in Ham’s F-12 media (Kaighn’s Modification) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. All other reagents were purchased from major commercial sources. Unless otherwise noted, 1H and 13C NMR spectra were determined in CDCl3 on a spectrometer operating at 300 and 75.5 MHz, respectively. All mass spectra were obtained by chemical ionization using methane as the ionizing gas. Chromatography refers to flash chromatography on silica gel, and Rf values were determined using silica gel-GF TLC plates (Merck) using the solvent system indicated.
2-(2’-Benzyloxyphenyl)-benzoxazole-4-carboxylic acid ethyl ester (6)
2-(2’-Benzyloxyphenyl)-benzoxazole-4-carboxylic acid, (4, 142 mg, 0.41 mmol) was dissolved in absolute EtOH (1.25 mL) in a flask fitted with a drying tube. Thionyl chloride (45 μl, 0.62 mmol) was then added drop-wise while stirring, and the reaction was left stirring overnight at room temperature. The reaction mixture was poured over ice, extracted with CH2Cl2, washed with sodium bicarbonate, 1% HCl, water, dried over anhydrous Na2SO4, and filtered. The product was then concentrated by roto-evaporation, and purified by column chromatography (20% EtOAc in hexanes) to give a white solid (103 mg, 69 % yield): m.p. 112-114 °C; Rf 0.30 (20% EtOAc in hexanes); 1H NMR 1.44 (t, 3H, J = 7.2 Hz), 4.49 (q, 2H, J = 7.2Hz), 5.27 (s, 2H), 7.07-7.12 (m, 2H), 7.27-7.49 (m, 5H), 7.63 (d, 2H, J = 7.8 Hz), 7.71 (dd, 1H, J = 8.4, 1.0 Hz), 8.00 (dd, 1H, J = 7.5, 1.2 Hz), 8.28 (dd, 1H, J = 8.1, 1.8 Hz); 13C NMR δ 14.30, 61.07, 70.47, 113.62, 114.38, 116.28, 120.94, 122.31, 123.95, 126.40, 126.69, 127.57, 128.37, 131.91, 133.05, 136.60, 141.49, 151.22, 157.59, 163.61, 165.12; CIMS m/z 374 (MH+); HRMS m/z calc for C23H20NO4: 374.1392, found 374.1389.
2-(2’-Benzyloxy-phenyl)-benzoxazole-4-carboxylic acid methylamide (7)
Methylaluminumchloride-methylamide [24] was prepared from MeNH2·HCl (675 mg, 10 mmol) and Me3Al (2 M solution in hexanes, 5 mL) in 10 mL dry benzene. Compound 5, (100 mg, 0.28 mmol) was dissolved in 2.8 mL dry benzene. To this solution was added 830 μL of the aluminum amide solution (0.67 M, 0.56 mmol), and the mixture heated under reflux overnight. The reaction mixture was then cooled to room temperature and quenched with 5% HCl. The organic layer was separated and the aqueous layer was extracted three times with EtOAc. The organic layers were combined, dried over MgSO4, and concentrated by rotory-evaporation. The product was re-crystallized from EtOAc/hexanes to give light pink needles (49 mg, 49 % yield): mp. 158.5-160 °C; Rf 0.74 (10% MeOH in CH2Cl2); 1H NMRδ 2.82 (d, 3H, J = 4.8 Hz), 5.24 (s, 2H), 7.11-7.17 (m, 2H), 7.32-7.44 (m, 4H), 7.50-7.55 (m, 3H), 7.65 (m, 1H, J = 8.0, 0.8 Hz), 8.16-8.19 (m, 2H), 8.90 (s, 1H); 13C NMR δ 26.20, 70.64, 113.15, 113.56, 115.50, 121.03, 123.89, 124.69, 125.29, 127.03, 128.00, 128.56, 131.29, 133.40, 136.29, 139.47, 150.01, 157.77, 161.84, 164.92; CIMS m/z 359 (MH+); HRMS m/z calc for C22H19N2O3: 359.1396, found 359.1398.
