Abstract
The overall mechanisms governing the role of laminins during osteogenic differentiation of human mesenchymal stem cells (hMSC) are poorly understood. We previously reported that laminin-332 induces an osteogenic phenotype in hMSC and does so through a focal adhesion kinase (FAK) and extracellular signal-related kinase (ERK) dependent pathway. We hypothesized that this is a result of integrin-ECM binding, and that it occurs via the known α3 LG3 integrin binding domain of laminin-332. To test this hypothesis we cultured hMSC on several different globular domains of laminin-332. hMSC adhered best to the LG3 domain, and this adhesion maximally activated FAK and ERK within 120 minutes. Prolonged culturing (8 or 16 days) of hMSC on LG3 led to activation of the osteogenic transcription factor Runx2 and expression of key osteogenic markers (osterix, bone sialoprotein 2, osteocalcin, alkaline phosphatase, extracellular calcium) in hMSC. LG3 domain binding did not increase matrix mineralization, demonstrating that the LG3 domain alone is not sufficient to induce complete osteogenic differentiation in vitro. We conclude that the LG3 domain mediates attachment of hMSC to laminin-332 and that this adhesion recapitulates most, but not all, of the osteogenic differentiation associated with laminin-5 binding to hMSC.
Keywords: LG domain, osteogenesis, laminin-332, mesenchymal stem cell, osterix, extracellular matrix
Introduction
Laminins are large molecular weight glycoproteins that are crucial for the assembly and maintenance of basement membranes in all tissue types [1]. Assembled from an α, β, and γ chain, they form a cross-like structure held together by a coiled-coil domain and disulfide bonds [2]. There have been 15 laminin isoforms identified [3], all of which can bind other extracellular matrix molecules. Several families of receptors bind laminins, including integrins. Through the binding via these receptors laminins and other matrix molecules laminins are able to regulate differentiation, cellular growth and migration, and survival [4,5].
Laminins contain functionally distinct domains which are shared amongst most laminins. There are numerous domains common to most or all laminins. The coiled-coil domain is responsible for joining the three subunits into a heterotrimer. Within the N-terminus of all laminin chains there are multiple epidermal growth factor repeats that mediate adhesive interactions and determine the amount of flexibility and spacing in laminin networks [6]. Also contained in the N terminus of most laminin chains is an LN type VI domain that is involved in the polymerization of laminin molecules [7]. The laminin type IV domain, also known as the laminin B domain, has an unknown function and is not found in short arm laminins.
A characteristic trait of laminin alpha chains is a tandem repeat of five globular domains (termed LG domains) located at the C terminus. They are relatively small (∼20 kDa) and have a wide variety of roles in adhesion, migration, signaling, and differentiation [8,9]. There is a short spacer sequence that separates the LG1−3 from the LG4−5 region and is often cleaved, affecting adhesion and migratory behavior [10]. The LG4 domain is essential for heparin and nidogen binding, while LG5 is only needed for heparin binding [11,12]. Dystroglycan typically binds to the LG4−5 area of laminins, though it also can bind to the LG1−3 domain of laminin-5 [13,14]. The LG domains are also crucial for cell attachment via integrins. The LG3 domain of α3 containing laminins has a motif PPFLMLLKGSTR that mediates integrin binding and cell adhesion [15].
Stem cells have great potential for the treatment and repair of connective tissues. Human mesenchymal stem cells (hMSC), derived from the bone marrow, are able to differentiate into a number of tissue types including nerve, cartilage, and bone [16]. They have shown a great deal of promise in animal studies and have even been used to treat arthritis [17]. Their ability to divide at higher rates than their differentiated counterparts [18] allows for a larger pool of potential cells to be transplanted at sites of injury. They can also be autologously transplanted with a diminished chance of immune rejection. Being able to transplant hMSC into a wound predisposed to becoming osteoblasts is crucial for bone repair. Most of the work done in vitro with hMSC and osteogenesis has been done using soluble factors [19] that would not be effective for transplant since they show decreased bone formation and immune suppression in vivo [20].
The transcription factor Runx2/Cbfa-1 is critical for connective tissue formation and is a downstream target of ERK [21]. Mice deficient in the Cbfa-1 gene die immediately after birth and exhibit maturational arrest in osteoblast differentiation resulting in a complete lack of endochondral and intramembranous bone formation [22]. The skeleton is present in these mice, but it is completely cartilaginous. These same mice also lack hypertrophic chondrocytes, implying Cbfa-1 as necessary for differentiation of these cells [23]. These two observations identify Cbfa-1 as a common regulator of both osteogenic and chondrogenic differentiation. Heterozygous Cbfa-1 mice display an absence of osteoblasts but not chondrocytes [24], showing a greater dependency of osteogenesis on Cbfa-1. The importance of Cbfa-1 for osteoblastic differentiation is underscored by the fact that forced expression of this gene in nonosteoblast cells induces osteogenic gene expression and ossification in areas where it typically does not occur [25].
Osterix (osx) is a marker for osteogenic differentiation downstream of Runx2. It is a zinc finger-containing transcription factor that is expressed in osteoblasts of all endochondral and intramembranous bone and essential for bone formation [26]. Formation of bone is ablated in osx deficient mice while activity and expression of Cbfa-1/Runx2 remain unaffected [27]. Cbfa-1 deficient mice do not produce osx. ERK 1/2 pathways also regulate osx expression [28], implying that integrin pathways may be critical for osteogenic differentiation.
Members of the Small Integrin-Binding Ligands with N-Linked Glycosylation (SIBLING) family play key roles in bone biology. This family includes bone sialoprotein I and II, dentin matrix protein-1 (DMP-1), dentin sialoprotein (DSP), dentin sialophosphoprotein (DPP), extracellular matrix glycoprotein (MEPE), and osteocalcin. They promote osteogenic signaling [29,30] and matrix mineralization [31]. A Cbfa-1 binding site is located in the promoters of several late osteoblastic markers, including osteocalcin and both bone sialoproteins [32,33].
There is a growing body of evidence that laminin-332 is present in [34], and controls the functions of, certain mesodermal tissues [5,35,36]. Recently we have described a function for laminin-332 in the commitment of hMSC to an osteogenic fate through an ERK 1/2 and FAK dependent pathway [37,38]. Miyazaki's group has also shown that laminin-332 suppresses the chondrogenic differentiation of these cells [39]. However, the role of specific functional domains in controlling the behavior or hMSC has not been well established. In this study, we examined the ability of globular domains of laminin-332 α3 chain to support osteogenic differentiation in vitro.
