Abstract
A series of small-molecule microbicides has been developed for vaginal delivery to prevent heterosexual HIV transmission, but results from human clinical trials have been disappointing. Protein-based microbicides, such as HIV-specific monoclonal antibodies, have been considered as an alternative approach. Despite their promising safety profile and efficacy, the major drawback of such molecules is the economy of large-scale production in mammalian cells, the current system of choice. Here, we show that an alternative biomanufacturing platform is now available for one of the most promising anti-HIV antibodies (2G12). Our data show that the HIV-neutralization capability of the antibody is equal to or superior to that of the same antibody produced in CHO cells. We conclude that this protein production system may provide a means to achieve microbicide ingredient manufacture at costs that would allow product introduction and manufacture in the developing world.
The number of people infected with HIV rose to just more than 40 million in 2006, an increase of over 2 million since 2004 (1). More than 60% of infected people live in sub-Saharan Africa, where at least 2 million deaths from HIV/AIDS occurred in 2006. Novel therapeutic strategies are urgently needed for deployment alongside conventional antiretroviral drugs, vaccines, and microbicides to prevent the spread of the disease. Broadly reactive human monoclonal antibodies (mAbs) against HIV could be used as both prophylactic and therapeutic modalities.
HIV-1 entry into susceptible cells is mediated by the envelope protein (Env), which comprises a trimer of gp120/gp41 heterodimers, with gp120 acting as the external surface subunit responsible for engaging cellular receptors, and gp41 as the transmembrane subunit that mediates membrane fusion (2). Infection occurs when gp120 interacts with cellular CD4 and then a coreceptor, usually CCR5 or CXCR4. Env is therefore an ideal target for neutralizing antibodies, and four mAbs with broad HIV-neutralizing activity have been characterized: the anti-gp120 antibodies b12 (3) and 2G12 (4) and the anti-gp41 antibodies 2F5 (5) and 4E10 (6).
The Env protein has evolved defenses to prevent neutralization, which few antibodies can overcome (7). The 2G12 antibody is an exception (8) and achieves neutralization by recognizing a unique gp120 epitope that although not directly associated with the receptor-binding sites (9) still prevents the virus interacting with its receptors (10). As well as neutralizing HIV-1 in vitro, 2G12 can protect macaques from simian-HIV challenge in passive immunization experiments, usually in combination with other antibodies (11). The crystal structure of the gp120 core shows that the epitope is highly conserved despite being located on the relatively variable surface of the outer domain, opposite to the CD4-binding site (12). The epitope contains high-mannose glycans to which the antibody probably binds (12).
The 2G12, 2F5 and 4E10 antibodies have been produced in Chinese hamster ovary (CHO) cells for prophylactic (13) and therapeutic (14) use, but the limitations of this platform (including high cost, low capacity, low scalability, and safety issues) (15) mean that large-scale production for use in developing countries is unfeasible (16). Among the alternative systems available for biopharmaceutical manufacture, only plants provide the scalability and economy required to meet the anticipated demand for such products in the HIV-endemic regions of Africa and Asia at a price the market can bear (17–20). Cereal seeds are likely to be the most suitable platform for deployment in such areas, because the infrastructure for large-scale cultivation and harvesting is already in place, and the dry seeds favor product stability (21). Maize is particularly attractive, because it has been developed as a commercial platform for recombinant protein production by a number of companies, and its success as a production system for pharmaceutical proteins is widely documented (22–24).
Because of the lack of direct comparative studies, it is unclear whether plant-derived antibodies are functionally comparable to antibodies produced in mammalian cells, particularly because of the differences in glycan structures between plants and mammals. Here, we show that a monoclonal antibody produced in maize seeds is not only comparable in terms of antigen-binding activity with its CHO-derived counterpart, but it is at least as active, if not superior, in HIV-neutralization assays. We also show that it is possible to isolate the antibody from maize seeds efficiently and to achieve 90% purity, using a simple two-step extraction process, which could be scaled up much more economically than the Protein A chromatography systems currently favored for industrial-scale antibody production in mammalian cells. Furthermore, by selecting a maize line with high-level antibody expression and subjecting it to a dedifferentiation–regeneration cycle, we were able to increase the accumulation of 2G12 to well over 100 μg per gram of dry seed weight, much higher than previously reported. We conclude that both the product and process can be economically advantageous when maize is used to express recombinant pharmaceutical proteins for humans.
Results
Transgenic Maize Plants.
A large number of transgenic events were generated in the elite maize cultivar M37W by particle bombardment with vectors pTRAGtiGH and pTRAGtiGL (containing secretable forms of the 2G12 heavy and light chain genes, respectively, under the control of an endosperm-specific promoter) and pTRAuxBar (containing the bar selectable marker gene, expressed under a constitutive promoter to facilitate phosphinothricin selection). The plants were either selfed or crossed with wild-type M37W, and Southern blot analysis was used to confirm transgene integration and stability. Most of the transgenic plants contained all three input transgenes linked within a single genetic locus, as anticipated (25, 26). Single genetic loci are beneficial, because they favor high-level and stable transgene expression over future generations (27–31).
2G12 Expression in Seeds and Selection of a Highly Expressing Plant Line.