General Procedure for CDI Coupling: 2-(2’-Benzyloxy-phenyl)-benzoxazole-4-carboxylic acid butylamide (8)
The benzoxazole acid imidazolide was prepared by dissolving carboxylic acid 4 (306 mg, 0.89 mmol) in dry CH2Cl2, approximately 6 mL/g, and adding portion-wise 1.3 equivalents of carbonyldiimidazole (CDI) with stirring for 1 hour at room temperature, until the evolution of CO2 ceased. Butylamine (215 mg, 3 mmol) was added to the reaction mixture, which was heated under reflux for 2 days. The reaction mixture was diluted with CH2Cl2 and washed with H2O (x2), 0.1N HCl (x2), H2O (x2), 0.1N NaOH, and H2O (x2). The organic layer was dried over anhydrous Na2SO4, and the solvent was evaporated. Compound 8 was obtained after column chromatography as white needle-like crystals (157 mg, 45 % yield): mp. 122-123°C; Rf0.22 (20% EtOAc in hexanes); 1H NMR δ 0.89 (t, 3H, J = 7.2 Hz), 1.34-1.42 (m, 2H), 1.49-1.57 (m, 2H), 3.42, (q, 2H, J = 6.4 Hz), 5.29 (s, 2H), 7.11-7.15 (m, 2H), 7.29-7.52 (m, 7H), 7.66 (dd, 1H, J = 8.0, 0.8 Hz), 8.16-8.20 (m, 2H), 9.16 (m, 1H); 13C NMR δ13.68, 20.16, 31.53, 39.31, 70.50, 113.11, 113.76, 115.72, 121.03, 124.08, 124.64, 125.35, 126.67, 127.86, 128.48, 131.36, 133.33, 136.38, 139.35, 150.21, 157.67, 162.11, 164.16; CIMS m/z 401 (MH+); HRMS m/z calc for C25H24N2O3: 401.1865, found 401.1870.
2-(2’-Benzyloxyphenyl)-benzoxazole-4-carboxylic acid (6-hydroxyhexyl)amide (9)
Following the general procedure described above, compound 3 (232 mg, 0.67 mmol) was coupled with 262 mg of aminohexanol (2.24 mmol). Compound 9 was obtained after column chromatography as white crystals (135 mg, 45 % yield): mp. 139-141°C; Rf 0.54 (10% MeOH in CH2Cl2); 1H NMR δ 1.35 (m, 4H), 1.52 (m, 4H), 2.47 (s, 1H), 3.38, (q, 2H, J = 6.6 Hz), 3.57, (t, 2H, J = 6.6 Hz), 5.22 (s, 2H), 7.09 (m, 2H), 7.26-7.50 (m, 7H), 7.60 (d, 1H, J = 7.5 Hz), 8.14 (t, 2H, J = 7.4 Hz), 9.16 (m, 1H); 13C NMR δ 25.19, 26.58, 29.37, 32.47, 39.34, 62.37, 70.45, 113.17, 113.70, 115.56, 120.99, 123.82, 124.62, 125.29, 126.64, 127.80, 128.44, 131.29, 133.35, 136.33, 139.28, 150.14, 157.60, 162.12, 164.27; CIMS m/z 445 (MH+); HRMS m/z calc for C27H28N2O4: 445.2127, found:445.2126
2-(2’-Benzyloxy-phenyl)-benzoxazole-4-carboxylic acid [2-(2’-hydroxy-ethoxy)-ethyl]-amide (10)
Following the general procedure described above, compound 3 (112 mg, 0.32 mmol) was coupled with 113 mg of 2-(2-aminoethoxy)ethanol (1.08 mmol). Compound 10 was obtained after chromatography a white solid (120 mg, 86 % yield): mp. 91-94 °C; Rf 0.68 (10% MeOH in CH2Cl2); 1H NMR δ 2.2 (s(br), 1H), 3.54-3.67 (m, 8H), 5.28 (s, 2H), 7.12-7.15 (m, 2H), 7.29-7.53 (m, 7H), 7.65-7.68 (m, 1H, J =8.0, 0.8Hz), 8.16-8.20 (m, 2H), 9.42 (m, 1H); 13C NMR δ 39.22, 61.69, 69.86, 70.77, 72.08, 113.47, 114.01, 115.80, 121.21, 123.80, 124.76, 125.53, 126.92, 127.96, 128.55, 131.56, 133.54, 136.45, 139.42, 150.38, 157.72, 162.49, 164.56; CIMS m/z 433 (MH+); HRMS m/z calc for C25H24N2O5: 433.1763, found: 433.1747.