Materials and Methods
Tissue culture media (DMEM) and penicillin G-streptomycin sulfate (GPS) were purchased from Mediatech (Cellgro, VA). Fetal bovine serum (FBS) was purchased from Gemini Bio-Products (Woodland, CA). Trypsin-EDTA and purified bovine COLL-I were obtained from Sigma Chemical Co. (St. Louis, MO). Laminin-332 LG1, LG2, LG4, and LG5 domains were cloned into a pET vector from Novagen (Madison, WI), expressed in BL21DE3 bacteria (Novagen), and purified using a nickel column. LG3 domain was cloned into a pGEX-4T-3 expression vector from Amersham (Piscataway, NJ) and purified using Ni-NTA agarose beads from Qiagen (Valencia, MD). Purified laminin-332 was generously provided by Desmos Inc. (San Diego, CA). The αvβ3 (catalog # MAB1976) and the α1−6 and β1−4 integrin function-blocking antibodies (Alpha integrin blocking and IHC kit, catalog # ECM 430; Beta integrin screening kit, catalog # ECM 440), rabbit polyclonal phosphothreonine (cat. #AB1607), and purified human plasma fibronectin (FN) were purchased from Chemicon International (Temecula, CA). Rabbit polyclonal IgG anti-G-actin (catalog # AAM01-A) antibody was from Cytoskeleton (Denver, CO). Rabbit polyclonal IgG antibodies against anti- ERK1/2 (catalog # AB3053), and phosphoserine (catalog # AB1603) were obtained from Chemicon International (Temecula, CA). Rabbit polyclonal IgG antibodies against anti- human FAK (cat. #AHO0502) and rabbit polyclonal IgG phosphospecific antibodies against anti-ERK 1/2 (pTpY185/187) (catalog # 44−680) and anti- FAK (pY397) (cat. # 44−624G) were obtained from Biosource International (Camarillo, CA). Mouse monoclonal IgG antibodies against anti-Runx2/Cbfa1 were purchased from MBL International (Watertown, MA). Goat polyclonal IgG for osterix (sc-22538) was obtained from Santa Cruz Biotechnologies (Santa Cruz, CA). Horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG, HRP-conjugated goat anti-rabbit IgG, and HRP-conjugated rabbit anti-goat IgG secondary antibodies were obtained from Jackson Immuno Research (West Grove, PA). Catch and Release® v2.0 Reversible Immunoprecipitation System (cat. #17−500) were from Upstate Cell Signaling Solutions (Lake Placid, NY). RNeasy isolation kit and QuantiTect® SYBR® Green One Step RT-PCR Kit were purchased from Qiagen (Valencia, CA). Reverse transcriptase-polymerase chain reaction (RT-PCR) primers were purchased from IDT Technologies (Coralville, Iowa). The protein assay kit was purchased from Pierce (Rockford, IL). Calcium Reagent Set was from Teco Diagnostics Inc. (Anaheim, CA). Unless otherwise specified, the other standard reagents were obtained from Fisher Scientific (Fair Lawn, NJ).
Cell culture
Cryopreserved hMSC were purchased from Cambrex Inc. (Walkersville, MD) and were grown according to the manufacturers' instructions. Briefly, cells were plated at 5 × 103 cells/cm2 in a T75 flask (75 cm2) for continuous passaging in DMEM medium supplemented with 10% FBS, 1% L-glutamine [29.2 mg/mL], penicillin G [10,000 units/mL] and streptomycin sulfate [10,000 μg/mL]. Medium was changed twice weekly and cells were detached by trypsin-EDTA and passaged into fresh culture flasks at a ratio of 1:3 upon reaching confluence. Cultures were incubated at 37°C in a humidified atmosphere containing 95% air and 5% CO2.
Laminin-332 matrix was isolated from rat bladder carcinoma 804G cells as described previously [40].
For in vitro osteogenic assays, hMSC were passaged three times before they were inducted and plated at densities of 3.1 × 103 cells/cm2 in 0.2 mL/cm2 of medium on 100 mm Falcon dishes (78.5 cm2). The following day (day 0), we replaced the culture medium with fresh control medium in the absence or presence of osteogenic supplements (OS) containing 0.1 μM Dexamethasone, 0.05 mM ascorbic acid-2-phosphate, and 10 mM β-glycerophosphate (Cambrex Inc). In each experiment, control and OS media-treated cells were processed in parallel. At days 8, 14, 16, and 21, cultures were assayed as described below.
Adhesion assays
Cell adhesion assays were performed as previously described using Sarstedt 96-well suspension cell culture plates [41]. Tissue culture plates were coated with purified ECM proteins at a concentration of 10 μg/ml (laminin-332), 20 μg/ml (fibronectin), 25 μg/ml (LG1, LG2, LG4, LG5), or 50 μg/ml (LG3) for 2 hour at 37°C. Wells were washed twice with PBS and incubated with nd-blotto (5% non-dairy creamer in PBS + 0.2% Tween 20) for 30 minutes prior to the assay. Cells were allowed to attach for 2 hours at 37°C and were subsequently fixed with 3% paraformaldehyde, washed twice in PBS, and incubated in crystal violet dye for 15 minutes. Wells were washed thoroughly with water and the violet dye was extracted with 10% SDS solution. Absorbance was measured using a TECAN SPECTAFluor spectrophotometer at 595 nm and relative adhesion was compared to cells attached to nd-blotto.
Integrin blocking adhesion assays were performed according to the procedure above, but the cells were incubated with a functional integrin-blocking antibody for 30 minutes at 37°C, with vortexing every 5 minutes prior to plating.
Immunoprecipitation of Runx2, Osterix, and Western blotting
Whole cell extracts were prepared by harvesting serum deprived cells overnight (DMEM + 0.1% FBS) in ice-cold RIPA buffer (150 mM NaCl, 50 mM Tris, 1% Triton-X, 0.3 mM Sodium vanadate, 1% Deoxycholic acid, 0.2% SDS, pH 7.4). For immunoprecipitation of Runx2 and osterix, the Catch and Release® v2.0 Reversible Immunoprecipitation System (Upstate) was employed. Briefly, 1× wash buffer, 500 ug of cell lysate, 4 μg of Runx2 or Osterix antibody, and antibody capture affinity ligand were added to a spin column at a final volume of 500 μl. Following two hour incubation on a rotator at 4°C, the column was centrifuged at 5000 rpm for 30 sec to discard all the non-labeled proteins, followed by repeated washes. Denatured Runx2 protein was eluted after the addition of 70 μl of 1× denaturing elution buffer containing β-ME and centrifugation. For other assays, proteins were diluted in 4× Laemmli's sample buffer, denatured at 100°C for 5 min, resolved by 8% SDS-PAGE, and electrophoretically transblotted to Trans-Blot® nitrocellulose membranes (0.2 μm) (Bio-Rad, Hercules, CA). The membranes were incubated with blocking solution (5% non-fat dried milk in 1× PBS + 0.2% Tween-20 (PBST) for 1 hr, then probed with various primary antibodies (1:500) overnight at 4°C. After three washes with PBST, membranes were incubated with HRP-conjugated secondary IgG (1:25,000) for 1 hr, followed by another three washes with PBST. Immunoreactive bands were detected using the SuperSignal® Chemiluminescent reagent (Pierce) and quantitatively analyzed in triplicate by normalizing band intensities to the controls on scanned films by IMAGEJ software. Blots to be reprobed were stripped with 200mM glycine pH 2.2 for 30 minutes and then treated as above.
Quantitative reverse transcriptase PCR
RNA was isolated from 10 × 108 hMSC cultured for 16 days on tissue culture plastic (± OS media), or on laminin-332, FN, G3, G4, or G5 in control media. Total RNA was isolated using the RNeasy mini kit (Qiagen, Valencia, CA). Primers used for amplification of differentiation marker genes were designed using OligoPerfect™ (Invitrogen, Carlsbad, CA) and are listed in Table 1. Quantitative reverse transcription RT-PCR was performed using the LightCycler® 480 Real-Time PCR System (Roche, Pleasonton, CA) and the QuantiTect® SYBR® Green RT-PCR Kit with HotStar Taq DNA Polymerase. 1× QuantiTect SYBR Green, 0.5μM Primer F and Primer R, 0.5μl/reaction QuantiTect RT Mix were combined with sample RNA to yield a 20μl reaction volume. RT-PCR was performed according to the following protocol defined by the manufacturer: RT 20min at 50°C 20°C/s ramp, PCR activation 15min at 95°C 20°C/s ramp, 35−55 cycles of [Denaturation 15s at 94°C 20/s ramp, Annealing 20−30s at 50−60°C 20°C /s ramp, Extension 30s 72°C 2°C/s ramp]. All samples were loaded in duplicate and normalized to total RNA content and to the performance of the housekeeping gene, GAPDH. Fold differences in gene expression were relative to hMSC cultured on tissue culture plastic and calculated using ΔΔCt method [42].