Transgenic seeds (T1 generation) were separated into embryo and endosperm and screened by dot-blot analysis [supporting information (SI) Fig. 3], and the amount of 2G12 in the endosperm was estimated by ELISA. Western blot analyses were carried out under reducing conditions to confirm the presence of the 50-kDa heavy chain and 25-kDa light chain. Embryos from such seeds with high 2G12 expression levels were germinated, and the resulting plants self-pollinated. The new seeds (T2 generation) were then analyzed by dot-blot analysis (SI Fig. 4) and ELISA, identifying event 3C as the highest expressing line. The endosperm tissue from 30 additional 3C seeds was tested by dot-blot analysis and all seeds were shown to be high expressers. Biacore surface plasmon resonance (SPR) spectroscopy showed considerable variation in the concentration of 2G12 among seeds tested from event 3C (SI Fig. 5). Immature zygotic embryos from event 3C were dedifferentiated in vitro, and numerous plants were regenerated from the resulting callus tissue. These plants accumulated even higher levels of 2G12 (see below).
Purification and Quantitation of 2G12.
Affinity purification with Protein A.
The 2G12 antibody was purified from maize by protein A affinity chromatography, which specifically binds the Fc portion of the antibody heavy chain. The flow-through and elution fractions were analyzed on-line by Biacore SPR spectroscopy. Light chain was detected in the flow-through fraction, which indicated that unassembled light chain was present in planta (i.e., the light chain was produced in excess). No heavy chain was detected in the flow-through, indicating that all of the heavy chain produced in planta was folded correctly (at least in the vicinity of the Fc region) and retained on the affinity matrix. Of the possible intact species that might be found in the elution fraction (H2L2, H2L, and H2), only H2L2 was detected, indicating highly efficient antibody assembly in planta. The antibody was eluted into a low-pH buffer to prevent aggregation and precipitation.
The fate of the antibody chains was also monitored by SDS/PAGE (Fig. 1A) and Western blot (Fig. 1B), confirming the presence of the heavy and light antibody chains (revealed as 50- and 25-kDa bands, respectively) before dialysis and concentration to 3.42 mg/ml (as determined by Biacore SPR spectroscopy using a protein L surface). Some degradation products (≈40 kDa) were visible in the pooled protein samples (indicated by red arrows in Fig. 1). The calculated extractable yield of 2G12 was 75 μg per gram of dry seed weight, 75% of which was recovered. Levels of 2G12 in seeds of plants regenerated from dedifferentiated immature zygotic embryos exceeded 100 μg per gram of dry seed weight. Furthermore, the dedifferentiation-regeneration cycle eliminated the wide variability in 2G12 expression we had observed previously in seeds of primary transformants and their progeny.
Fig. 1.
Analysis of the 2G12 purification process. (A) Coomassie staining after SDS/PAGE (reducing conditions) of fractions taken during 2G12 purification by protein A chromatography. (B) Western blot (reducing conditions) of the 2G12 heavy and light chains from fractions taken during the same purification process. The presence of the heavy chain (HC), light chain (LC), and degradation products (D) are shown with arrows. An excess of light chain can be seen in the flow-through lane. Lanes are marked as follows: C, control; M, markers; L, load; F, flow-through; and W, wash. Numbers represent elution fractions 6, 7, 8–12 pooled, 13, and 14.
Non-protein A purification.
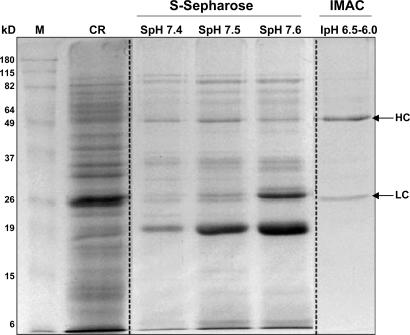
The 2G12 antibody was also purified by using S-Sepharose FF cation exchange chromatography, followed by immobilized metal affinity chromatography (IMAC) on Zn2+- iminodiacetic acid (IDA)-agarose. The antibody was adsorbed at pH 6.0 and subsequently eluted in a stepwise pH gradient (pH 7.0–8.0). Bulk maize endosperm protein (acidic pI) either did not bind to the cation exchanger or eluted at pH <7.2 (Fig. 2). Elution fraction corresponding to pH values between 7.4 and 7.6 in the first step separation favor the final obtainment of a highly pure antibody preparation. This procedure resulted in 10- to 15-fold purification of 2G12 with >70% recovery. Pooled eluted fractions (pH 7.4, 7.5, and 7.6) were loaded directly onto Zn2+-IDA-agarose without any pretreatment and 2G12 was eluted in a stepwise pH gradient (pH 6.5 and 6.0) with step-recovery of ≈80%. Analysis of the eluted fractions by SDS/PAGE (Fig. 2) showed that the antibody was ≈90% pure. The overall recovery was 50–60%.
Fig. 2.
SDS/PAGE of selected fractions from non-protein A purification procedure. Lanes are marked as follows: M, markers; CR, transgenic maize crude extract; SpH 7.4-SpH 7.6, pool of eluted fractions at pH 7.4–7.6 from S-Sepharose FF cation exchanger; and IpH 6.5–6.0, pool of eluted fractions at pH 6.5–6.0 from IMAC.
