Abstract
Acute myeloid leukemia (AML) carrying NPM1 mutations and cytoplasmic nucleophosmin (NPMc+ AML) accounts for about one-third of adult AML and shows distinct features, including a unique gene expression profile. MicroRNAs (miRNAs) are small noncoding RNAs of 19–25 nucleotides in length that have been linked to the development of cancer. Here, we investigated the role of miRNAs in the biology of NPMc+ AML. The miRNA expression was evaluated in 85 adult de novo AML patients characterized for subcellular localization/mutation status of NPM1 and FLT3 mutations using a custom microarray platform. Data were analyzed by using univariate t test within BRB tools. We identified a strong miRNA signature that distinguishes NPMc+ mutated (n = 55) from the cytoplasmic-negative (NPM1 unmutated) cases (n = 30) and includes the up-regulation of miR-10a, miR-10b, several let-7 and miR-29 family members. Many of the down-regulated miRNAs including miR-204 and miR-128a are predicted to target several HOX genes. Indeed, we confirmed that miR-204 targets HOXA10 and MEIS1, suggesting that the HOX up-regulation observed in NPMc+ AML may be due in part by loss of HOX regulators-miRNAs. FLT3-ITD+ samples were characterized by up-regulation of miR-155. Further experiments demonstrated that the up-regulation of miR-155 was independent from FLT3 signaling. Our results identify a unique miRNA signature associated with NPMc+ AML and provide evidence that support a role for miRNAs in the regulation of HOX genes in this leukemia subtype. Moreover, we found that miR-155 was strongly but independently associated with FLT3-ITD mutations.
Keywords: FLT3-ITD, HOX, NPM1
Acute myeloid leukemia (AML) arises from multiple and sequential genetic alterations involving hematopoietic precursors (1). In ≈25% of cases, specific chromosomal translocations like the t(8;21), inv(16) or t(15;17) represent the initial events leading to malignant transformation (1) and are associated with a good outcome. In contrast, 40–50% of AMLs have normal karyotype by conventional banding analysis and are characterized by great molecular and clinical heterogeneity (2). Recent work has identified novel molecular abnormalities in normal karyotype AML (NK-AML) that has improved the classification and risk stratification of this large subgroup of patients. Among them, internal tandem duplications in the juxta-membrane domain or mutations in the second tyrosine kinase domain (TKD) of the FLT3 gene have been found in 30–45% of NK-AML (3). Both types of mutations constitutively activate FLT3 and FLT3-ITD mutations have been associated with increased risk of relapse (4). Mutations in the myeloid transcription factor CEBPA have been detected in 10–15% of NK-AML (5) and are associated with favorable prognosis (5, 6).
Mutations of the nucleophosmin (NPM1) gene, usually occurring at exon-12 (7) and more rarely at exon-11 (8) represent the most common genetic alteration in AML-NK (50–60% of cases) and account for about one-third of all adult AML (7). This gene encodes for a ubiquitously expressed nucleolar protein (NPM1 or B23) that shuttles between the nucleus and cytoplasm and is implicated in multiple functions, including ribosomal protein assembly and transport, control of centrosome duplication and regulation of Arf tumor suppressor gene integrity (9). NPM1 mutations result in the relocalization of NPM1 from the nucleus into the cytoplasm (7), hence the term NPMc+ (cytoplasmic-positive) AML. NPMc+ AML with displays distinctive features including mutual exclusion with AML with recurrent genetic abnormalities, multilineage involvement, unique gene expression profile [up-regulation of Homeobox (HOX) genes and CD34 negativity], increased frequency of FLT3-ITD mutations, and favorable prognosis (in the absence of FLT3-ITD) (10).
Despite this progress, little is known about how NPM1 mutants promote leukemia. The integration of a whole genomic approach including non coding RNAs may lead to an improved understanding of the NPM1 biology. MicroRNAs (MiRNAs) are noncoding RNAs of 19–25 nucleotides in length that regulate gene expression by inducing cleavage or translational inhibition of their targets mRNA through base pairing to partially complementary sites (11). MiRNAs are involved in controlling cell development and differentiation, as well as apoptosis and proliferation (11). Recently, miRNAs expression has been linked to hematopoiesis and cancer (12–15). Here, we asked whether NPMc+ (mutated) and NPMc− (unmutated) AML cases may differ in their miRNA signature and whether the distinctive gene expression profiling of NPMc+ AML (CD34 negativity and HOX genes overexpression) (7, 16, 17) may be dictated by a specific miRNA signature. The miRNAs expression in AML with FLT3-ITD and FLT3 active loop mutations was also explored. Hereby, we report a unique miRNA signature associated with NPMc+ AML and provide evidence that support a role for miRNAs in the regulation of HOX genes in this leukemia subtype. Moreover, we found that miR-155 was strongly but independently associated with FLT3-ITD mutations.
Results
A Unique miRNA Signature Is Associated with NPMc+ AML.