Table 1.
Primers used for quantitative RT-PCR
| Gene Name | Primer Sequences |
|---|---|
| IBSP (Bone sialoprotein 2) NM_004967 | Forward 5′ - CGACCACTTTGTCAAGCTCA -3′ |
| |
Reverse 5′ - GATTGCTTCCTCTGGCAGTC - 3′ |
| BGLAP (Osteocalcin) NM-199173 | Forward 5′ - GACTGTGACGAGTTGGCTGA- 3′ |
| |
Reverse 5′ - CTGGAGAGGAGCAGAACTGG -3′ |
| SP7 (Osterix) NM_152860 | Forward 5′ - GCCAGAAGCTGTGAAACCTC - 3′ |
| |
Reverse 5′ - GCTGCAAGCTCTCCATAACC - 3′ |
| CBFA1 (Core binding factor alpha 1) NM_004348 | Forward 5′ - TTACTTACACCCCGCCAGTC - 3′ |
| |
Reverse 5′ - CACTCTGGCTTTGGGAAGAG - 3′ |
| GAPDH (Glyceraldehyde-3-phosphate dehydrogenase) NM_002046 | Forward 5′ - CGACCACTTTGTCAAGCTCA - 3′ |
| |
Reverse 5′ - AGGGGTCTACATGGCAACTG - 3′ |
| ALP (Alkaline Phosphatase) NM_000478 | Forward 5′ - TGGAACATTCTGGATCTGAC - 3′ |
| Reverse 5′ - GAGTGAGTGAGTGAGCAAGG -3′ |
Histological staining of alkaline phosphatase activity and calcium precipitation
For detection of alkaline phosphatase activity, a solution of naphthol AS-MX phosphate and fast blue RR dissolved in dH2O was poured into 100 mm dishes on day 14 according to the manufacturer's instructions contained in Sigma Kit #85 (Sigma). Cellular specimens were scored according to the quantity and intensity of the precipitated dye as determined by IMAGEJ software and was normalized to control values. For the detection of a calcium-phosphate-containing matrix after 21 days, cell layers were stained by the alizarin red-S method. Specimens were washed three times with PBS and fixed in ice-cold 70% ethanol for 1 hr. Afterwards, cells were incubated with a 0.4% alizarin red-S solution in water (pH 4.2) for 15 min at room temperature. Cells layers were washed thoroughly with dH2O five times and left in PBS for 15 min. Cellular specimens were scored according to the quantity and size of precipitated granules.
Calcium Assay
Cultured monolayers of hMSC were washed twice with PBS, extracted with 0.5 N HCl, and the resulting lysate was shaken for 5 h at 4°C, followed by centrifugation at 2,000g for 10 minutes. The supernatant was utilized for calcium determination, according to the manufacturer's instructions contained in Teco Diagnostics Calcium Reagent Set. Total calcium was calculated from absorbance at 570 nm using standard solutions prepared in parallel as calibration points.
Mineralized bone: Fourier transform infrared (FTIR) analysis
The presence of apatite in cell matrix was detected by FTIR of ground powders. Cell layers, collected in 50 mM ammonium bicarbonate (pH 8.0) after 21 days, were lyophilized and analyzed as potassium bromide (KBr) pellets on a Bio-Rad FTS 40-A spectrometer (Bio-Rad, Cambridge, MA). The data was collected under nitrogen purge, and the spectral baseline corrected and analyzed using GRAMS/386 software (Galactic Industries, Salem, NH) as previously described [43]. The mineral content was calculated based on the spectrally derived mineral-to-matrix ratio (the integrated areas of the phosphate absorbance (900−1200 cm−1) and protein amide I band (1585−1720 cm−1)).
Statistical analysis
All experiments were repeated a minimum of three times and the representative data were presented as mean ± S.E. Statistical analyses were preformed using Student's unpaired t test, and a p value less than 0.05 was considered significant.
Results
hMSC adhere greatest to the LG3 domain of Ln-332 and do so through the α3β1 integrin
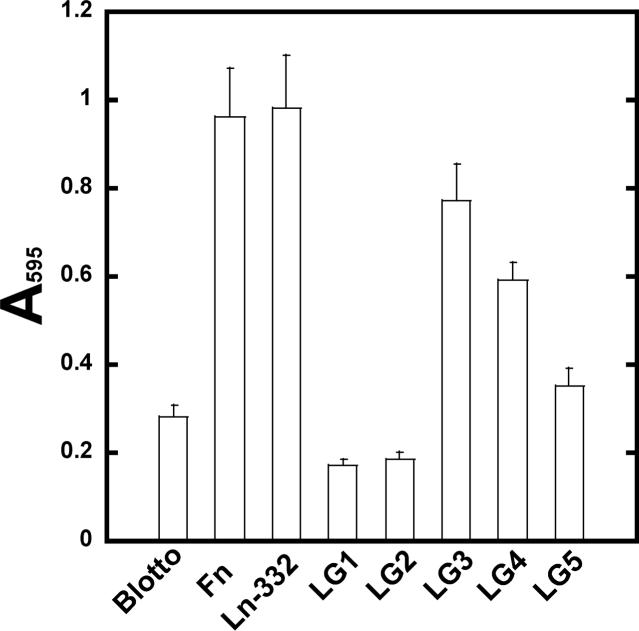
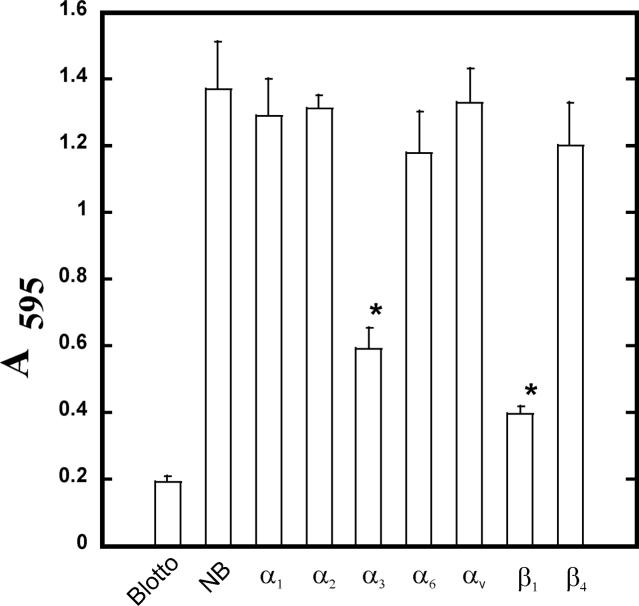
To test whether or not specific domains of laminin-332 can recapitulate its osteogenic effects, we first plated hMSC on purified α3 LG domains for two hours to assess their effectiveness at binding. Because LG1 and LG2 did not support adhesion, they were not further characterized in this study. In contrast, the three terminal LG domains all supported varied hMSC adhesion, with LG3 > LG4 > LG5 (Figure 1). This data is consistent with other studies showing α3 integrin binding sites in the LG3 domain and dystroglycan interactions in the LG4/LG5 domains. Adhesion to the LG3 domain was blocked using antibodies to the α3 and β1 integrin subunits (Figure 2). Adhesion to LG4 and LG5 was unaffected by integrin blocking antibodies.
Figure 1. hMSC adhere best to the LG3 domain of laminin-332.
hMSC were allowed to adhere to blotto (nfdm in PBST), 20 ug/ml FN, 10 ug/ml LN-332, 25 ug/ml LG1, LG2, LG4, LG5, 50 ug/ml LG3 for 2 hours. Non-adherent cells were removed by washing and adherent cells were stained with crystal violet, then solubilized in SDS and absorbance determined at 570nm.