Antigen Binding Assays.
Biacore SPR spectroscopy was used to compare the antigen-binding properties of the maize-derived antibody with its CHO-derived counterpart. Three surfaces were used: protein A (detects any species containing a correctly folded heavy chain), protein L (detects any species containing a correctly folded light chain), and gp120 (detects the assembled antibody only, not the free chains). The three surfaces also detect related degradation products with intact binding domains, although the signal would be weaker because of the lower molecular mass. By measuring ratios between the signals, i.e., the protein L/protein A ratio (SI Fig. 6A), the gp120/protein A ratio (SI Fig. 6B), and the gp120/protein L ratio (SI Fig. 6C), it is possible to characterize the binding properties of each antibody accurately. The results (Table 1) show that the gp120/protein L ratios of the maize and CHO 2G12 antibodies are the same, whereas the protein L/protein A and gp120/protein A ratios differ by the same relative amount. This deviation is caused by an increased protein A signal, meaning that the maize-derived 2G12 preparation contains an excess of heavy chain. Because we have already established that an excess of light chain is produced in planta and that partially assembled intact antibody forms are not eluted, the most likely explanation for the excess heavy chain in the eluted antibody preparation is the degradation products visible in Fig. 1. These heavy-chain degradation products do not bind to the antigen, and the gp120/protein L ratio confirms that the intact assembled antibody has full antigen-binding activity.
Table 1.
Biacore characterization of the antigen-binding properties of the maize and CHO antibodies
| Binding ratios | CHO2G12 | zm2G12 | CHO, % |
|---|---|---|---|
| protein-L/protein-A | 0.7114 ± 0.0051 | 0.6147 ± 0.0038 | 86.4 ± 0.5 |
| gp120/protein-A | 0.2849 ± 0.0018 | 0.2473 ± 0.0017 | 86.8 ± 0.6 |
| gp120/protein-L | 0.4002 ± 0.0023 | 0.4022 ± 0.0022 | 100.5 ± 0.6 |
Protein-L/protein-A measures the presence of degradation products and unassembled or partially assembled antibody chains. gp120 surfaces/protein-A and gp20/protein-L measure the specific antigen binding activity.
Glycan Analysis.
Tryptic glycopeptides from maize 2G12 were analyzed by ESI-Q-TOF mass spectrometry, yielding valuable additional data on the protein sequence. The heavy chain from the protein A-purified antibody contained the expected, correctly processed N-terminal peptide EVQLVESGGGLVK, which was absent from the degradation product. The full-size heavy chain partially lacked the C-terminal lysine residue, as was recently found in the IGN314 antibody produced in moss (32), whereas this residue was absent from most of the degradation product. The continuous peptide map of the degradation product ran from L123 to K472, with the proteolytic cleavage site presumably located between G119 and L123.
Tryptic in-gel digestion of the heavy chains yielded two, a fully and an incompletely cleaved tryptic glycopeptides (i.e., EEQYN297STYR and TKPREEQYN297STYR, respectively), which both represent the same glycan acceptor site. There were striking differences in the glycan profiles of the two heavy chains (Table 2). More than half (54.5%) of the full-sized chains contained a single GlcNAc residue at the acceptor site, whereas only 17% of the degraded product carried this residue. Approximately 6% of the full-size product carried high-mannose glycans, whereas 28% carried complex type glycans, including fucose and xylose residues. In contrast, no high-mannose glycans were detected on the degraded heavy chain, and 67% carried complex glycans. A similar proportion of both types of heavy chain were nonglycosylated (11.5% for the full-size chain, 16% for the degradation product).
Table 2.
Q-TOF analysis of tryptic glycopeptides from the purified, assembled 2G12 antibody isolated from maize seed endosperm and the 40-kDa degradation product thereof
| Structure | Intensity |
|
|---|---|---|
| Full-size 2G12 | Degraded 2G12 | |
| Nonglycosylated | 11.5 | 16.1 |
| Oligomannose-type N-glycans | 5.9 | nd |
| Complex-type N-glycans | 27.9 | 67.0 |
| Single GlcNAc | 54.6 | 16.9 |
The group of oligomannose-type glycans consisted of the N-glycans Man5GlcNAc2 to Man9GlcNAc2 in similar orders of magnitude. Complex-type N-glycans comprised the structures MUXF3, MMXF3, GnMXF3/MGnXF3, and GnGnXF3 in roughly similar amounts in both samples. nd, none detected.
Virus Neutralization Assays.
Virus-neutralization assays were performed with matched preparations of 2G12 derived from maize and CHO cells, i.e., preparations matched in terms of the amount of antibody capable of binding to the antigen, as established by Biacore analysis. HIV-1 neutralization was determined by using a syncytium inhibition assay. The 50% inhibiting concentration (IC50) of maize-derived 2G12 was 2.88 μg/ml, whereas that of the control (CHO) antibody was 8.11 μg/ml, demonstrating that the maize 2G12 preparation was nearly three times as potent as its CHO counterpart. This apparent increase in neutralization efficiency probably reflects the presence of dimers and other aggregates in the maize preparation, which are known to have higher neutralization ability than monomeric antibody forms (33).