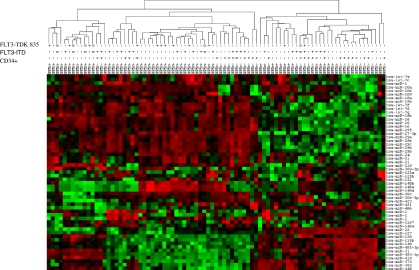
To identify miRNAs associated with NPMc+ AML we profiled 85 de novo AML patients (55 NPMc+ and 30 NPMc−) [see supporting information (SI) Table 2 for patient characteristics] by using our previously validated miRNA platform (18) (SI Table 2). We first compared NPMc+ to NPMc− AML patients using the univariate t test within BRB tools (Class comparison). We found 36 up-regulated and 21 down-regulated miRNAs in NPMc+ NK-AML patients (Table 1). Among the up-regulated miRNAs in NPMc+ samples, we identified miR-10a and -b, several let-7 and miR-29 family members along with the miR-15a-16-1 and miR-17-18a-19a-20a cluster. Additionally, to validate the results of the microarray platform we performed qRT-PCR for the most differentially expressed miRNAs (miR-10a, miR-10b, and miR-22) in 44 AML patients randomly chosen from the initial cohort (28 NPMc+ and 16 NPMc−). As shown in SI Fig. 5, the qRT-PCR data reproduced the chip results with accuracy. Unsupervised analyses of the data revealed that the AML samples segregated mainly in two main clusters (Fig. 1). Samples within the cluster 1 (Fig. 1 Left) had a higher frequency of NPMc+ than cluster 2 (Fig. 1 Right) (χ2 P = 0.007). It is noteworthy that miR-10a and miR-10b expression clearly differentiates NPMc+ vs. NPMc− cases.
Table 1.
miRNAs differentially expressed between NPMc+ (mutated) vs. NPMc− (unmutated) AML patients
| miRNA | Parametric P value | FDR* | Fold change |
|---|---|---|---|
| Up-regulated in NPMc+ | |||
| hsa-miR-10a | <1e-07 | <1e-07 | 20 |
| hsa-miR-10b | <1e-07 | <1e-07 | 16.67 |
| hsa-miR-100 | <1e-07 | <1e-07 | 4.35 |
| hsa-let-7a-3 | <1e-07 | <1e-07 | 3.45 |
| hsa-miR-21 | 3.16E-05 | 4.24E-04 | 3.33 |
| hsa-let-7f | 2.63E-04 | 1.97E-03 | 3.33 |
| hsa-let-7c | 4.00E-07 | 1.45E-05 | 3.33 |
| hsa-miR-16b | 2.14E-04 | 1.80E-03 | 3.23 |
| hsa-miR-16a | 7.21E-04 | 4.16E-03 | 2.94 |
| hsa-let-7a-2 | 2.00E-07 | 8.50E-06 | 2.94 |
| hsa-miR-19b | 1.29E-04 | 1.31E-03 | 2.94 |
| hsa-miR-18a | 8.60E-06 | 1.97E-04 | 2.86 |
| hsa-miR-29c | 2.59E-04 | 1.97E-03 | 2.78 |
| hsa-miR-29a | 3.97E-04 | 2.59E-03 | 2.78 |
| hsa-let-7a-1 | 1.60E-06 | 4.52E-05 | 2.71 |
| hsa-miR-16-1 | 5.07E-04 | 3.07E-03 | 2.63 |
| hsa-miR-29b | 1.11E-03 | 5.64E-03 | 2.56 |
| hsa-miR-24 | 2.32E-04 | 1.84E-03 | 2.51 |
| hsa-miR-20 | 1.21E-05 | 2.20E-04 | 2.44 |
| hsa-miR-17 | 3.83E-03 | 1.52E-02 | 2.33 |
| hsa-miR-369 | 1.23E-03 | 6.11E-03 | 2.27 |
| hsa-let-7 g | 1.02E-03 | 5.30E-03 | 2.27 |
| hsa-let-7d | 7.60E-06 | 1.93E-04 | 2.13 |
| hsa-miR-19a | 9.30E-06 | 1.97E-04 | 2.13 |
| hsa-miR-106 | 5.52E-05 | 6.48E-04 | 2.13 |
| hsa-miR-16-2 | 3.35E-04 | 2.30E-03 | 2.04 |
| hsa-miR-195 | 1.50E-03 | 7.32E-03 | 2.04 |
| hsa-miR-102 | 8.10E-03 | 2.66E-02 | 2.00 |
| hsa-miR-152 | 3.17E-03 | 1.30E-02 | 1.89 |
| hsa-miR-9 | 2.03E-03 | 9.09E-03 | 1.85 |
| hsa-miR-142 | 2.04E-03 | 9.09E-03 | 1.82 |
| hsa-miR-378 | 3.14E-03 | 1.30E-02 | 1.82 |
| hsa-miR-98 | 1.69E-03 | 8.12E-03 | 1.64 |
| hsa-miR-374 | 3.15E-04 | 2.22E-03 | 1.64 |
| hsa-miR-15a | 1.91E-03 | 8.99E-03 | 1.61 |
| hsa-miR-155 | 6.78E-03 | 2.39E-02 | 1.54 |
| Down-regulated in NPMc+ | |||
| hsa-miR-22 | 1.10E-05 | 2.15E-04 | 0.31 |
| hsa-miR-192 | 2.43E-05 | 3.86E-04 | 0.67 |
| hsa-miR-128a | 2.93E-05 | 4.24E-04 | 0.65 |
| hsa-miR-383 | 9.20E-05 | 1.02E-03 | 0.52 |
| hsa-miR-373 | 2.07E-04 | 1.80E-03 | 0.57 |
| hsa-miR-324 | 2.20E-04 | 1.80E-03 | 0.58 |
| hsa-miR-127 | 2.74E-04 | 1.99E-03 | 0.45 |
| hsa-miR-373* | 4.61E-04 | 2.85E-03 | 0.58 |
| hsa-miR-139 | 5.73E-04 | 3.38E-03 | 0.49 |
| hsa-miR-193b | 9.32E-04 | 5.01E-03 | 0.55 |
| hsa-miR-145 | 9.38E-04 | 5.01E-03 | 0.65 |
| hsa-miR-498 | 2.23E-03 | 9.78E-03 | 0.56 |
| hsa-miR-135a | 2.40E-03 | 1.02E-02 | 0.57 |
| hsa-miR-299 | 2.42E-03 | 1.02E-02 | 0.66 |
| hsa-miR-429 | 3.90E-03 | 1.52E-02 | 0.63 |
| hsa-miR-493 | 3.97E-03 | 1.53E-02 | 0.53 |
| hsa-miR-326 | 4.65E-03 | 1.76E-02 | 0.63 |
| hsa-miR-204 | 4.89E-03 | 1.80E-02 | 0.61 |
| hsa-miR-198 | 8.20E-03 | 2.66E-02 | 0.65 |
| hsa-miR-486 | 8.23E-03 | 2.66E-02 | 0.45 |