Figure 2. Adhesion of hMSC to the LG3 domain of laminin-332 occurs through α3β1.
hMSC were incubated with blocking antibodies to the indicated integrins and allowed to adhere to purified LG3 domain. Non-adherent cells were removed by washing and adherent cells were stained with crystal violet, then solubilized in SDS and absorbance determined at 570nm.
Adhesion to the LG3 domain induces phosphorylation of both FAK and ERK 1/2
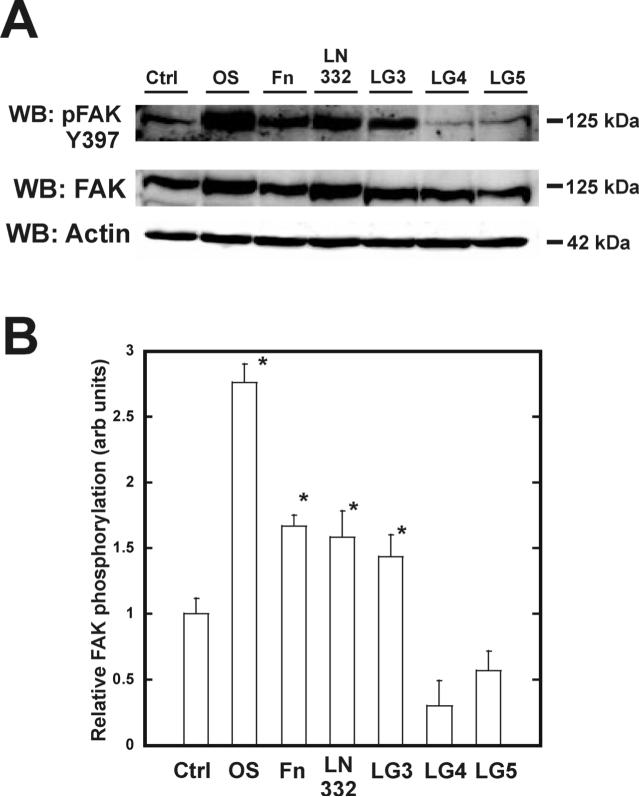
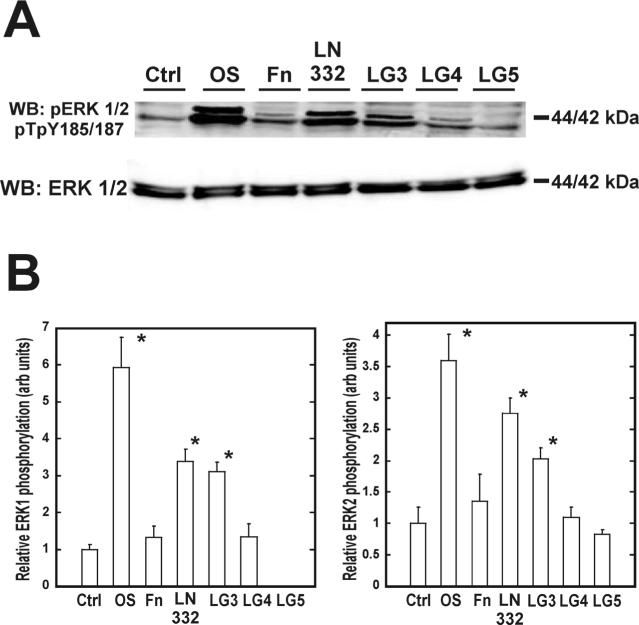
FAK was maximally phosphorylated on Y397 after 120 minutes in hMSC (Figure 3A). Specifically, cells plated on the LG3 domain exhibited a 1.4-fold greater phosphorylation of FAK than cells plated on the non-specific substrate poly-L-lysine (Figure 3B). This activation was comparable to that seen in hMSC plated on intact laminin-332, which exhibited a 1.6-fold greater phosphorylation of FAK than control cells. LG4 and LG5 failed to phosphorylate FAK on Y397 during this time course. Adhesion of hMSC to LG3 after two hours also resulted in a similar activation of ERK (Figure 4A). Laminin-332 exhibited a 3.4-fold and 2.8-fold increase in phosphorylation of ERK 1 and 2, respectively, while LG3 displayed a 3.1-fold and 2.0-fold increase (Figure 4B+C). The LG4 domain induced comparatively low ERK 1/2 activation; 1.4-fold and 1.1-fold respectively. The LG5 domain failed to activate these signaling proteins.
Figure 3. FAK Y397 phosphorylation occurs upon binding to the LG3 domain of laminin-332.
hMSC were allowed to adhere for two hours on the indicated substrates and assayed for phosphorylated pY397 FAK (A). Densitometric measures of band intensities for pY397 FAK are shown in (B). Values were normalized to total protein and the two hour ctrl lane.
Figure 4. Adhesion of hMSC to the LG3 domain of laminin-332 induces phosphorylation of ERK 1/2.
hMSC were allowed to adhere for two hours on the indicated substrates and assayed for phosphorylated pTpY185/187 ERK 1/2 (A). Densitometric measure of band intensities for ERK 1 and ERK 2 are shown in (B) and (C), respectively. ERK 1 and 2 bands were normalized to the intensity of ERK 1 and 2 in the two hour ctrl lane.
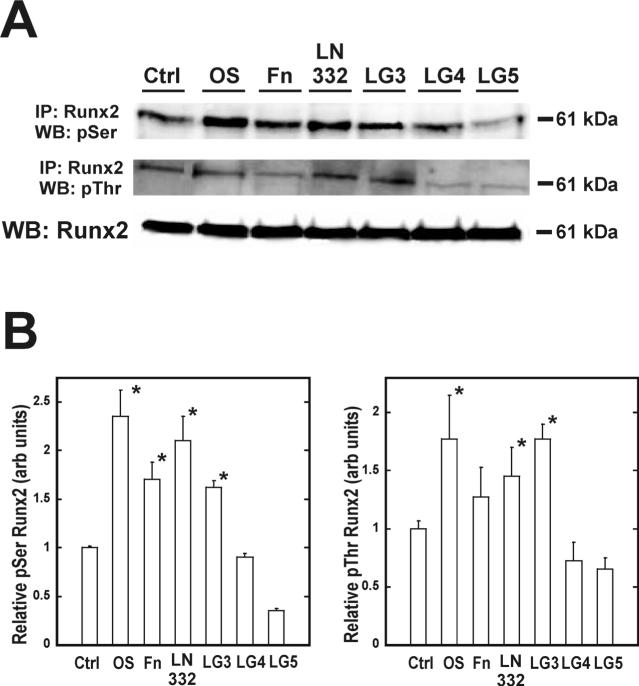
The LG3 domain stimulates phosphorylation and expression of the osteogenic transcription factors Runx2/Cbfa-1 and Osterix respectively
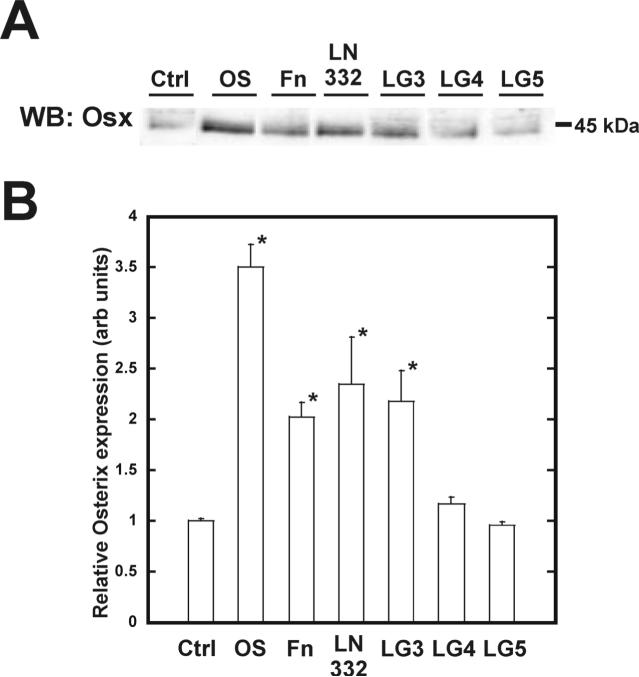
To determine if the LG domains are responsible for laminin-332-induced Runx2 phosphorylation, hMSC were plated on the LG domains for eight days. Runx2 protein was then immunoprecipitated and assayed for phosphoserine and phosphothreonine by western blot analysis (Figure 5A). LG3 was the only domain that induced phosphorylation of Runx2 during this time. Phosphoserine increased 1.6-fold while phosphothreonine went up 1.8-fold (Figure 5B+C). Intact laminin-332 induced a 2.1-fold and 1.5-fold increase in phosphoserine and phosphothreonine on Runx2 respectively. Protein levels of osterix exhibited similar trends as assessed by immunoblotting (Figure 6A). Laminin-332 stimulated a 2.3-fold increase in total osterix while LG3 induced 2.0-fold rise in the amount of osterix protein (Figure 6B).