2-(2’-Benzyloxyphenyl)-benzoxazole-4-carboxylic acid 2-{2-[2-(2’-hydroxy-ethoxy)-ethoxy]-ethoxy}-ethyl ester (11)
Following the general procedure described above, compound 3 (112 mg, 0.32 mmol) was coupled with 210 mg of tetraethyleneglycol (1.08 mmol). Compound 11 was obtained after column chromatography as a transparent oil (86 mg, 51% yield): Rf 0.76 (10% MeOH in CH2Cl2); 1H NMR δ 2.6 (s(br), 1H), 3.52-3.54 (m, 2H), 3.58-3.70 (m, 8H), 3.72-3.74 (m, 2H), 3.87 (t, 2H, J = 4.8 Hz), 4.58 (t, 2H, J = 4.8 Hz), 5.27 (s, 2H), 7.06-7.11 (m, 2H), 7.28-7.48 (m, 5H), 7.61 (d, 2H, J = 7.2 Hz), 7.71 (dd, 1H, J = 8.0, 0.8 Hz), 8.01 (dd, 1H, J = 7.6, 0.8 Hz), 8.27 (dd, 1H, J = 8.2, 1.8 Hz); 13C NMR δ 61.54, 64.18, 69.08, 70.16, 70.44, 70.48, 70.58, 72.37, 113.60, 114.61, 116.23, 120.95, 121.80, 123.96, 126.57, 126.68, 127.59, 128.40, 131.97, 133.12, 136.57, 141.57, 151.27, 157.59, 163.78, 164.86; CIMS m/z 522 (MH+); HRMS m/z calc for C29H31NO8: 521.2050, found: 521.2036.
General Procedure for Hydrogenolysis: 2-(2’-Hydroxyphenyl)-benzoxazole-4-carboxylic acid ethyl ester (12)
The benzyl ether 6 (103 mg, 0.28 mmol) compounds was dissolved in absolute EtOH (4 mL/mmol), and an equal weight of 10% palladium on activated carbon was added. The reaction mixture was placed under a balloon of hydrogen for approximately 2 h. After verifying by TLC that the reaction had gone to completion, the solution was filtered through Celite and washed with EtOAc and CH2Cl2. The solution was then evaporated under reduced pressure conditions, and the compound purified by column chromatography (10% EtOAc in hexanes) to obtain compound 12 as white crystals (75 mg, 96% yield): mp. 116-118 °C; Rf 0.56 (20% EtOAc in hexanes); 1H NMR (400 MHz) δ 1.51 (t, 3H, J = 7.2 Hz), 4.46 (q, 2H, J = 7.2Hz), 7.00 (ddd, 1H, J = 8.0, 7.4, 1.2 Hz), 7.12 (dd, 1H, J = 8.4, 0.8 Hz), 7.41-7.46 (m, 2H), 7.70 (dd, 1H, J = 8.4, 1.0 Hz), 8.01 (dd, 1H, J = 7.8, 1.4 Hz), 8.06 (dd, 1H, J = 7.8, 2.0 Hz), 11.80 (s(br), 1H); 13C NMR δ 14.23, 61.34, 109.82, 114.66, 117.51, 119.43, 121.39, 124.59, 127.02, 127.24, 134.05, 139.20, 149.57, 159.30, 164.06, 165.03; CIMS m/z 284 (MH+); HRMS m/z calc for C16H14NO4: 284.0923, found: 284.0926; EA calc for C16H13NO4: C, 67.84; H, 4.63; N, 4.94; found: C, 67.63; H, 4.52; N, 4.91.