MiRNAs are sorted by P value of the univariate test (BRB tools). The first 56 genes are significant at the nominal 0.01 level of the univariate test.
*FDR, False discovery rate or q value is the expected percentage of genes identified by chance.
Fig. 1.
Hierarchical cluster analysis of NK-AML patients. Overview of two-way (genes against samples) hierarchical cluster (Euclidean distance) of 85 AML samples using the genes that varies the most between samples. Mean centered signal intensities of gene-expression are depicted by a log-transformed (2 scale). Color areas indicate relative expression of each gene with respect to the gene median expression (red above, green below the median value and black, samples with signal intensity to background of 2 or less). Some miRNAs are represented by more than one probe. As shown, the clustering is mainly determined by the presence of cytoplasmic nucleophosmin (NPMc+). Among the NPMc+ positive samples, we identified also many subgroups. Patients with FLT3-ITD+ segregated among the different clusters.
MiR-10a Correlates Positively with HOXB4 Expression.
Notable the two most up-regulated miRNAs in NPMc+ AML (miR-10a and miR-10b) are embedded in HOX gene clusters (miR-10a is located between the HOXB4 and HOXB5 gene in chromosome 17q21 and mir-10b between the HOXD3 and HOXD4 gene in chromosome 2q31). It has been reported that in mouse embryos, miR-10a and -10b expression closely follows their host HOX cluster expression during development (19), suggesting that these miRNAs may be regulated by the same CIS elements that also regulates HOX genes. To investigate whether miR-10a correlated with its flanking HOXB4 gene, we measured miR-10a and HOXB4 in 18 AML patients from the original cohort (NPMc+ = 9 and NPMc− = 9) by qRT-PCR. Indeed, we identified a positive correlation between miR-10a and HOXB4 expression (R = 0.57, P = 0.01, Pearson correlation test). However, no correlation was found between miR-10b and its flanking HOXD3 gene (data not shown).
MiR-204 Targets HOXA10 and MEIS1.
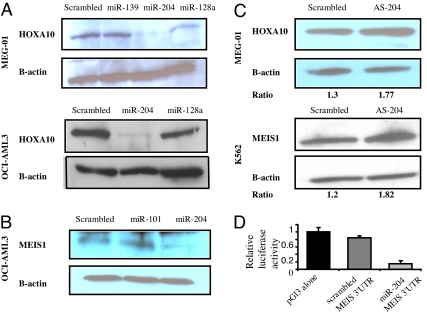
Among the down-regulated miRNAs in patients with NPMc+ AML, there are several miRNAs predicted to interact with HOX genes according to three available “in silico” target prediction software [Targetscan (20), Pictar (21), and RNAhybrid (22)] (SI Table 3). Indeed, few miRNAs have been shown to regulate HOX genes and play important roles during early development (14, 19). We hypothesize that the high HOX genes expression in NPMc+ AML may be due in part to the down-regulation of HOX regulators miRNAs in this subgroup. To start unraveling the role for miRNAs in HOX regulation in NPMc+ AML, we validated the predicted HOX targets for the down-regulated miRNAs in NPMc+ AML by performing Western blotting using HOXA9, HOXA10, and MEIS1 antibodies in AML cell lines after transfection of the candidate oligonucleotides miRNAs or scrambled oligonucleotides (SI Table 3 and SI Fig. 6). To perform this screening, we used the OCI-AML3 cell line (which harbors a NPM1 mutation) (23) and the MEG-01 cell line (both cell lines with high expression of HOXA9, HOXA10, and MEIS1). As shown in Fig. 2A and B, there was a robust down-regulation of HOXA10 and MEIS1 protein in both AML cells lines transfected with miR-204 but not with the other miRNAs or scrambled oligonucleotides. As an additional control, we transfected antisense oligonucleotides against miR-204 in K562 and MEG-01 cells and measured MEIS1 and HOXA10 protein expression after 48 h (Fig. 2C). We observed up-regulation of MEIS1 and HOXA10 in both cell lines after transfection with miR-204 antisense oligonucleotides with respect to scrambled oligonucleotides. To validate further these interactions, we cloned the 3′ untranslated region (3′ UTR) of MEIS1 predicted to interact with miR-204 into a luciferase reporter vector and cotransfected with miR-204 or scrambled oligonucleotides into the MEG-01 cell line. A marked reduction in the luciferase/Renilla ratio was seen for MEIS1 constructs transfected with miR-204 (Fig. 2D).