Discussion
Protein microbicides against HIV are promising alternatives to the current generation of small-molecule drugs and have performed well in preclinical and clinical studies (34). However, producing and distributing such drugs in key HIV-endemic areas is difficult and expensive because of the lack of fermenter capacity and the absence of a cold chain, meaning that those people most likely to benefit from the microbicides are effectively excluded on economic grounds. Many therapeutic heterologous proteins have been expressed in plants, including a large number of vaccine candidates and various recombinant antibody formats for veterinary and human use (35). The advantages of plants over fermenter-based systems are described in refs. 18, 19, and 36, but, in the context of our study, the most important benefits of using maize seeds include the enhanced stability of recombinant proteins accumulating in the endosperm, which means that a cold chain is not necessary for product distribution (17, 37, 38), and the economy of large-scale production both in terms of the upstream phase and downstream processing, which will make the product affordable without detracting from its safety or performance (17, 21).
One often-cited limitation of plant-based production systems is the low yield of recombinant protein (21). We have previously demonstrated that the methylation clock in plants can be reset by dedifferentiation, resulting in significant increases in levels of transgene expression (39). By subjecting immature zygotic embryos from one 2G12 highly expressing line to dedifferentiation and subsequent regeneration, we were able to not only boost accumulation levels by 30–40% compared with plants restricted to the sexual reproduction cycle but also to eliminate most of the seed-to-seed variation.
Another criticism of plant-based platforms for pharmaceutical protein production is that, although plants process recombinant proteins in a very similar way to mammalian cells, differences in glycosylation result in plant-specific glycan structures that could modify the biological properties of recombinant human glycoproteins produced in plants. High mannose glycans are the same in plants and mammalian cells, but the processing of complex glycans is distinct, resulting in the inability of plants to incorporate sialic acid on N-linked glycans and the introduction of fucose and xylose linkages that are not found in mammalian cells (40). For injectable products, the presence of plant glycans might result in immunogenic reactions, but because our product is intended for topical application, the potential immunogenicity should not be an issue (21).
To investigate these differences functionally, we set out to produce a well characterized therapeutic antibody in maize and test its efficacy against the same antibody produced in CHO cells. The 2G12 heavy and light chain genes were introduced into maize, and one homozygous line (3C) was identified after screening over two generations that expressed 2G12 at levels exceeding 75 μg per gram of dry seed weight, which is considerably higher than the typical levels for pharmaceutical proteins expressed from the plant nuclear genome.
The first step was to obtain a preparation of the antibody equivalent in terms of purity to the product from CHO cells. The downstream processing of antibodies produced in plants plays an important role in the technical and economical feasibility of large-scale applications. For this reason, in addition to the use of a standard protein A affinity chromatography purification method, we evaluated a two-step non-protein A method as an alternative. Protein A binds with great affinity and specificity to the Fc portion of full-length antibodies and is widely used in commercial antibody production. However, the resin is very expensive (particularly when used in industrial scale chromatography columns) and can undergo degradation (41) and leach into the final product, causing toxicity (42). However, conventional chromatographic methods are suitable for large-scale processes because they are less expensive, easier to use, and largely resistant to chemical and biological degradation (43, 44). Therefore, we developed a two-step strategy that exploited two intrinsic properties of IgG1 monoclonal antibodies: their pI and their ability to bind metal ions because of the presence of a surface-accessible histidine-rich region near the C terminus. The procedure consisted of an initial S-Sepharose FF cation exchange chromatography step, followed by an IMAC step, using Zn2+-IDA-agarose, yielding 2G12 at 90% purity and achieving ≈50–60% recovery. As well as using inexpensive media, the antibody could be recovered under mild nondenaturing conditions, a significant advantage for therapeutic proteins.
Once purified, the maize 2G12 was characterized by SDS/PAGE and Western blot analysis. The heavy and light chains comigrated with their counterparts from the CHO antibody (50 kDa for the heavy chain, 25 kDa for the light chain) confirming their structural integrity and approximately equivalent molecular masses. Western blot analysis suggested that the light chain was produced in excess (visible in the flow-through lane in Fig. 1B) as has been reported for other full-length antibodies (45, 46).
A comparison of the functional properties of maize 2G12 and its CHO counterpart was carried out by performing antigen binding assays, using SPR spectroscopy and virus-neutralization assays based on syncytium inhibition. We found no significant difference between the two intact antibodies in terms of their binding activity as determined through Biacore analysis, although binding to the protein A surface was higher in the case of the maize antibody because of the presence of heavy chain degradation products that were unable to bind the antigen. We anticipated that the antibodies would also be equivalent in terms of their neutralization activity, but we found that the maize antibody was three times more efficient than the commercially available CHO 2G12. The most likely explanation for this is the presence of aggregates in the maize-derived 2G12, because aggregates are known to have greater virus neutralizing activity. The absence of aggregates and degradation products in the CHO preparation reflects the addition processing steps that have been carried out on this antibody, specifically the gel filtration step, which removes products with identical physicochemical and functional properties but distinct molecular masses.