2-(2’-Hydroxyphenyl)-benzoxazole-4-carboxylic acid methylamide (13)
Following the general procedure for hydrogenolysis, compound 7 (44 mg, 0.12 mmol) was de-protected to give compound 13, which was purified by column chromatography (10% MeOH inCH2Cl2), and re-crystallized from MeOH in CH2Cl2 to afford white crystals (27.87 mg, 85% yield): mp. 225.5-226.5 °C; Rf 0.74 (10% MeOH in hexanes); 1H NMR δ 3.14 (d, 3H, J = 7.2 Hz), 7.06 (ddd, 1H, J = 8.0, 7.2, 1.0 Hz), 7.13 (dd, 1H, J = 8.7, 0.9 Hz), 7.46-7.53 (m, 2H), 7.73 (dd, 1H, J = 7.8, 0.9 Hz), 7.79 (s(br), 1H), 8.06 (dd, 1H, J = 8.1, 1.5 Hz), 8.20 (dd, 1H, J = 7.5, 0.9 Hz), 10.45 (s(br), 1H); 13C NMR δ 26.69, 109.89, 113.55, 117.45, 120.25,123.96, 125.48, 126.48, 127.75, 134.57, 137.18, 148.95, 158.27, 163.39, 164.54 CIMS m/z 269 (MH+); HRMS m/z calc for C15H13N2O3: 269.0926, found 269.0926; EA calc for C15H12N2O3+(H2O)1/2: C, 66.52; H, 4.57; N, 10.34; found: C, 66.88, H, 4.42, N, 9.96.
2-(2’-Hydroxyphenyl)-benzoxazole-4-carboxylic acid butylamide (14)
Following the general procedure for hydrogenolysis, compound 8 (150 mg, 0.37 mmol) was de-protected to give compound 14, which was recrystallized from EtOAc/hexanes to give white-flake like crystals (105.9 mg, 91% yield): mp. 181-183 °C; Rf 0.22 (20% EtOAc in hexanes); 1H NMR δ 0.99 (t, 3H, J = 7.2 Hz), 1.45-1.52 (m, 2H), 1.66-1.72 (m, 2H), 3.58 (q, 2H, J = 7.2 Hz), 7.04 (t. 1H, J = 8.4 Hz), 7.12 (d, 1H, J = 8 Hz), 7.42-7.46 (m, 2H), 7.72 (d, 1H, J = 8.4 Hz), 7.84 (s(br), 1H), 8.04 (dd, 1H, J = 8.2, 1.4 Hz), 8.15 (d, 1H, J = 8.1 Hz), 10.37 (s, 1H); 13C NMR δ 13.72, 20.23, 31.53, 39.60, 109.81, 113.41, 117.43, 120.14, 124.08, 125.42, 126.40, 127.65, 134.50, 137.15, 148.88, 158.33, 163.31, 163,70; CIMS m/z 311 (MH+); HRMS m/z calc for C18H19N2O3: 311.1396, found: 311.1397; EA calc for C18H18N2O3+(H2O)1/4: C, 68.69; H, 5.92; N, 8.90; found: C, 68.68; H, 5.73; N, 8.69.
2-(2’-Hydroxyphenyl)-benzoxazole-4-carboxylic acid (6-hydroxyhexyl)amide (15)
Following the general procedure for hydrogenolysis, compound 9 (120 mg, 0.27 mmol) was de-protected to give compound 15, which was purified by column chromatography (10% MeOH in CH2Cl2), to afford an off-white solid (87 mg, 90% yield): mp. 145.5-147 °C; Rf 0.54 (10% MeOH in CH2Cl2); 1H NMR (400 MHz) δ 1.47-1.49 (m, 4H), 1.59-1.62 (m, 2H), 1.70-1.74 (m, 2H), 1.82 (s(br), 1H), 3.58 (dt, 2H, J = 6.8, 6.0 Hz), 3.64 (t, 2H, J = 6.4 Hz), 7.03 (t, 1H, J = 7.4 Hz), 7.10 (d, 1H, J = 8.4 Hz), 7.44-7.48 (m, 2H), 7.70 (d, 1H, J = 8.0 Hz), 7.83 (m, 1H), 8.03 (dd, 1H, J = 8.0, 1.2 Hz), 8.16 (d, 1H, J = 7.6 Hz), 10.44 (s, 1H); 13C NMR δ 25.30, 26.72, 29.55, 32.54, 39.71, 62.62, 109.89, 113.53, 117.46, 120.27, 124.04, 125.51, 126.48, 127.74, 134.60, 137.20, 148.95, 158.30, 163.40, 163.84; CIMS m/z 355 (MH+); HRMS m/z calc for C20H23N2O4: 355.1658, found: 355.1660; EA calc for C20H22N2O4: C, 67.78; H, 6.26; N, 7.90; found: C, 67.64; H, 6.33; N, 7.48.