Fig. 2.
MiR-204 targets HOXA10 and MEIS1 in AML. (A) Western blotting showing HOXA10 protein using whole cell lysates from MEG-01 and OCI-AML3 cell lines transfected with scrambled oligonucleotides or miR-139, miR-128a, and miR-204. The blots were stripped and reprobed with β-actin for loading control. (B) Western blotting showing MEIS1 protein expression in OCI-AML3 cell lines transfected with scrambled oligonucleotides or sense miR-204 and miR-101. (C) Western blotting showing HOXA10 and MEIS1 protein expression in MEG-01 and K562 cells transfected with scrambled oligonucleotides or antisense oligonucleotides against miR-204. The numbers below the immunoblot images represent the intensity of the HOXA-10/MEIS bands relative to the β-actin expression. (D) Relative luciferase activity in MEG01 cells transiently cotransfected with the luciferase reporter vector containing the 3′ UTR of MEIS1 with miR-204 or scrambled oligonucleotides. The results are presented as a fold difference in the luciferase/Renilla ratios with respect to the luciferase reporter constructs transfected with scrambled oligonucleotides.
CD34 Regulation by miRNAs.
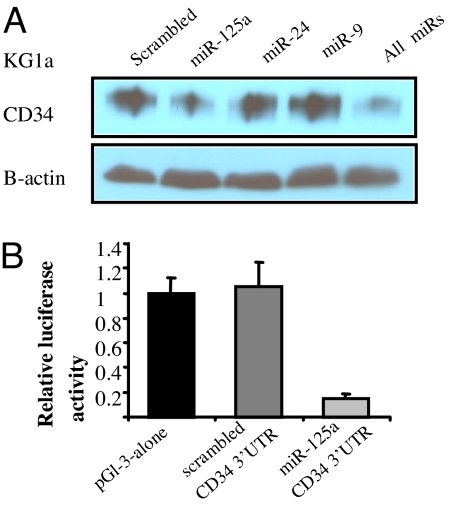
We have previously shown that AML blasts from NPMc+ patients are frequently CD34 negative (7, 16, 17). Interestingly, many miRNAs up-regulated in NPMc+ AML are predicted to target CD34, including miR-9, miR-24, and miR-125a. To confirm these interactions, we transfected miR-9, miR-24, and miR-125a oligonucleotides or control oligonucleotides into the CD34+ AML cell line KG1a using nucleoporation and measured CD34 protein expression using Western blotting. As shown in Fig. 3A, a marked reduction of CD34 protein was evident in the miR-125a transfected cells compared with the scrambled oligonucleotides. To further validated this interaction, we performed luciferase reporter assays, where the 3′ UTR of the CD34 gene predicted to interact with miR-125a, was cloned in to the luciferase reporter assay and cotransfected with miR-125a oligonucleotide or scrambled oligonucleotides into MEG-01 cell lines using lipofectamine. As shown in Fig. 3B, 85% of reduction in the luciferase normalized ratios was observed in the cells transfected with miR-125a compared with the controls.
Fig. 3.
MiR-125a targets CD34. (A) Western blotting showing CD34 protein expression in KG1a cells 48 h after transfection with scrambled oligonucleotides or sense miR-125a, mIR-24, miR-9, or all combined oligonucleotides (B) Relative luciferase activity in KG1a cells transiently cotransfected with the luciferase reporter vector containing the 3′ UTR of CD34 predicted to interact with miR-125a or scrambled oligonucleotides. The results are presented as a fold difference in the luciferase/Renilla ratios with respect to the empty reporter (pGl3 alone).
MiR-155 Is Up-Regulated in FLT3-ITD+ AML.