Figure 5. hMSC adhesion to LG3 domain increases phosphorylation of the master bone transcription factor, Runx2/Cbfa-1.
Samples were incubated for 8 days, subsequently lysed, and Runx2/Cbfa-1 proteins immunoprecipitated with a Runx2/Cbfa-1 specific antibody. Immunoprecipitated proteins were separated by SDS-PAGE and blotted for phosphorylated serine and threonine, which is indicated by the 61 kDa band (A). Total Runx2/Cbfa-1 from each lysate was detected by western blot as a loading control. Densitometric measure of the ratio of band intensities for 61 kDa phosphoserine and phosphothreonine to total Runx2/Cbfa-1 are shown in (B) and (C), respectively.
Figure 6. The LG3 domain of laminin-332 amplifies the expression of the osteogenic transcription factor osterix.
Samples were cultured as per figure 5 and blotted with an osterix specific antibody (A). Densitometric measure of band intensities for osterix are shown in (B).
LG domains of laminin-332 promote osteogenic gene transcription and matrix mineralization
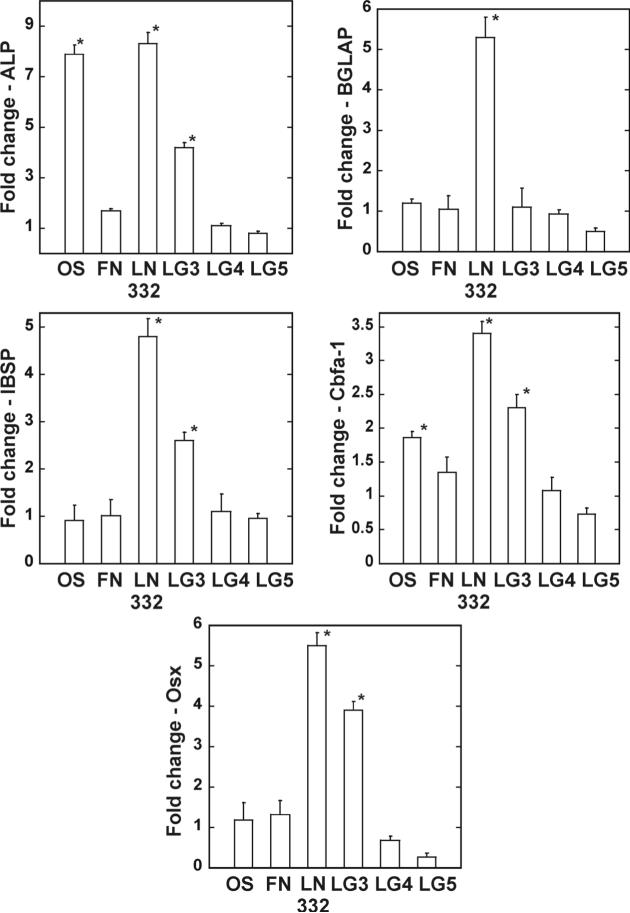
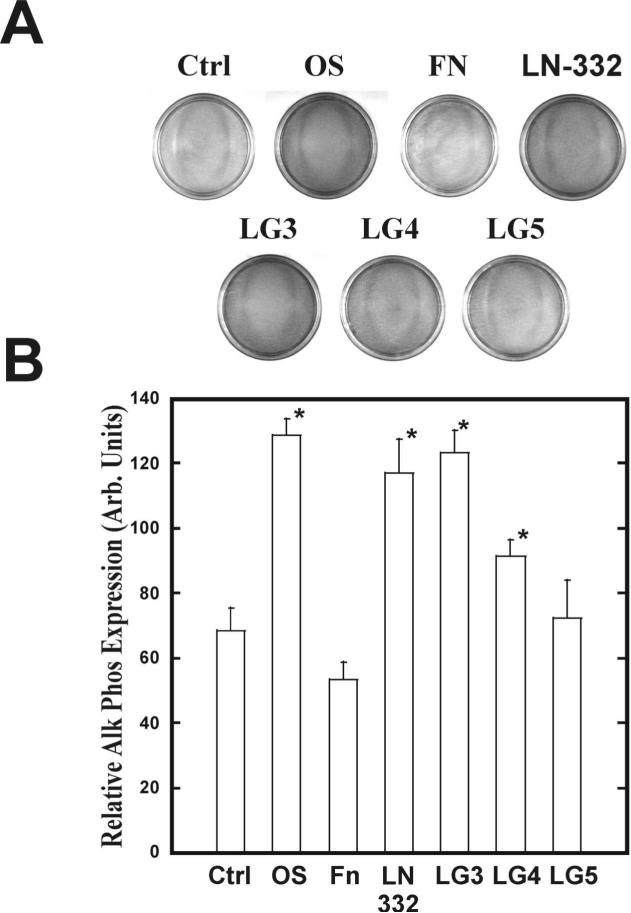
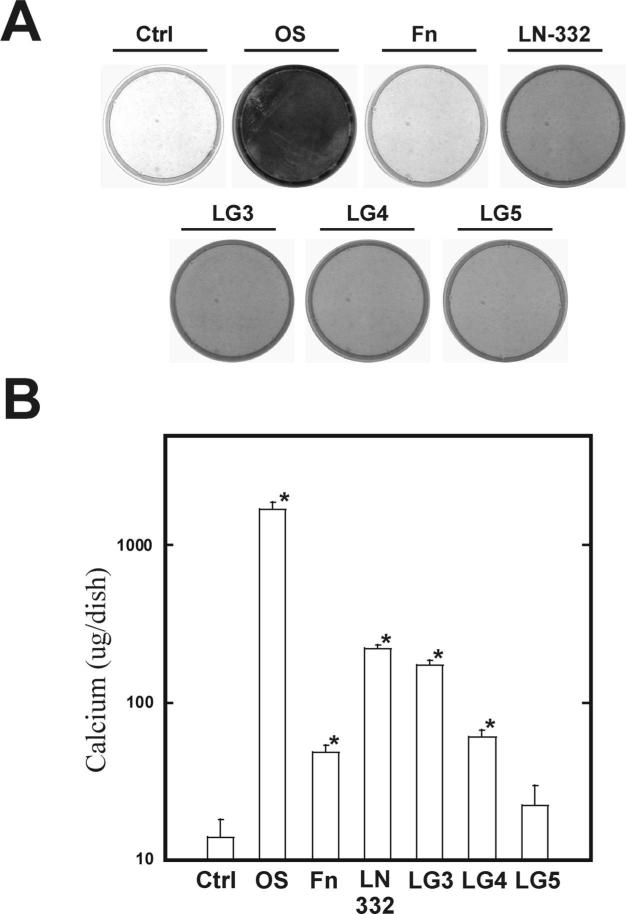
In order to evaluate the effects of the LG domains on osteogenic gene expression and promotion of an osteogenic phenotype, hMSC were cultured for either14, 16, or 21, days. Quantitative RT-PCR analysis was performed on mRNA isolated after 16 days (Figure 7). Cells plated on both laminin-332 and LG3 exhibited a greater than 4-fold increase in alkaline phosphatase and osterix gene levels as compared to unstimulated hMSC (Figure 7). There was also a greater than 2-fold increase in the amounts of Cbfa-1 and bone sialoprotein in these two conditions as compared to the control hMSC. This is consistent with the results shown in Figure 6 for an increase in osterix in those experimental conditions. There was no appreciable transcription of any gene in both the LG4 and LG5 conditions. Stem cells cultured on LG3 or LG4 domains for 14 days demonstrated positive staining for alkaline phosphatase activity (Figure 8). There was a 1.7-fold increase for both laminin-332 and LG3 while LG4 exhibited an increase of 1.4-fold (Figure 8B). All three LG domains displayed positive staining for extracellular calcium as compared to untreated hMSC (Figure 9A) and also exhibited a greater level of total calcium (Figure 9B). As assessed by FTIR, which quantifies the amount of insoluble phosphate embedded in the ECM, the degree of mineralization in hMSC plated on LG3 was not significantly different from control cultures (Table 2).