2-(2’-Hydroxyphenyl)-benzoxazole-4-carboxylic acid [2-(2’-hydroxy-ethoxy)-ethyl]-amide (16)
Following the general procedure for hydrogenolysis, compound 10 (111 mg, 0.27 mmol) was de-protected to give compound 16, which was purified by column chromatography (10% MeOH in CH2Cl2), affording white crystals (56 mg, 63 % yield): mp. 157-159 °C; Rf 0.68 (10% MeOH in CH2Cl2); 1H NMR δ 2.65 (s(br), 1H), 3.67 (m, 2H), 3.74 (m, 2H, J), 3.80-3.84 (m, 4H), 7.05 (ddd, 1H, J = 8.1, 6.9, 1.2 Hz), 7.12 (dd, 1H, J = 8.4, 0.8 Hz), 7.45-7.50 (m, 2H), 7.72 (dd, 1H, J = 8.0, 0.8 Hz), 8.04 (dd, 1H, J = 8.2, 1.4 Hz), 8.19 (dd, 1H, J = 7.4, 1.0 Hz), 8.38 (m, 1H), 10.80 (s, 1H); 13C NMR δ39.41, 61.93, 70.08, 73.24, 109.93, 113.70, 117.59, 120.62, 123.87, 125.56, 126.54, 127.71, 134.69, 137.29, 149.01, 157.72, 163.35, 163.64; CIMS m/z 343 (MH+); HRMS m/z calc for C18H19N2O5: 343.1294, found: 343.1285; EA calc for C18H18N2O5+(H2O)1/3: C, 62.06; H, 5.40; N, 8.04; found: C, 62.24; H, 5.19; N, 7.69.
2-(2’-Hydroxyphenyl)-benzoxazole-4-carboxylic acid 2-{2-[2-(2’-hydroxy-ethoxy)-ethoxy]-ethoxy}-ethyl ester (17)
Following the general procedure for hydrogenolysis, compound 11 (65 mg, 0.12 mmol) was de-protected to give compound 17, which was purified by column chromatography (10% MeOH in CH2Cl2), affording a yellow oil (46mg, 85% yield): Rf 0.76 (10% MeOH in CH2Cl2); 1H NMR (400 MHz) δ 2.60 (s(br), 1H), 3.52-3.54 (m, 2H), 3.59-3.71 (m, 8H), 3.74-3.77 (m, 2H), 3.91 (t, 2H, J = 4.8 Hz), 4.58, (t, 2H, J = 4.8 Hz), 6.98 (t. 1H, J = 7.6 Hz), 7.09 (d, 1H, J = 8 Hz), 7.39-7.46 (m, 2H), 7.76 (dd, 1H, J = 8.0, 0.8 Hz), 7.98 (dd, 1H, J = 8.0, 1.6 Hz), 8.05 (dd, 1H, J = 7.8, 0.6 Hz), 11.82 (s, 1H); 13C NMR δ 61.64, 64.50, 69.10, 70.24, 70.53, 70.61, 72.44, 109.88, 114.90, 117.59, 119.49, 121.16, 124.68, 127.13, 127.46, 134.12, 139.42, 149.67, 159.34, 164.19, 165.04; CIMS m/z 432 (MH+); HRMS m/z calc for C22H26NO8: 432.1675, found 432.1658; EA calc for C22H25NO8+(H2O)1/3: C, 60.40; H, 5.91; N, 3.26; found: C, 60.53; H, 5.92; N, 3.10.
Metal Ion Binding Studies
Stock solutions of UK-1 and analogs, Mg(NO3)2, and Cu(NO3)2, were prepared fresh on the day of the spectrophotometric titrations. The fraction of complex formed was measured by using solutions of increasing mole fraction of metal ion, keeping the concentration of ligand constant, at 10 μM, using as a reference the absorbance of a solution containing the same concentration of metal ion without the ligand. The change in absorbance was monitored from 600 to 200 nm for each sample, as a function of the concentration of metal ion. The maximum absorbance change for the ligand-metal complex was recorded at each metal-ion concentration.