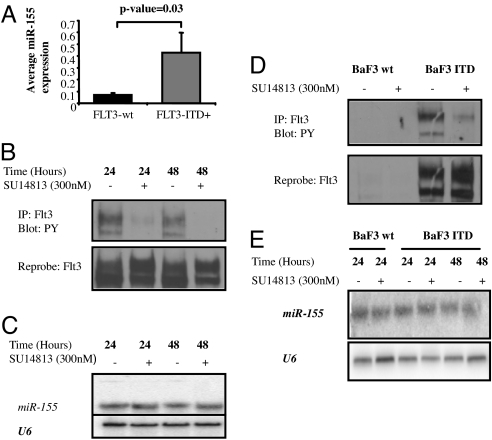
Using the univariate t test (BRB), we identified three up-regulated miRNAs (miR-155, miR-302a, and miR-133a) in FLT3-ITD+ as compared with FLT3-wt AML patients. This signature had a FDR of 0% at a significance level of P < 0.01. Then, we compared the miRNAs expression in FLT3-ITD+ to FLT3-wt in NPMc+ patients. Only miR-155 was up-regulated at a significance level of P < 0.01. We validated the miR-155 up-regulation in FLT3-ITD+ patients by measuring miR-155 using a different method (qRT-PCR) in a group of 32 randomly chosen AML patients from the original cohort (Fig. 4A). AML patients with FLT3-TKD mutations were not associated with any distinctive miRNA signature. To further obtain insights about the relationship among FLT3-ITD and miR-155, we carried out an experiment to investigate whether blocking FLT3 signaling using a potent FLT3 inhibitor (SU14813) in two FLT3-ITD positive AML cell lines (MV4-11 and Molt-13) will impact on miR-155 expression (Fig. 4B). As shown in Fig. 4C and SI Fig. 7, the miR-155 expression remained unchanged after 48 h of successful FLT3 signaling inhibition in the two cell lines tested. Furthermore we performed an additional control experiment where we generated stable Baf3 cell clones harboring the human FLT3 gene with ITD by using retroviral infection, and measured miR-155 expression in Baf3 wt and in Baf3 FLT3-ITD by Northern blotting (Fig. 4 D and E). We did not observe any significant difference in miR-155 expression between Baf3 wt and Baf3 FLT3-ITD. In addition, miR-155 expression did not change after treatment of Baf3 FLT3-ITD cells with the FLT3 inhibitor (SU14813) for 48 h (Fig. 4 D and E).
Fig. 4.
MiR-155 is overexpressed in FLT3-ITD+ AML. (A) Average miR-155 expression in 32 NK-AML patients with FLT3-ITD+ (n = 10) and FLT3-wt (n = 22) after normalization with 18s and 2ΔCt calculations. The average miR-155 expression values between the two groups were compared by using t test. (B) IP/Western blot. MV4-11 cells were incubated for 24 and 48 h with SU14813 (300 nM) or DMSO (vehicle control). Lysates were generated and an immunoprecipitation was performed with an antibody for FLT3. SDS/PAGE was performed, followed by a Western blot with an antibody for phospho-tyrosine. Subsequently, the blot was stripped and reprobed with anti-FLT3. (C) Northern blotting of miR-155 in MV4-11 cells incubated for 24 and 48 h with SU14813 (300 nM) or DMSO (vehicle control). Total RNA was obtained with TRIzol and Northern blotting was performed with a probe antisense to miR-155. The blot was stripped and reprobed with U6 for loading control. (D) IP/Western blot. BaF3 and BaF3 FLT3-ITD cells were incubated for 24 h with SU14813 (300 nM) or DMSO (vehicle control). Lysates were generated and an immunoprecipitation was performed with an antibody for FLT3. SDS/PAGE was performed, followed by a Western blot with an antibody for phospho-tyrosine. Subsequently, the blot was stripped and reprobed with anti-FLT3. (E) Northern blotting for miR-155 in BaF3 and Baf3 FLT3-ITD clones incubated for 24 and 48 h with SU14813 or DMSO (control) as described. The blot was stripped and reprobed with U6 for loading control.
Discussion
Employing a microarray platform, we systematically analyzed the miRNA expression of AML patients with known subcellular localization/mutation status of NPM1 and identified a strong signature associated with the presence of cytoplasmic mutated nucleophosmin. Among the miRNAs up-regulated in NPMc+ AML, there were three families of tumor suppressor miRNAs: miR-15-a/miR-16-1, miR-29s (a/b/c), and the let-7 (let-7a, let-7b, and let-7f). Low level of let-7a has been associated with short survival in lung cancer after surgery (24), and low level of miR-29b has been associated with shorter survival in CLL (25). Previous reports have shown that NK-AML with NPM1-mutated/FLT-3-ITD negative genotype have a good prognosis (reviewed in ref. 10). This clinical observation is in keeping with our finding of several up-regulated tumor suppressors miRNAs in NPMc+ AML.
Consistent with previous reports, we have shown that HOXB4 expression correlated positively with its embedded miRNA miR-10a, suggesting that this miRNA may be regulated by CIS elements that also regulate HOX genes (14, 19, 26). However, no correlation was found for miR-10b and its flanking HOXD3 gene. This observation raises the question about the possibility of microarray cross hybridization, because the two miRNAs differ only in one nucleotide. Nevertheless, miR-10b up-regulation in NPMC+ was also detected by qRT-PCR, which is more specific. Finally, whether the HOX embedded miRNAs are innocent bystanders or have a critical role during leukemogenesis remained to be explored.