Figure 7. Osteogenic gene expression is upregulated in hMSC grown on the LG3 domain of laminin-332.
hMSC were plated as indicated and total mRNA isolated after day 16. Quantitative RT-PCR was performed on each sample using primers specific for the amplification of the indicated genes and normalized to GAPDH levels and gene expression from hMSC grown on tissue culture plastic.
Figure 8. Adhesion to LG domains increases osteogenic-specific marker expression of alkaline phosphatase in hMSC.
hMSC were plated for 14 days as indicated and stained for alkaline phosphatase activity (A). Densitometry of plates shown in panel A (B).
Figure 9. The LG domains of laminin-332 increase the osteogenic-specific marker of calcified matrix in hMSC.
Cells were cultured for 21 days on indicated substrates and stained for calcium using Alizarin Red (A). Colorimetric assessment of total calcium in hMSC (B). Cells were plated for 21 days as for panel A, then lysed and total calcium detected colorimetrically. Measurements were read at 570 nm.
Table 2. Peptides of laminin-332 are not sufficient to cause matrix mineralization.
hMSC were cultured for 21 days as indicated. The mineral:matrix (number) is computed by comparing the area of the phosphate peak (mineral) to the amide peak (protein) Osteogenic differentiation of hMSC plated on indicated substrates for 21 days, as assessed by indicated measurements of mineral to matrix ratio.
| hMSC culture conditions for 21 days | Mineral to matrix ratio |
|---|---|
| TCP | 1.83 +/− 0.2 |
| OS | 4.85 +/− 0.5 |
| Fn | 2.07 +/− 0.03 |
| LN-332* | 2.78 +/− 0.18 |
| LG3 | 1.85 +/− 0.08 |
| LG4 | 2.04 +/− 0.04 |
| LG5 | 1.98 +/− 0.13 |
Discussion
We have previously demonstrated that hMSC express laminin-332 both in vitro and in vivo, and that it is a pro-osteogenic stimulus [37]. Here, we used new tools (individual LG domains) to dissect out the mechanism. Our results strongly suggest that domains of laminin-332 recapitulate many of the pro-osteogenic effects of intact laminin-332 on hMSC. However, not all domains behaved identically. The differential activity is likely not due to denaturation, as these peptides support adhesion of other cell types [44,45]. As expected, the LG3 domain supported α3β1 integrin mediated adhesion and activated both FAK and MAPK pathways. These pathways are critical for osteogenesis, especially ECM induced [46,46]. Surprisingly, although LG3 could induce major differentiation pathways, it alone did not induce matrix mineralization by hMSC. The range for mineralization spans from ∼1.7 for naïve hMSC to between 4 and 5 for normal bone tissue. Stem cells cultured on laminin-332 have a mineral/matrix ratio of ∼2.85, closer to mature bone than stem cells, but stem cells cultured on LG3 exhibit a mineralized matrix closer to that of untreated hMSC. The lack of matrix mineralization occurring in LG3 cultures may be indicative of a requirement for other domains of laminin-332, either other LG domains or perhaps domains from the other two subunit chains of laminin-332. Another possible reason for the differences is that these domains were isolated from a prokaryotic expression system, and are therefore not glycosylated.
Direct comparison with other osteogenic proteins reveals that laminin-332 and the LG3 domain are comparable with regards to signaling and osteogenic gene expression. Laminin-332 is as potent an activator of the early signaling kinases, FAK and ERK 1/2, as both collagen-1 and vitronectin [38,46]. It also induces osteogenic signaling and gene expression in equal or greater amounts than the other ECM proteins. In this study we demonstrate that the LG3 domain signals much like that of the intact protein, although it does not provide similar mineralization. It is worth noting that in long term cultures these domains may be inducing expression of other matrix proteins that possibly participate in osteogenic differentiation. This suggests that both laminin-332 and LG3 are efficient at inducing most if not all aspects of osteogenesis, but not as universally potent as collagen-I.
Most proteins are composed of multiple functional domains, each with a specialized task. One of the canonical proteins studied with respect to domains and individual function is the glycoprotein fibronectin. There are 3 major internal repeats found in fibronectin: I, II, III. The fibronectin domain II binds to collagen while domains I and III interact with fibrin/heparin and DNA/heparin respectively [47]. Within fibronectin is a tripeptide sequence Arg-Gly-Asp (RGD) which mediates cell adhesion and migration [48,49]. None of these pieces however is able to recapitulate all of intact fibronectin's activities. The notion that fibronectin is a network of smaller protein domains working in coordination holds true for ECM proteins like laminin-332 as well. Laminin-332 is also comprised of numerous domains, all of which have specific functions for interacting with cells or other matrix molecules [50]. Like fibronectin, no one domain of laminin-332 can function as the intact protein. For osteogenesis, we have shown that LG3 is very important for this process, though it is not sufficient.
How then, are LG4 and LG5 able to regulate specific extracellular markers (alkaline phosphatase activity and calcium deposition) of osteogenesis? The dystroglycan complex, which interacts with the C-terminus of the α3 laminin chain, is a potential candidate. It acts as a transmembrane connection between the extracellular matrix and the cytoskeleton, including cytoplasmic adapter proteins such as Grb2 that activate the Ras-MAPK pathway [51]. Indeed, blocking ERK 1/2 signaling decreases these markers in our cells [37]. This raises the possibility of a subset of ERK signaling pathways controlling distinct stages of osteogenic differentiation. A second possibility is that LG4 and LG5 bind and potentially sequester heparin, which stimulates osteogenic differentiation in vitro [52].
Peptide fragments of larger proteins have shown promise as therapeutic targets for the treatment of a variety of afflictions. Endostatin, a 20-kDa C-terminal fragment from collagen XVIII, suppresses tumor growth by blocking angiogenesis [53]. The C-terminal domain of perlecan, termed endorepellin, is another inhibitor of angiogenesis [54]. A thrombin peptide chrysalin, unlike endorepellin and endostatin, promotes vascularization. It also accelerates the rate of fracture repair in rats [55]. These studies set precedence for the use of peptide fragments in the treatment of diseases and in tissue engineering. Therefore, the LG domains of laminin-332 may be used in the future for their pro-osteogenic potential in the regeneration of bone tissue using mesenchymal stem cells.