Spectrophotometric data was analyzed by equations (1) and (2). Binding constants were calculated by non-linear regression of the experimental points.
| [1] |
| [2] |
Where:
Kd is the dissociation constant, C0 is the initial concentration of the UK-1 analog, Cm is the concentration of the metal ion, ΔA is the increase in absorption at the wavelength of maximum absorbance for the ligand-metal ion complex (~410 nm) upon addition of each metal ion, and ΔAmax is the same parameter when the ligand is totally bound to the metal on.
Cytotoxicity Assays
Cell culture cytotoxicity assays were carried out as described previously [6]. Briefly, aliquots of 100 μl cell suspension (1-3*103 cells) were placed in microtiter plates in an atmosphere of 5% CO2 at 37 °C. After 24 h, 100 μL of culture media and 2 μl of the compound in DMSO were added to each well in duplicate, and the plates incubated an additional 72 h at 37 °C. There was no effect on the growth of cells compared to that of cells in culture media alone at this DMSO concentration. Compounds, along with mitomycin-C as a positive control were evaluated at final concentrations ranging from 0.001 to 50 μM.
After the 72 h incubation, the culture media was removed from each well, and 200 μL of fresh media and 20 μL of AlamarBlue reagent were added, followed by additional 6 h incubation. Cell viability was detected by fluorescent intensity using a Beckman Coulter DTX880 plate reader with excitation at 530 nm and emission at 590 nm.
The fluorescence data obtained from the cytotoxicity studies was used to calculate the percent growth according to the following equation:
| [3] |
Where:
Mean Ftime0 = the averaged measured fluorescent intensities of alamarBlue reagent at the time just before the exposure of the cells to the test substance.
Mean Ftest = the averaged measured fluorescent intensities of alamarBlue reagent after 72 exposure of the cells to the test substance at a particular concentration.
Mean Fcrtl = the averaged measured fluorescent intensities of alamarBlue reagent after 72 h exposure of the cells to the vehicle without the test substance.
The IC50, the compound concentration for which the growth of treated cells from time0 was only 50% as much as the vehicle-control was determined by non-linear regression fitting the data to equation (4):
| [4] |
Where:
Min= the minimum response plateau (0%Growth)
Max= the maximum response plateau (100% Growth)
y= % Growth at each test compound concentration
n is a fitted parameter (the Hill slope coefficient)
Table 1.
Cytotoxicity of UK-1 Analogs against MCF-7 and A549 cell lines
| Compound | IC50 for CF7 (μM)a | IC50 for A549 (μM)a |
|---|---|---|
| UK-1 | 1.4 ± 0.9 | 2.0 ± 0.5 |
| 1 | 4±2 | 12±3 |
| 2 | >50 | >50 |
| 3 | 7±1 | 35±4 |
| 12 | 15 ± 5 | 40 ± 10 |
| 13 | 10 ± 8 | 11 ± 1 |
| 14 | 30 ± 10 | 14 ± 4 |
| 15 | 31 ± 7 | 39 ±17 |
| 16 | > 50 | 41 ±17 |
| 17 | 13 ± 2 | 11 ± 1 |
| MMCb | 0.20 ± 0.07 | 0.4 ± 0.1 |
determined using the AlamarBlue assay. See Experimental Section for details.
Mitomycin C.
