Abstract
Nearly all neurodegenerative diseases are associated with abnormal accumulation of ubiquitin (Ub) conjugates within neuronal inclusion bodies. To directly test the hypothesis that depletion of cellular Ub is sufficient to cause neurodegeneration, we have disrupted Ubb, one of four genes that supply Ub in the mouse. Here, we report that loss of Ubb led to a progressive degenerative disorder affecting neurons within the arcuate nucleus of the hypothalamus. This neurodegenerative cytopathology was accompanied by impaired hypothalamic control of energy balance and adult-onset obesity. Ubb was highly expressed in vulnerable hypothalamic neurons and total Ub levels were selectively reduced in the hypothalamus of Ubb-null mice. These findings demonstrate that maintenance of adequate supplies of cellular Ub is essential for neuronal survival and establish that decreased Ub availability is sufficient to cause neuronal dysfunction and death.
Keywords: hypothalamus, ubiquitin, energy homeostasis
Ubiquitin (Ub) is a highly conserved small protein that functions as a key signaling molecule in multiple proteolytic and nonproteolytic pathways in all eukaryotic cells (1, 2). Ub signaling is initiated by covalent ligation of one or more Ub molecules to amino groups on target proteins and is terminated by hydrolysis of these isopeptide bonds (1–3). Cellular Ub, therefore, exists in a dynamic equilibrium between pools of free (activated or monomeric) Ub and polymeric Ub–substrate conjugates, governed by the opposing activities of enzymes that catalyze covalent Ub conjugation and deconjugation. Ub is an abundant protein in eukaryotic cells (4–7) and, because of its crucial roles in so many different signaling processes, its levels are tightly regulated. Although steady-state Ub pools are thought to be largely maintained by enzymatic recycling of Ub by deubiquitinating enzymes (DUBs) associated with the 26S proteasome, a fraction of Ub is consumed during proteolysis (8) and must be replenished by de novo synthesis.
Mammalian Ub is encoded by two polyubiquitin genes, Ubb and Ubc, composed, respectively, of three or four and nine or ten tandem-repeat Ub coding units arranged in a head-to-tail spacerless array, and two Uba genes, Uba52 and Uba80 (also known as Rps27a), comprised of linear fusions between Ub and small ribosomal proteins (9–14). Although disruption of murine Ubc results in embryonic lethality, owing to a requirement of this polyubiquitin gene for fetal liver development (15), mice lacking Ubb are born at the expected Mendelian frequency (16).
A substantial body of genetic evidence suggests that the Ub system is essential for neuronal development, function, and survival. Ub-dependent protein degradation is required for axon remodeling during development and after injury (17), and Ub conjugation is emerging as a regulator of synaptic function (18). Recessive mutations in genes encoding the E3 Ub ligase Parkin underlie the pathology in autosomal recessive juvenile parkinsonism (19). Loss-of-function mutations in the DUBs Usp14 and UchL1 cause synaptic dysfunction and degeneration of neurons in the ataxia (axJ) (20) and gracile axonal dystrophy (gad) (6) mice, respectively, suggesting that maintenance of Ub pool homeostasis is essential for neuronal function and survival. The finding that cytoplasmic inclusion bodies, the defining cytopathological features of most late-stage neurodegenerative diseases, are heavily enriched in Ub conjugates has long suggested that disruption of Ub pool dynamics could be a common neuropathogenic mechanism (21, 22). Although this hypothesis has been strongly bolstered by the recent demonstration of global changes to Ub conjugate pools early in disease progression in a mouse model of Huntington's disease (23), whether changes to Ub pools and consequent chronic decreased Ub availability are causes or effects of underlying pathology has remained untested. In this study, we directly tested this hypothesis by disrupting Ubb in the mouse.
Results
Adult-Onset Obesity in Ubb−/− Mice.