High expression of HOX genes is one of the most distinguishing features of NPMc+ AML (16, 17, 27). Remarkably, the pattern of perturbed HOX gene dysregulation in NPM1-mutated AML clearly differ from that observed in AML carrying rearrangements of MLL gene, because up-regulation of group-B HOX genes (especially HOXB2 and B6) is observed in NPM1-mutated but not MLL-rearranged AML (17). This finding suggests that dysregulation of HOX gene expression in NPM1 mutated AML occurs via a different mechanism than in AML with MLL rearrangement. Up-regulation of HOX expression in MLL-rearranged leukemia has been related to the direct binding of MLL fusion proteins to HOX gene promoters (28), but this is unlikely to be the case for NPM1-mutated AML. A major goal of our study was to assess whether miRNAs may contribute to the HOX up-regulation in NPMc+ AML, and, indeed, we demonstrated that miR-204, which is down-regulated in NPMc+ AML targets HOXA10 and MEIS-1. Over-expression of HOXA10 in murine hematopoietic stem cells perturbs myeloid differentiation and leads to AML (29). Likewise, enforced expression of HOXA9 and MEIS1 in mice induces AML after a short latency (30). This report links miRNAs in the regulation of HOX genes involved in leukemia. However, it remains to be clarified whether this finding reflects derivation from hematopoietic progenitors that physiologically tune HOX gene expression through their miRNA apparatus, or whether the NPM1 mutant protein contributes to leukemogenesis by inducing down-regulation of specific miRNAs that control HOX gene expression. Hypothetically, the mutant could target a myeloid committed progenitor and confer to this cell self-renewal capability through up-regulation of HOX genes, thus explaining why, despite frequent multilineage involvement (8), lymphoid lineage is typically not involved in NPMc+ AML (31).
It is of interest that several miRNAs up-regulated in NPMc+ AML are predicted to target CD34, a gene whose expression is frequently down-modulated in these leukemias (7, 16). Of the several miRNAs up-regulated in NPMc+ AML, we have validated that only miR-125a targets CD34. However, miR-125a is not consistently up-regulated in NPMc+ AML (only identified in some cases by using the unsupervised clustering), and its contribution in the CD34 regulation of NPMc+ AML remains to be clarified.
The analysis of miRNA expression in the whole group of NK-AML patients with known FLT3 status revealed a distinct signature. A surprising result was the identification of miR-155 as overexpressed in FLT3-ITD+ patients. Previous reports have established that miR-155 overexpression halts myeloid development (32) and induced B cell lymphoma/leukemia in a transgenic mice model (33). Here, we also demonstrated, by inhibiting FLT3 signaling in FLT3-ITD+ cell lines, that miR-155 expression seems to be independent from FLT3 signaling. Previous work showed that transgenic mice models for FLT3-ITD do not develop acute leukemia, but a chronic myeloproliferative disorder (34). Given the clear independent association of miR-155 with FLT3-ITD mutations in AML and the known myeloid blocking effects that result from miR-155 overexpression, it is tempting to hypothesize that, perhaps, miR-155 could play a role in leukemogenesis.
Our findings have also potential clinical and therapeutic implications. The demonstration that NPMc+ AML carries a distinct microRNA signature further supports the view that NPM1 mutations identify a distinct AML genetic entity (showing a normal karyotype in >90% of cases). Finally, our studies identify at least one miRNA (miR-204) to be explored for therapeutic intervention.
Patients and Methods
Patient Samples.
The study was carried out on diagnostic bone marrow samples from 85 adult AML patients: 25 patients from the Institute of Hematology, University of Perugia; 29 cases from the Gruppo Italiano Malattie Ematologiche dell' Adulto (GIMEMA); and 31 cases from the Munich Leukemia Laboratory (MLL). The characteristics of these patients are shown in SI Table 2. All patients gave informed consent for the bone marrow biopsy (used for immunohistochemistry) and aspirate for cryopreservation (used for molecular studies). Approval was obtained from the institutional board review of each institution.
NPM1 and FLT3 Mutation Analysis.
NPM1 mutations status was assessed by using immunohistochemistry/Western blotting, or mutation analysis. A bone marrow biopsy was available in 54 cases: 37 showed aberrant cytoplasmic NPM (NPMc+) that is fully predictive of NPM1 mutations (35) whereas 19 cases showed the nucleus-restricted reactivity (NPMc−) typical of cases carrying a wild-type NPM1 gene (35). In 19 of 54 cases, the immunohistochemical findings were also confirmed by Western blotting with specific anti-NPM mutants antibodies (36). NPM1 mutations analysis carried out as previously described was available in 59 cases, including 28 patients who were previously evaluated by immunohistochemistry (37). FLT3-ITD and FLT3-TKD mutations were investigated in 79 cases, as described in ref. 37.
MiRNA Microarrays Experiments.
RNA extraction and miRNA microchip experiments were performed as described in detail elsewhere (18). The miRNA microarray is based on a one-channel system (18). Five micrograms of total RNA was used for hybridization on the OSU custom miRNA microarray chips (OSU_CCC version 3.0), which contains ≈1,100 miRNA probes, including 345 human and 249 mouse miRNA genes, spotted in duplicates.
Real-Time Quantification of microRNAs.