We conclude that 2G12, an HIV-neutralizing monoclonal antibody, has been expressed successfully in maize endosperm at a mean concentration of ≈75 μg per gram of dry seed weight. The antibody can be recovered efficiently from maize kernels at 90% purity, using two distinct processing strategies, one based on protein A affinity chromatography and the other based on a more economical two-step process, using ion exchange chromatography and IMAC, which should be applicable on a large scale. The purified maize-derived 2G12 was physically identical to its CHO-derived counterpart with the exception of its glycan structure, and in functional terms it showed identical antigen-binding activity but nearly three times the efficacy in HIV-neutralization assays, probably because of the presence of aggregates. Further antibodies produced in maize are currently undergoing testing to see whether the encouraging results achieved with 2G12 can be repeated for other molecules. If so, this would represent a remarkable opportunity to produce therapeutic antibodies inexpensively and on a massive scale, making it much more likely that antibody-based therapeutics could be made more widely available at a much lower cost than currently possible.
Methods
Transformation Vectors.
All transformation constructs were based on the binary vector pTRA, a derivative of pPAM (GenBank accession no. AY027531) containing two tobacco RB7 scaffold attachment regions flanking the expression cassette (47). The coding regions of the 2G12 heavy and light chains (obtained from Polymun) contained an N-terminal signal sequence targeting the polypeptide to the secretory pathway. The expression cassette comprised the endosperm-specific rice glutelin-1 promoter, the Tobacco Etch Virus 5′ leader, the coding region, and the CaMV 35S terminator, resulting in final constructs pTRAgtiGH and pTRAgtiGL. The third construct (pTRAuxbar) contained the bar gene between the constitutive maize ubiquitin-1 promoter and 35S terminator. All expression cassettes contained the maize ubiquitin-1 first intron.
Transformation, Selection, and Regeneration of Transgenic Plants.
Immature zygotic embryos of the South African elite white maize genotype M37W were transformed by particle bombardment at 10–14 days after pollination as described in ref. 26. Bombarded embryos were transferred in the dark every 2 weeks to fresh N6-based medium containing 3 mg/liter phosphinothricin (PPT). Four to 6 weeks after bombardment, pieces of PPT-resistant embryogenic type I callus were transferred to regeneration medium containing 3 mg/liter PPT for 2–4 weeks with a 16-h/8-h (day/night) photoperiod. Developing plantlets with well formed shoots and roots were hardened off in soil. All experiments were performed at 25°C. Embryos from a lead event (3C) were subjected to a dedifferentiation regeneration cycle by reintroduction into callus culture following the procedure described above for transformation and regeneration, in the absence of selection. Numerous regenerants were recovered, and all were shown by DNA analysis to be clones of the original transformant 3C.
Screening Seeds for 2G12 Expression.
We analyzed five T1 seeds per transgenic event by removing and germinating the embryos and extracting total soluble protein from the endosperm, using three volumes of PBS. After centrifugation, 4 μl of supernatant was spotted individually onto a nitrocellulose membrane and blocked for 1 h. The membrane was then incubated at room temperature overnight with an alkaline phosphatase-labeled antibody [goat anti-human IgG Fc chain (Sigma A9544) or goat anti-human kappa chain (Sigma A3813)]. Blots were washed three times with PBS buffer supplemented with 2% Tween-20 and the signal detected with Sigma FAST BCIP/NBT dissolved in distilled water.
SDS/PAGE, Western blot, and ELISAs.
Proteins were separated by SDS/PAGE in a 12% denaturing gel, and Western blots were carried out by using standard methods and the same antibodies as used in the dot-blot anaylses described above. For quantitation of 2G12 expression, 10 T2 seeds each from events 3C (high expression) and 1G (low expression) were analyzed by ELISA. The capture antibody [goat anti-human kappa chain (Sigma K3502) or goat anti-human IgG Fc chain (Sigma K2136) at 1:200 dilution] was coated directly onto 96-well Nunc-Immuno Maxisorp surface plates (Nalge Nunc) and incubated overnight at 4°C. Plates were blocked with 5% nonfat milk for 2 h and then washed with PBS containing 0.2% Tween-20. Samples diluted in this buffer were added to the wells and incubated for 2–3 h at room temperature. After washing, the alkaline phosphatase-labeled antibodies used for dot-blot analysis were added to the plates at 1:1,000 dilution and incubated at room temperature for 2 h before the signal was detected with p-nitrophenyl phosphate. After 4–5 min, the signal was quantified by measuring the absorbance at 415 nm.
Protein A Affinity Purification.
Maize seeds were crushed to a fine powder and extracted overnight at 4°C in five volumes of buffer [1× PBS, 5 mM EDTA, and 1 mM 2-mercaptoethanol (pH 7.4)]. Insoluble material and fat deposits were removed by centrifuging twice at 8,000 × g for 30 min at 4°C. The supernatant was filtered and the antibody concentration determined by Biacore analysis. The sample was loaded onto a protein A affinity column (ceramic HyperDF) at a flow rate of 2 ml/min. The column was washed with PBST and PBS, and the antibody was eluted with 100 mM glycine (pH 3.6) containing 100 mM fructose, and buffered with one-tenth volume of 1 M acetate/sodium acetate (pH 4.75). Antibody-containing fractions were identified by the droplet Bradford method, and 2G12 concentrations were determined by Biacore. Fractions containing >50 μg/ml were pooled and dialyzed for 2 days against 10 mM acetate/sodium acetate (pH 4.75) containing 1 mM EDTA and 0.1 mM 2-mercaptoethanol (omitted in final dialysis step). The dialyzed antibody was concentrated by ultrafiltration, using spin-columns with a 30-kDa molecular mass cut-off. Biacore was used to monitor all steps. The final antibody concentration was determined by Biacore, using CHO-2G12 as standard.