Acknowledgments
Contract grant sponsor: National Institutes of Health (to G.E.P.); Contract grant number: Grant #1R01EB002197
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Engvall E. Laminin variants: why, where and when? Kidney Int. 1993;43:2–6. doi: 10.1038/ki.1993.2. [DOI] [PubMed] [Google Scholar]
- 2.Malinda KM, Kleinman HK. The laminins. Int.J.Biochem.Cell Biol. 1996;28:957–959. doi: 10.1016/1357-2725(96)00042-8. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen NM, Senior RM. Laminin isoforms and lung development: all isoforms are not equal. Dev.Biol. 2006;294:271–279. doi: 10.1016/j.ydbio.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 4.Stahl S, Weitzman S, Jones JC. The role of laminin-5 and its receptors in mammary epithelial cell branching morphogenesis. J.Cell Sci. 1997;110(Pt 1):55–63. doi: 10.1242/jcs.110.1.55. [DOI] [PubMed] [Google Scholar]
- 5.Kingsley K, Huff JL, Rust WL, Carroll K, Martinez AM, Fitchmun M, Plopper GE. ERK1/2 mediates PDGF-BB stimulated vascular smooth muscle cell proliferation and migration on laminin-5. Biochem.Biophys.Res.Commun. 2002;293:1000–1006. doi: 10.1016/S0006-291X(02)00331-5. [DOI] [PubMed] [Google Scholar]
- 6.Balzar M, Briaire-de Bruijn IH, Rees-Bakker HA, Prins FA, Helfrich W, de Leij L, Riethmuller G, Alberti S, Warnaar SO, Fleuren GJ, Litvinov SV. Epidermal growth factor-like repeats mediate lateral and reciprocal interactions of Ep-CAM molecules in homophilic adhesions. Mol.Cell Biol. 2001;21:2570–2580. doi: 10.1128/MCB.21.7.2570-2580.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yurchenco PD, Cheng YS. Self-assembly and calcium-binding sites in laminin. A three-arm interaction model. J.Biol.Chem. 1993;268:17286–17299. [PubMed] [Google Scholar]
- 8.Timpl R, Tisi D, Talts JF, Andac Z, Sasaki T, Hohenester E. Structure and function of laminin LG modules. Matrix Biol. 2000;19:309–317. doi: 10.1016/s0945-053x(00)00072-x. [DOI] [PubMed] [Google Scholar]
- 9.Kitagawa Y, Yamashita H. [The role of laminin alpha 4 G domain in peripheral tissues]. Seikagaku. 2001;73:1227–1233. [PubMed] [Google Scholar]
- 10.Hirosaki T, Mizushima H, Tsubota Y, Moriyama K, Miyazaki K. Structural requirement of carboxyl-terminal globular domains of laminin alpha 3 chain for promotion of rapid cell adhesion and migration by laminin-5. J.Biol.Chem. 2000;275:22495–22502. doi: 10.1074/jbc.M001326200. [DOI] [PubMed] [Google Scholar]
- 11.Yurchenco PD, Sung U, Ward MD, Yamada Y, O'Rear JJ. Recombinant laminin G domain mediates myoblast adhesion and heparin binding. J.Biol.Chem. 1993;268:8356–8365. [PubMed] [Google Scholar]
- 12.Sung U, O'Rear JJ, Yurchenco PD. Localization of heparin binding activity in recombinant laminin G domain. Eur.J.Biochem. 1997;250:138–143. doi: 10.1111/j.1432-1033.1997.00138.x. [DOI] [PubMed] [Google Scholar]
- 13.Gee SH, Blacher RW, Douville PJ, Provost PR, Yurchenco PD, Carbonetto S. Laminin-binding protein 120 from brain is closely related to the dystrophin-associated glycoprotein, dystroglycan, and binds with high affinity to the major heparin binding domain of laminin. J.Biol.Chem. 1993;268:14972–14980. [PubMed] [Google Scholar]
- 14.Ido H, Harada K, Futaki S, Hayashi Y, Nishiuchi R, Natsuka Y, Li S, Wada Y, Combs AC, Ervasti JM, Sekiguchi K. Molecular dissection of the alpha-dystroglycan- and integrin-binding sites within the globular domain of human laminin-10. J.Biol.Chem. 2004;279:10946–10954. doi: 10.1074/jbc.M313626200. [DOI] [PubMed] [Google Scholar]
- 15.Kim JM, Park WH, Min BM. The PPFLMLLKGSTR motif in globular domain 3 of the human laminin-5 alpha3 chain is crucial for integrin alpha3beta1 binding and cell adhesion. Exp.Cell Res. 2005;304:317–327. doi: 10.1016/j.yexcr.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 16.Alhadlaq A, Mao JJ. Mesenchymal stem cells: isolation and therapeutics. Stem Cells Dev. 2004;13:436–448. doi: 10.1089/scd.2004.13.436. [DOI] [PubMed] [Google Scholar]
- 17.Augello A, Tasso R, Negrini SM, Cancedda R, Pennesi G. Cell therapy using allogeneic bone marrow mesenchymal stem cells prevents tissue damage in collagen-induced arthritis. Arthritis Rheum. 2007;56:1175–1186. doi: 10.1002/art.22511. [DOI] [PubMed] [Google Scholar]
- 18.Smith E, Redman RA, Logg CR, Coetzee GA, Kasahara N, Frenkel B. Glucocorticoids inhibit developmental stage-specific osteoblast cell cycle. Dissociation of cyclin A-cyclin-dependent kinase 2 from E2F4-p130 complexes. J.Biol.Chem. 2000;275:19992–20001. doi: 10.1074/jbc.M001758200. [DOI] [PubMed] [Google Scholar]
- 19.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 20.Ng PC, Lam CW, Wong GW, Lee CH, Cheng PS, Fok TF, Chan IH, Wong E, Cheung K, Lee SY. Changes in markers of bone metabolism during dexamethasone treatment for chronic lung disease in preterm infants. Arch.Dis.Child Fetal Neonatal Ed. 2002;86:F49–F54. doi: 10.1136/fn.86.1.F49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao G, Gopalakrishnan R, Jiang D, Reith E, Benson MD, Franceschi RT. Bone morphogenetic proteins, extracellular matrix, and mitogen-activated protein kinase signaling pathways are required for osteoblast-specific gene expression and differentiation in MC3T3-E1 cells. J.Bone Miner.Res. 2002;17:101–110. doi: 10.1359/jbmr.2002.17.1.101. [DOI] [PubMed] [Google Scholar]
- 22.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 23.Kim IS, Otto F, Zabel B, Mundlos S. Regulation of chondrocyte differentiation by Cbfa1. Mech.Dev. 1999;80:159–170. doi: 10.1016/s0925-4773(98)00210-x. [DOI] [PubMed] [Google Scholar]
- 24.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 25.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 26.Tai G, Christodoulou I, Bishop AE, Polak JM. Use of green fluorescent fusion protein to track activation of the transcription factor osterix during early osteoblast differentiation. Biochem.Biophys.Res.Commun. 2005;333:1116–1122. doi: 10.1016/j.bbrc.2005.05.195. [DOI] [PubMed] [Google Scholar]
- 27.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 28.Celil AB, Campbell PG. BMP-2 and insulin-like growth factor-I mediate Osterix (Osx) expression in human mesenchymal stem cells via the MAPK and protein kinase D signaling pathways. J.Biol.Chem. 2005;280:31353–31359. doi: 10.1074/jbc.M503845200. [DOI] [PubMed] [Google Scholar]
- 29.Tartaix PH, Doulaverakis M, George A, Fisher LW, Butler WT, Qin C, Salih E, Tan M, Fujimoto Y, Spevak L, Boskey AL. In vitro effects of dentin matrix protein-1 on hydroxyapatite formation provide insights into in vivo functions. J.Biol.Chem. 2004;279:18115–18120. doi: 10.1074/jbc.M314114200. [DOI] [PubMed] [Google Scholar]