Acknowledgments
We would like to thank Chad M McKee and Dr. John Richburg (The University of Texas at Austin) for their assistance in the cell culture work, and financial support from the National Institutes of Health (R01 GM65956), the US–Egypt Science and Technology Joint Fund, administered through the USDA (BIO9-002-015), and the Robert Welch Foundation (F-1298).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ukei M, Ueno K, Miyadoh S, Abe K, Shibata K, Taniguchi M, Oi S. J Antibiot. 1993;46:1089–1094. doi: 10.7164/antibiotics.46.1089. [DOI] [PubMed] [Google Scholar]
- 2.Ueki M, Taniguchi M. J Antibiot. 1997;50:788–790. doi: 10.7164/antibiotics.50.788. [DOI] [PubMed] [Google Scholar]
- 3.Sato S, Takayuki K, Noguchi M, Takehana K, Kobayashi T, Tsuji T. J Antibiot. 2001;54:102–104. doi: 10.7164/antibiotics.54.102. [DOI] [PubMed] [Google Scholar]
- 4.Reynolds MB, DeLuca MR, Kerwin SM. Bioorg Chem. 1999;27:326–337. [Google Scholar]
- 5.DeLuca MR, Kerwin SM. Tetrahedron Lett. 1997;38:199–202. [Google Scholar]
- 6.Kumar D, Jacob MR, Reynolds MB, Kerwin SM. Bioorg Med Chem. 2002;10:3997–4004. doi: 10.1016/s0968-0896(02)00327-9. [DOI] [PubMed] [Google Scholar]
- 7.Wang BB, Maghami N, Goodlin VL, Smith PJ. Bioorg Med Chem Lett. 2004;14:3221–3226. doi: 10.1016/j.bmcl.2004.03.095. [DOI] [PubMed] [Google Scholar]
- 8.Huang ST, Hsei IJ, Chen C. Bioorg Med Chem. 2006;14:6106–6119. doi: 10.1016/j.bmc.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 9.Reyzer ML, Brodbelt JS, Kerwin SM, Kumar D. Nucl Acids Res. 2001;29:e103. doi: 10.1093/nar/29.21.e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oehlers L, Mazzitelli CL, Brodbelt J, Rodriguez M, Kerwin S. J Am Soc Mass Spec. 2004;15:1593–1603. doi: 10.1016/j.jasms.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 11.Thompson KH, Orvig C. Dalton Trans. 2006;6:761–764. doi: 10.1039/b513476e. [DOI] [PubMed] [Google Scholar]
- 12.Vergne AF, Waltz AJ, Miller MJ. Nat Prod Rev. 2000;17:99–116. doi: 10.1039/a809397k. [DOI] [PubMed] [Google Scholar]
- 13.Rovig C, Abrams MJ. Chem Rev. 1999;9:2201–2203. doi: 10.1021/cr980419w. and entire issue. [DOI] [PubMed] [Google Scholar]
- 14.Huang R, Wallqvist A, Covell DG. Biochem Pharmacol. 2005:1009–1039. doi: 10.1016/j.bcp.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Boerner LJK, Zaleski JM. Curr Opin Chem Biol. 2005;9:135–144. doi: 10.1016/j.cbpa.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Hou M, Wang AH. Nucl Acids Res. 2005;33:1352–1361. doi: 10.1093/nar/gki276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Astrid Sigel A, Sigel H, editors. Metal Ions in Biological Systems. Vol. 33. Marcel Dekker: New York; 1996. [Google Scholar]
- 18.Sissi C, Adreolli M, Cecchetti V, Fravolini A, Gatto B, Palumbo M. Bioorg Med Chem. 1998;6:1555–1561. doi: 10.1016/s0968-0896(98)00086-8. [DOI] [PubMed] [Google Scholar]
- 19.Fan JY, Sun D, Yu H, Kerwin SM, Hurley LH. J Med Chem. 1995;38:408–424. doi: 10.1021/jm00003a003. [DOI] [PubMed] [Google Scholar]
- 20.Ihara T, Ikegami T, Fujii T, Kitamura Y, Sueda S, Takagi M, Jyo A. J Inorg Biochem. 2006;100:1744–1754. doi: 10.1016/j.jinorgbio.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Fang YY, Ray BD, Claussen CA, Lipkowitz KB, Long EC. J Am Chem Soc. 2004;126:5403–5412. doi: 10.1021/ja049875u. [DOI] [PubMed] [Google Scholar]
- 22.Zhang XB, Peng J, He CL, Shen GL, Yu RQ. Anal Chim Acta. 2006;567:189–195. [Google Scholar]
- 23.Mazzitelli CL, Rodriguez M, Kerwin SM, Brodbelt JS. Evaluation of Metal-Mediated DNA Binding of Benzoazole Ligands by Electrospray Ionization Mass Spectrometry. J Am Soc Mass Spectrom. doi: 10.1016/j.jasms.2007.05.009. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levin JI, Turos E, Weinreb S. M Syn Comm. 1982;12:989–993. [Google Scholar]