Non-Protein A Purification.
Maize seeds (5 g) were extracted by blending for 10 min in 20 ml of 20 mM sodium phosphate buffer (pH 6.0) followed by rotary mixing for 1 h at room temperature. After centrifuging at 10,000 × g for 30 min, the supernatant was collected and passed through a 0.45-μm filter. Cation exchange chromatography was carried out on 1 ml of filtered extract, using S-Sepharose FF (0.5 ml of moist adsorbent) at 4°C after equilibration with 20 mM sodium phosphate buffer (pH 6.0). The column was then washed with a series of buffers of increasing pH (20 mM sodium phosphate at pH 6.0, then pH 6.5, then pH 6.75), and the antibody was eluted by using stepwise increases in pH from pH 7.0 to 8.0. All chromatographic steps were performed at a flow rate of 0.5 ml/min. Collected fractions (1.5 ml) were analyzed by ELISA, and total protein was determined by the Bradford assay. Pooled eluted fractions at pH 7.4–7.6 (4.5 ml) were loaded onto IDA-agarose resin (0.5 ml of moist adsorbent) at 4°C charged with Zn2+, using a solution of zinc chloride (2 ml, 0.1 M) and equilibrated with sodium phosphate buffer (50 mM, pH 7.5) containing 1 M NaCl. The column was washed with 10 ml of equilibration buffer, and elution was achieved by using a stepwise pH gradient (pH 6.5–5.0 in 50 mM sodium phosphate buffer containing 1 M NaCl). Collected fractions were analyzed by ELISA, and total protein was determined by the Bradford assay.
Surface Plasmon Resonance (SPR) spectroscopy.
SPR assays were performed at 25°C with a flow rate of 30 μl/min, using a BIACORE 2000 (GE Healthcare), a CM5-rg sensor chip and HBS-EP as the running buffer. Ligands were immobilized by using the standard EDC/NHS protocol. The first flow cell was activated/deactivated and used for blank subtraction. Immobilization buffers and levels were 4,200 resonance units (RU) for protein A [200 μg/ml and 10 mM sodium acetate (pH 4.5)], 2,300 RU for protein L [100 μg/ml and 10 mM sodium acetate (pH 4.5)] and 12,000 RU for gp120 [(10 mM sodium acetate (pH 4.75)]. Protein A and protein L surfaces were regenerated with a 30-s pulse of 30 mM HCl, whereas the gp120 surface was regenerated with 0.5 M citrate, pH 3.0. CHO-derived 2G12 was used as standard. Triplicate dilution series were injected simultaneously over the four flow cells. The blank-subtracted endpoint signals were used to evaluate the quality of the preparations. All surfaces exhibited high binding capacities and strong mass-transport limitation, resulting in linear dose-response relationships and constant binding rates up to an antibody concentration of 1 μg/ml. Data were evaluated by using BIAevaluation software, Version 4.0 and Microcal Origin software, Version 5.0 software. The binding signal ratios were derived from the endpoint responses for extracts or determined by pair wise plotting of the binding signals obtained at different dilutions and linear regression analysis for purified preparations.
N-Glycan Analysis.
Antibody bands from Coomassie-stained SDS/PAGE were destained, carbamidomethylated, digested with trypsin, and extracted from gel pieces as described in ref. 44. The subsequent fractionation of the peptides by capillary reversed phase chromatography with detection by a Q-TOF Ultima Global (Waters Micromass) mass spectrometer was performed as described in refs. 48 and 49.
The MS-data from the tryptic peptides were analyzed as described in ref. 49 and compared with datasets generated by in silico tryptic digestion of the 2G12 coding, using the PeptideMass program (available at www.expasy.org/tools/peptide-mass.html). Based on the tryptic peptide datasets, tryptic glycopeptide datasets were generated by the addition of glycan mass increments to the masses of the two identified glycopeptides.
Neutralization Assays.
HIV-1 neutralization was assessed by using a syncytium inhibition assay. Ten twofold serial dilutions (start concentration: 100 μg/ml) of Zm2G12, CHO2G12, and a nonneutralizing control were preincubated with HIV-1 RF at 10 half-maximum tissue culture infectious dose (2, 42) per milliliter for 1 h at 37°C. CD4-positive human AA-2 cells were added at a density of 4 × 105 cells per milliliter and incubated for a further 5 days. Experiments were performed with eight replicates per antibody dilution step. The presence of one or more syncytium per well after 5 days was scored as positive for infection. The 50% inhibiting concentrations (IC50) were calculated by the method of Reed and Muench (50), using the concentrations present during the antibody-virus preincubation step.
Supplementary Material
ACKNOWLEDGMENTS.