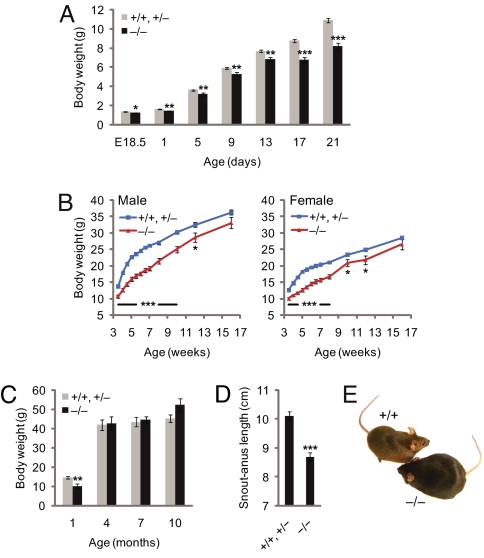
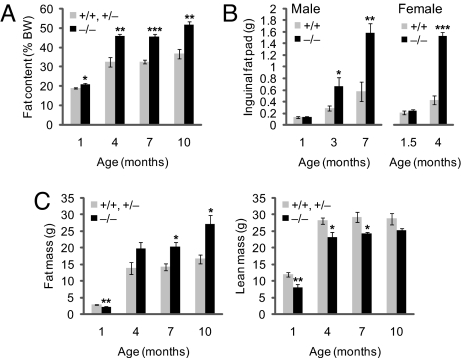
Ubb−/− mice are smaller than wild-type or heterozygous littermates (Fig. 1A) and exhibit subtle perinatal linear growth retardation (Fig. 1B) but are otherwise indistinguishable from wild-type mice in gross appearance. By 16 weeks of age, however, body weights of Ubb−/− mice stabilized at a level similar to that of littermate controls (Fig. 1C). Strikingly, adult Ubb−/− mice could be readily identified by their short stature (Fig. 1D) and obese appearance (Fig. 1E). Body fat content, assessed by dual-energy x-ray absorptiometry (DEXA) (Fig. 2A) or by direct measurement of dissected inguinal and reproductive fat pad mass (Fig. 2B and data not shown), was significantly elevated in adult Ubb−/− mice. This increased body fat content was not accompanied by an increase in total body mass (Fig. 1C) but was associated instead with reduced lean body mass and correspondingly increased fat mass (Fig. 2C). The adult-onset obesity was not accompanied by disturbances in blood glucose [supporting information (SI) Fig. 6A] or serum insulin (SI Fig. 6B) levels, indicating that the increased fat content was not a consequence of impaired pancreatic endocrine function or glucose homeostasis. Serum leptin levels were within the normal range in young Ubb−/− mice but were significantly elevated in adults (SI Fig. 6C), even after food deprivation (SI Fig. 6D). Because leptin is secreted by adipocytes, it is probable that this adult-onset hyperleptinemia is a direct consequence of the increased fat content of the adult Ubb−/− mice.
Fig. 1.
Abnormal growth of Ubb−/− mice. (A) Newborn Ubb−/− (−/−) (n = 18) mice are smaller and exhibit reduced perinatal weight gain compared with littermate controls (+/+ and +/−) (n = 39). Smaller size of Ubb−/− mice is also evident at E18.5 (+/+ and +/−, n = 13; −/−, n = 3). (B) Growth curve of male and female mice after weaning. Body weights of Ubb−/− (male, n = 19; female, n = 17) mice are significantly lower than those of their littermate controls (male, n = 51; female, n = 76) before 16 weeks of age. At 16 weeks of age, there is no significant difference in weight between Ubb−/− mice and littermate controls. (C) Total body mass of male mice determined by DEXA (+/+ and +/−, n = 4–12; −/−, n = 3 or 4). (D) Snout–anus length of 4-month-old mice (+/+ and +/−, n = 18; −/−, n = 13). (E) Representative photographs of adult Ubb+/+ (+/+) and Ubb−/− littermates. All data are expressed as means ± SEM from the indicated number of mice. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001 for Ubb−/− vs. littermate controls.
Fig. 2.