The single tube TaqMan miRNA assays were used to detect and quantify mature miRNAs as described in ref. 38, using PCR 9700 Thermocycler ABI Prism 7900HT and the sequence detection system (Applied Biosystems). Normalization was performed with 18s. Comparative real-time PCR was performed in triplicate, including no-template controls. Relative expression was calculated by using the comparative 2ΔCt method (39).
Data Analysis.
Microarray images were analyzed by using GenePix Pro. Average values of the replicate spots of each miRNA were background-subtracted. Quantiles normalization was implemented by using the Bioconductor package/function. Differentially expressed microRNAs were identified by using the univariate t test within the BRB tools (http://linus.nci.nih.gov/BRB-ArrayTools.html). Cluster and Java TreeView were used to build the unsupervised tree. The filtering was based on the variance for the gene across the arrays. Additional details are reported in SI Methods. The microarray dataset is deposited in Array-Express. Fisher's exact test, t test, and χ2 were used to compare baseline patient characteristics and average miRNA expression between groups of patients. All reported P values were two-sided and obtained by using the SPSS software package (SPSS 10.0).
Cell culture, transfections, and luciferase reporter assays are described in detail in SI Methods.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Catherine Heaphy for assistance in performing experiments, Dorothee Wernicke-Jameson for research supervision, Sharon Palko for administrative support, and the following GIMEMA investigators and centers in Italy that provided samples for the analysis: Istituto do Ematologia, Università la “Sapienza,” Rome (n = 10); Dr. N. Cantore, Azienda Ospedaliera S. G. Moscati, Avellino (n = 3); Prof. F. Nobile, Dipartimento di Emato-Oncologia, Azienda Ospedaliera Bianchi-Melacrino-Morelli, Reggio Calabria (n = 3); Dr. A. M. D'Arco, Istituto di Medicina Interna, Nocera Inferiore (n = 3); Dr. G. Specchia, Istituto di Ematologia, Università di Bari (n = 2); Dr. M. Sborgia, Divisione di Ematologia, Azienda USL, Pescara (n = 1); Dr. F. Fabbiano, Divisione di Ematologia, Ospedale V. Cervello, Palermo (n = 1); Prof. P. Leoni, Istituto di Ematologia, Università di Ancona, Ancona (n = 1); Prof. M. Petrini, Istituto di Ematologia, Università di Pisa (n = 1); Dr. L. Camba, Divisione di Ematologia, Fondazione Centro S. Raffaele, Milan (n = 1); Dr. E. Angelucci, Istituto di Ematologia, Ospedale A. Businco, Cagliari (n = 1); Dr. A. Camera, Istituto di Ematologia, Università Federico II, Naples (n = 1); and Dr. F. Ricciuti, Divisione di Ematologia, Ospedale S. Carlo, Potenza (n = 1). This work was supported by National Cancer Institute Grants PO1CA76259 and PO1CA81534 (to C.M.C.), Lauri Strauss Discovery Grants (to R.G), and by Associazione Italiana per la Ricerca sul Cancro (AIRC) (B.F. and M.F.M.).
Footnotes
B.F. and C.M. have applied for a patent on clinical use of NPM1 mutants. The authors declare no other conflict of interest.
Data deposition: The microarray dataset has been deposited in the ArrayExpress database, www.ebi.ac.uk/arrayexpress (accession no. E-TABM-429).
This article contains supporting information online at www.pnas.org/cgi/content/full/0800135105/DC1.
References
- 1.Lowenberg B, Downing JR, Burnett A. Acute Myeloid leukemia. N Engl J Med. 1999;341:1051–1062. doi: 10.1056/NEJM199909303411407. [DOI] [PubMed] [Google Scholar]
- 2.Burnett AK. Current controversies: Which patients with acute myeloid leukemia should receive bone marrow transplantation? An adult theater's view. Br J Haematol. 2002;118:357–364. doi: 10.1046/j.1365-2141.2002.03698.x. [DOI] [PubMed] [Google Scholar]
- 3.Kiyoi H, et al. Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia. Blood. 1999;93:3074–3080. [PubMed] [Google Scholar]
- 4.Shih LY, et al. Internal tandem duplication of FLT3 in relapsed acute myeloid leukemia: A comparative analysis of bone marrow samples from 108 adult patients at diagnosis and relapse. Blood. 2000;100:2387–2392. doi: 10.1182/blood-2002-01-0195. [DOI] [PubMed] [Google Scholar]
- 5.Frohling S, et al. CEBPA mutations in younger adults with acute myeloid leukemia and normal cytogenetics: Prognostic relevance and analysis of cooperating mutations. J Clin Oncol. 2004;22:624–633. doi: 10.1200/JCO.2004.06.060. [DOI] [PubMed] [Google Scholar]
- 6.Preudhomme C, et al. Favorable prognostic significance of CEBPA mutations in patients with de novo acute myeloid leukemia: A study from the Acute Leukemia French Association (ALFA). Blood. 2002;100:2717–2723. doi: 10.1182/blood-2002-03-0990. [DOI] [PubMed] [Google Scholar]
- 7.Falini B, et al. Cytoplasmatic nucleophospmin in acute myeloid leukemia with normal karyotype. N Engl J Med. 2005;352:254–261. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 8.Albiero E, et al. Identification and functional characterization of a cytoplasmic nucleophosmin leukaemic mutant generated by a novel exon-11 NPM1 mutation. Leukemia. 2007;21:1099–1103. doi: 10.1038/sj.leu.2404597. [DOI] [PubMed] [Google Scholar]