This work was supported by European Union Framework 6 Program—The Pharma-Planta Integrated Project Grant LSH-2002–1.2.5–2; Acciones Complementarias (MEC) Grant BIO2005–24826-E; Generalitat de Catalunya Grant 2005SGR118; the Ramon y Cajol Program; and Center CONSOLIDER, Ministerio de Educación y Ciencia, Spain.
Footnotes
The author declares no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708841104/DC1.
References
- 1.WHO/UNAIDS. AIDS Epidemic Update: Special Report on HIV/AIDS, December 2006. Geneva: UN/WHO; 2006. [Google Scholar]
- 2.Poignard P, Saphire EO, Parren PW, Burton DR. Annu Rev Immunol. 2001;19:253–274. doi: 10.1146/annurev.immunol.19.1.253. [DOI] [PubMed] [Google Scholar]
- 3.Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, et al. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 4.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conley AJ, Kessler JA, II, Boots LJ, Tung JS, Arnold BA, Keller PM, Shaw AR, Emini EA. Proc Natl Acad Sci USA. 1994;91:3348–3352. doi: 10.1073/pnas.91.8.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, et al. J Virol. 2001;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure. 2000;8:1329–1339. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- 8.Trkola A, Pomales AB, Yuan H, Korber B, Maddon PJ, Allaway GP, Katinger H, Barbas CF, Burton DR, Ho DD, et al. J Virol. 1995;69:6609–6617. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ugolini S, Mondor I, Parren PW, Burton DR, Tilley SA, Klasse PJ, Sattentau QJ. J Exp Med. 1997;186:287–1298. doi: 10.1084/jem.186.8.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fouts TR, Binley JM, Trkola A, Robinson JE, Moore JP. J Virol. 1997;71:2779–2785. doi: 10.1128/jvi.71.4.2779-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, Beary H, Hayes D, Frankel SS, Birx DL, et al. Nature Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 12.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 13.Hofmann-Lehmann R, Vlasak J, Rasmussen RA, Jiang S, Li PL, Baba TW, Montefiori DC, Bernacky BJ, Rizvi TA, Schmidt R, et al. J Virol. 2001;75:7470–7480. doi: 10.1128/JVI.75.16.7470-7480.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stiegler G, Armbruster C, Vcelar B, Stoiber H, Kunert R, Michael NL, Jagodzinski LL, Ammann C, Jäger W, Jacobson J, et al. AIDS. 2002;16:2019–2025. doi: 10.1097/00002030-200210180-00006. [DOI] [PubMed] [Google Scholar]
- 15.Chu L, Robinson D K. Curr Opin Biotechnol. 2001;12:180–187. doi: 10.1016/s0958-1669(00)00197-x. [DOI] [PubMed] [Google Scholar]
- 16.WHO/UNAIDS. Progress in Scaling up Access to HIV Treatment in Low and Middle-income Countries (June Fact Sheet) Geneva: WHO/UNAIDS; 2006. [Google Scholar]
- 17.Stoger E, Ma JKC, Fischer R, Christou P. Curr Opin Biotechnol. 2005;16:167–173. doi: 10.1016/j.copbio.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Ma JKC, Drake PMW, Christou P. Nat Rev Genet. 2003;4:794–805. doi: 10.1038/nrg1177. [DOI] [PubMed] [Google Scholar]
- 19.Fischer R, Stoger E, Schillberg S, Christou P, Twyman RM. Curr Opin Plant Biol. 2004;7:1–7. doi: 10.1016/j.pbi.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Stoger E, Sack M, Fischer R, Christou P. Curr Opin Biotechnol. 2002;13:161–166. doi: 10.1016/s0958-1669(02)00303-8. [DOI] [PubMed] [Google Scholar]
- 21.Twyman RM, Schillberg S, Fischer R. Expert Opin Emerg Drugs. 2005;10:185–218. doi: 10.1517/14728214.10.1.185. [DOI] [PubMed] [Google Scholar]
- 22.Hood EE, Witcher DR, Maddock S, Meyer T, Baszczynski C, Bailey M, Flynn P, Register J, Marshall L, Bond D, et al. Mol Breeding. 1997;3:291–306. [Google Scholar]
- 23.Witcher DR, Hood EE, Peterson D, Bailey M, Bond D, Kusnadi A, Evangelista R, Nikolov Z, Wooge C, Mehigh R, et al. Mol Breeding. 1998;4:301–312. [Google Scholar]
- 24.Hood EE, Woodard SL, Horn ME. Curr Opin Biotechnol. 2002;13:630–635. doi: 10.1016/s0958-1669(02)00351-8. [DOI] [PubMed] [Google Scholar]
- 25.Nicholson L, Gonzalez-Melendi P, van Dolleweerd C, Tuck H, Perrin Y, Ma JK, Fischer R, Christou P, Stoger E. Plant Biotechnol J. 2005;3:115–127. doi: 10.1111/j.1467-7652.2004.00106.x. [DOI] [PubMed] [Google Scholar]