- 30.Jadlowiec JA, Zhang X, Li J, Campbell PG, Sfeir C. ECM-mediated signaling by dentin phosphophoryn involves activation of the Smad pathway independent of BMP. J.Biol.Chem. 2005 doi: 10.1074/jbc.M506158200. [DOI] [PubMed] [Google Scholar]
- 31.Bianco P, Fisher LW, Young MF, Termine JD, Robey PG. Expression of bone sialoprotein (BSP) in developing human tissues. Calcif.Tissue Int. 1991;49:421–426. doi: 10.1007/BF02555854. [DOI] [PubMed] [Google Scholar]
- 32.Javed A, Gutierrez S, Montecino M, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Multiple Cbfa/AML sites in the rat osteocalcin promoter are required for basal and vitamin D-responsive transcription and contribute to chromatin organization. Mol.Cell Biol. 1999;19:7491–7500. doi: 10.1128/mcb.19.11.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Javed A, Barnes GL, Jasanya BO, Stein JL, Gerstenfeld L, Lian JB, Stein GS. runt homology domain transcription factors (Runx, Cbfa, and AML) mediate repression of the bone sialoprotein promoter: evidence for promoter context-dependent activity of Cbfa proteins. Mol.Cell Biol. 2001;21:2891–2905. doi: 10.1128/MCB.21.8.2891-2905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siler U, Rousselle P, Muller CA, Klein G. Laminin gamma2 chain as a stromal cell marker of the human bone marrow microenvironment. Br.J.Haematol. 2002;119:212–220. doi: 10.1046/j.1365-2141.2002.03800.x. [DOI] [PubMed] [Google Scholar]
- 35.Kingsley K, Rust WL, Huff JL, Smith RC, Plopper GE. PDGF-BB enhances expression of, and reduces adhesion to, laminin-5 in vascular smooth muscle cells. Biochem.Biophys.Res.Commun. 2002;294:1017–1022. doi: 10.1016/S0006-291X(02)00592-2. [DOI] [PubMed] [Google Scholar]
- 36.Kingsley K, Plopper GE. Platelet-derived growth factor modulates rat vascular smooth muscle cell responses on laminin-5 via mitogen-activated protein kinase-sensitive pathways. Cell Commun.Signal. 2005;3:2. doi: 10.1186/1478-811X-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klees RF, Salasznyk RM, Kingsley K, Williams WA, Boskey A, Plopper GE. Laminin-5 induces osteogenic gene expression in human mesenchymal stem cells through an ERK-dependent pathway. Mol.Biol.Cell. 2005;16:881–890. doi: 10.1091/mbc.E04-08-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salasznyk RM, Klees RF, Boskey A, Plopper GE. Activation of FAK is necessary for the osteogenic differentiation of human mesenchymal stem cells on laminin-5. J.Cell Biochem. 2007;100:499–514. doi: 10.1002/jcb.21074. [DOI] [PubMed] [Google Scholar]
- 39.Hashimoto J, Ogawa T, Tsubota Y, Miyazaki K. Laminin-5 suppresses chondrogenic differentiation of murine teratocarcinoma cell line ATDC5. Exp.Cell Res. 2005;310:256–269. doi: 10.1016/j.yexcr.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 40.Plopper G, Falk-Marzillier J, Glaser S, Fitchmun M, Giannelli G, Romano T, Jones JC, Quaranta V. Changes in expression of monoclonal antibody epitopes on laminin-5r induced by cell contact. J.Cell Sci. 1996;109(Pt 7):1965–1973. doi: 10.1242/jcs.109.7.1965. [DOI] [PubMed] [Google Scholar]
- 41.Plopper GE, Domanico SZ, Cirulli V, Kiosses WB, Quaranta V. Migration of breast epithelial cells on Laminin-5: differential role of integrins in normal and transformed cell types. Breast Cancer Res.Treat. 1998;51:57–69. doi: 10.1023/a:1006086218174. [DOI] [PubMed] [Google Scholar]
- 42.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 43.Kato Y, Boskey A, Spevak L, Dallas M, Hori M, Bonewald LF. Establishment of an osteoid preosteocyte-like cell MLO-A5 that spontaneously mineralizes in culture. J.Bone Miner.Res. 2001;16:1622–1633. doi: 10.1359/jbmr.2001.16.9.1622. [DOI] [PubMed] [Google Scholar]
- 44.Mizushima H, Takamura H, Miyagi Y, Kikkawa Y, Yamanaka N, Yasumitsu H, Misugi K, Miyazaki K. Identification of integrin-dependent and -independent cell adhesion domains in COOH-terminal globular region of laminin-5 alpha 3 chain. Cell Growth Differ. 1997;8:979–987. [PubMed] [Google Scholar]
- 45.Shang M, Koshikawa N, Schenk S, Quaranta V. The LG3 module of laminin-5 harbors a binding site for integrin alpha3beta1 that promotes cell adhesion, spreading, and migration. J.Biol.Chem. 2001;276:33045–33053. doi: 10.1074/jbc.M100798200. [DOI] [PubMed] [Google Scholar]
- 46.Salasznyk RM, Klees RF, Williams WA, Boskey A, Plopper GE. Focal adhesion kinase signaling pathways regulate the osteogenic differentiation of human mesenchymal stem cells. Exp.Cell Res. 2007;313:22–37. doi: 10.1016/j.yexcr.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calaycay J, Pande H, Lee T, Borsi L, Siri A, Shively JE, Zardi L. Primary structure of a DNA- and heparin-binding domain (Domain III) in human plasma fibronectin. J.Biol.Chem. 1985;260:12136–12141. [PubMed] [Google Scholar]
- 48.Pierschbacher MD, Ruoslahti E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature. 1984;309:30–33. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- 49.Ruoslahti E, Pierschbacher MD. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238:491–497. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 50.Schenk S, Quaranta V. Tales from the crypt[ic] sites of the extracellular matrix. Trends Cell Biol. 2003;13:366–375. doi: 10.1016/s0962-8924(03)00129-6. [DOI] [PubMed] [Google Scholar]
- 51.Yang B, Jung D, Motto D, Meyer J, Koretzky G, Campbell KP. SH3 domain-mediated interaction of dystroglycan and Grb2. J.Biol.Chem. 1995;270:11711–11714. doi: 10.1074/jbc.270.20.11711. [DOI] [PubMed] [Google Scholar]
- 52.Benoit DS, Durney AR, Anseth KS. The effect of heparin-functionalized PEG hydrogels on three-dimensional human mesenchymal stem cell osteogenic differentiation. Biomaterials. 2007;28:66–77. doi: 10.1016/j.biomaterials.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 53.Folkman J. Antiangiogenesis in cancer therapy--endostatin and its mechanisms of action. Exp.Cell Res. 2006;312:594–607. doi: 10.1016/j.yexcr.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 54.Fjeldstad K, Kolset SO. Decreasing the metastatic potential in cancers--targeting the heparan sulfate proteoglycans. Curr.Drug Targets. 2005;6:665–682. doi: 10.2174/1389450054863662. [DOI] [PubMed] [Google Scholar]
- 55.Ryaby JT, Sheller MR, Levine BP, Bramlet DG, Ladd AL, Carney DH. Thrombin peptide TP508 stimulates cellular events leading to angiogenesis, revascularization, and repair of dermal and musculoskeletal tissues. J.Bone Joint Surg.Am. 2006;88(Suppl 3):132–139. doi: 10.2106/JBJS.F.00892. [DOI] [PubMed] [Google Scholar]