- 9.Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- 10.Falini B, Nicoletti I, Martelli FM, Mecucci M. Acute myeloid leukemia carrying cytoplasmic/mutated nucleophosmin (NPMc+ AML): Biological and clinical features. Blood. 2007;109:874–885. doi: 10.1182/blood-2006-07-012252. [DOI] [PubMed] [Google Scholar]
- 11.Bartel D. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNAs expression and function in cancer. Trends Mol Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 13.Chen CZ, Li L, Lodish H. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 14.Garzon R, et al. MicroRNAs fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci USA. 2006;103:5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garzon R, et al. MicroRNA gene expression during retinoic acid-induced differentiation of human acute promyelocytic leukemia. Oncogene. 2007;26:4148–4157. doi: 10.1038/sj.onc.1210186. [DOI] [PubMed] [Google Scholar]
- 16.Alcalay M, et al. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood. 2005;106:899–902. doi: 10.1182/blood-2005-02-0560. [DOI] [PubMed] [Google Scholar]
- 17.Mullighan CG, et al. Pediatric acute myeloid leukemia with NPM1 mutations is characterized by a gene expression profile with dysregulated HOX gene expression distinct from MLL-rearranged leukemias. Leukemia. 2007;21:2000–2009. doi: 10.1038/sj.leu.2404808. [DOI] [PubMed] [Google Scholar]
- 18.Liu CG, et al. An oligonucleotide microchip for genomic–wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanzer A, Ameniya C, Kim CB, Stadler PF. Evolution of microRNAs located in HOX gene clusters. J Exp Zool B Mol Dev Evol. 2005;304:75–85. doi: 10.1002/jez.b.21021. [DOI] [PubMed] [Google Scholar]
- 20.Lewis B, Shih I, Jones-Rhoades M, Bartel D, Burge C. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 21.Krek A, et al. Combinatorial microRNA target prediction. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 22.Rehmsmeier M, Steffen P, Höchsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quentmeier H, et al. Cell line OCI/AML3 bears exon-12 NPM gene mutation-A and cytoplasmic expression of nucleophosmin. Leukemia. 2005;19:1760–1767. doi: 10.1038/sj.leu.2403899. [DOI] [PubMed] [Google Scholar]
- 24.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 25.Calin GA, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 26.Debernardi S, et al. MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia. 2007;21:912–916. doi: 10.1038/sj.leu.2404605. [DOI] [PubMed] [Google Scholar]
- 27.Verhaak RGW, et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): Association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005;106:3747–3754. doi: 10.1182/blood-2005-05-2168. [DOI] [PubMed] [Google Scholar]
- 28.Hess JL. MLL: A histone methyltransferase disrupted in leukemia. Trends Mol Med. 2004;10:500–507. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Thorsteinsdottir U, Sauvageau G, Hough MR. Over-expression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol Cell Biol. 1997;17:495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kroon E, Thorsteinsdottir U, Mayotte N, Nakamura T, Sauvageau G. NUP98-HOXA9 expression in hemopoietic stem cells induces chronic and acute myeloid leukemias in mice. EMBO J. 2001;20:350–361. doi: 10.1093/emboj/20.3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martelli PM, et al. Absence of nucleophosmin leukaemic mutants in B, T cells from AML with NPM1 mutations: Implications for the cell of origin of NPMc+ AML. Leukemia. 2008;22:195–198. doi: 10.1038/sj.leu.2404857. [DOI] [PubMed] [Google Scholar]
- 32.Georgantas G, et al. CD34+ hematopoietic stem-cell progenitor cell microRNA expression and function: A circuit diagram of differentiation control. Proc Natl Acad Sci USA. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costinean S, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in Eμ-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelly LM, et al. FLT3 in tandem duplication mutations associated with human acute myeloid leukemias induce myeloproliferative disease in a murine bone marrow transplant model. Blood. 2002;99:310–318. doi: 10.1182/blood.v99.1.310. [DOI] [PubMed] [Google Scholar]
- 35.Falini B, et al. Immunohistochemistry predicts nucleophosmin (NPM) mutations in acute myeloid leukemia. Blood. 2006;108:1999–2005. doi: 10.1182/blood-2006-03-007013. [DOI] [PubMed] [Google Scholar]
- 36.Falini B, et al. Both carboxy-terminus NES motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc+ AML. Blood. 2006;107:4514–4523. doi: 10.1182/blood-2005-11-4745. [DOI] [PubMed] [Google Scholar]
- 37.Schnittger S, et al. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005;106:3733–3739. doi: 10.1182/blood-2005-06-2248. [DOI] [PubMed] [Google Scholar]
- 38.Chen C, et al. Real-time quantification of microRNAs by tem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR, the 2(-Delta Delta C (T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.