- 26.Drakakaki G, Marcel S, Glahn RP, Lund EK, Pariagh S, Fischer R, Christou P, Stoger E. Plant Mol Biol. 2005;59:869–880. doi: 10.1007/s11103-005-1537-3. [DOI] [PubMed] [Google Scholar]
- 27.Kohli A, Leech M, Vain P, Laurie DA, Christou P. Proc Natl Acad Sci USA. 1998;95:7203–7208. doi: 10.1073/pnas.95.12.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohli A, Gahakwa D, Vain P, Laurie DA, Christou P. Planta. 1999;208:88–97. [Google Scholar]
- 29.Abranches R, Santos AP, Wegel E, Williams S, Castilho A, Christou P, Shaw PJ, Stöger E. Plant J. 2000;24:713–723. doi: 10.1046/j.1365-313x.2000.00908.x. [DOI] [PubMed] [Google Scholar]
- 30.Kohli A, Twyman RM, Abranches R, Wegel E, Stoger E, Christou P. Plant Mol Biol. 2003;52:247–258. doi: 10.1023/a:1023941407376. [DOI] [PubMed] [Google Scholar]
- 31.Kohli A, Gonzales-Melendi P, Abranches R, Capell T, Stoger E, Christou P. Plant Signal Behav. 2006;1:185–195. doi: 10.4161/psb.1.4.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuster M, Jost W, Mudde GC, Wiederkum S, Schwager C, Janzek E, Altmann F, Stadlmann J, Stemmer C, Gorr G. Biotechnol J. 2007;2:700–708. doi: 10.1002/biot.200600255. [DOI] [PubMed] [Google Scholar]
- 33.Wolbank S, Kunert R, Stiegler G, Katinger H. J Virol. 2003;77:4095–4103. doi: 10.1128/JVI.77.7.4095-4103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Clercq E. Nat Rev Drug Discov. 2007;6:1001–1018. doi: 10.1038/nrd2424. [DOI] [PubMed] [Google Scholar]
- 35.Floss DM, Falkenburg D, Conrad U. Transgen Res. 2007;16:315–332. doi: 10.1007/s11248-007-9095-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoger E, Sack M, Perrin Y, Vaquero C, Torres E, Twyman R, Christou P, Fischer R. Mol Breeding. 2002;9:149–158. [Google Scholar]
- 37.Stoger E, Vaquero C, Torres E, Sack M, Nicholson L, Drossard J, Williams S, Keen D, Perrin Y, Christou P, et al. Plant Mol Biol. 2000;42:583–590. doi: 10.1023/a:1006301519427. [DOI] [PubMed] [Google Scholar]
- 38.Twyman RM, Stoger E, Schillberg S, Christou P, Fischer R. Trends Biotechnol. 2003;21:570–578. doi: 10.1016/j.tibtech.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Kohli A, Prynne MQ, Miro B, Pereira A, Twyman RM, Capell T, Christou P. Mol Breeding. 2004;13:177–191. [Google Scholar]
- 40.Bakker H, Bardor M, Molthoff JW, Gomord V, Elbers I, Stevens LH, Jordi W, Lommen A, Faye L, Lerouge P, et al. Proc Natl Acad Sci USA. 2001;98:2899–2904. doi: 10.1073/pnas.031419998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Platis D, Sotriffer CA, Labrou NE. J Chromatogr A. 2006;1128:138–151. doi: 10.1016/j.chroma.2006.06.051. [DOI] [PubMed] [Google Scholar]
- 42.Terman DS, Bertram JH. Eur J Cancer Clin Oncol. 1985;21:1115–1122. doi: 10.1016/0277-5379(85)90001-x. [DOI] [PubMed] [Google Scholar]
- 43.Labrou NE. J Chromatogr B. 2003;790:67–78. doi: 10.1016/s1570-0232(03)00098-9. [DOI] [PubMed] [Google Scholar]
- 44.Shukla AA, Hubbard B, Tressel T, Guhan S, Low D. J Chromatogr B. 2007;841:28–39. doi: 10.1016/j.jchromb.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 45.Giritch A, Marillonnet S, Engler C, van Eldik G, Botterman J, Klimyuk V, Gleba Y. Proc Natl Acad Sci USA. 2006;103:14701–14706. doi: 10.1073/pnas.0606631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Law RD, Russell DA, Thompson LC, Schroeder SC, Middle CM, Tremaine MT, Jury TP, Delannay X, Slater SC. Biochim Biophys Acta. 2006;1760:1434–1444. doi: 10.1016/j.bbagen.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 47.Sack M, Paetz A, Kunert R, Bomble M, Hesse F, Stiegler G, Fischer R, Katinger H, Stoeger E, Rademacher T. FASEB J. 2007;21:1655–1664. doi: 10.1096/fj.06-5863com. [DOI] [PubMed] [Google Scholar]
- 48.Kolarich D, Altmann F. Anal Biochem. 2000;285:64–75. doi: 10.1006/abio.2000.4737. [DOI] [PubMed] [Google Scholar]
- 49.Van Droogenbroeck B, Cao J, Stadlmann J, Altmann F, Colanesi S, Hillmer S, Robinson DG, Van Lerberge E, Terryn N, Van Montagu M, et al. Proc Natl Acad Sci USA. 2006;104:1430–1435. doi: 10.1073/pnas.0609997104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reed LJ, Muench H. Am J Hyg. 1938;27:493–497. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.