Altered body composition in adult Ubb−/− mice. (A) Whole-body fat content of male Ubb−/− mice (n = 3 or 4) and littermate controls (n = 4–12) measured by DEXA. Fat content is expressed as a percentage of body weight. (B) Inguinal fat pad weight of male (+/+, n = 6–11; −/−, n = 6–11) and female (+/+, n = 5 or 7; −/−, n = 6) mice. (C) Fat and lean mass of male Ubb−/− mice (n = 3 or 4) and littermate controls (n = 4–12) measured by DEXA. All data are expressed as means ± SEM from the indicated number of mice. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001 for Ubb−/− vs. littermate controls or Ubb+/+.
Impaired Compensatory Refeeding and Hypothalamic Neuropeptide Expression in Ubb−/− Mice.
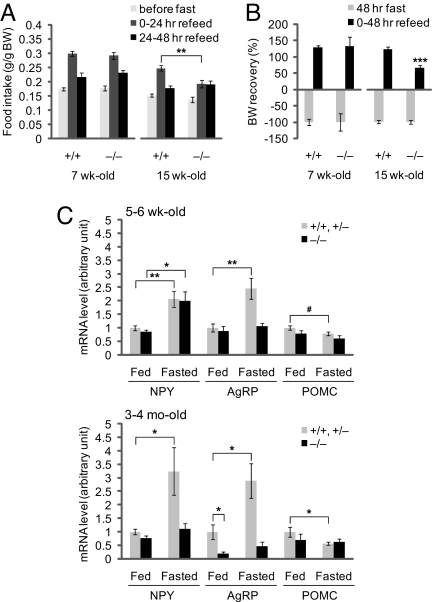
To determine whether the adult-onset obesity phenotype of Ubb−/− mice is associated with an underlying abnormality in the hypothalamic circuitry required for maintenance of peripheral energy stores, we examined whether Ubb−/− mice can increase food intake acutely, as in Ubb+/+ mice, after a period of food deprivation. Although young Ubb+/+ and Ubb−/− mice displayed a similar response to food deprivation, this compensatory hyperphagia was substantially blunted in adult Ubb−/− mice (Fig. 3A). Likewise, whereas young Ubb+/+ and Ubb−/− mice were able to fully recover their prefasting body weight within 48 h of refeeding, adult Ubb−/− mice were significantly impaired in body weight recovery after fasting (Fig. 3B). This defect in body weight recovery can, at least in part, be explained by the impaired compensatory hyperphagia observed in adult Ubb−/− mice (Fig. 3A). These findings indicate that the hypothalamic circuitry responsible for maintaining energy homeostasis is disrupted in adult Ubb−/− mice.
Fig. 3.
Defective central regulation of body weight and energy homeostasis in Ubb−/− mice. (A) Ad libitum food intake during the 24 h preceding fasting for 48 h and the two 24-h intervals after refeeding was measured and expressed as per gram body weight in 7-week-old (+/+, n = 9; −/−, n = 11) and 15-week-old (+/+, n = 11; −/−, n = 11) mice. (B) Recovery of body weight after fasting for 48 h and refeeding for 48 h in 7-week-old (+/+, n = 9; −/−, n = 11) and 15-week-old (+/+, n = 11; −/−, n = 11) mice. Loss of body weight after fasting was arbitrarily assigned as −100% in each genotype of mice. (C) Abnormal expression of hypothalamic neuropeptides in adult Ubb−/− mice. Normalized mRNA levels, measured by quantitative real-time RT-PCR from hypothalamus of young (5–6 weeks old) and adult (3–4 months old) ad libitum fed (+/+ and +/−, n = 12; −/−, n = 5 or 6) or 48-h fasted (+/+ and +/−, n = 12 or 16; −/−, n = 6) mice. All data are expressed as means ± SEM from the indicated number of mice. In A and C: ∗, P < 0.05; ∗∗, P < 0.01; #, P = 0.07. In B: ∗∗∗, P < 0.001 vs. 15-week-old Ubb+/+ mice after refeeding for 48 h.
Leptin receptors (Ob-R) on hypothalamic neurons transduce hormone binding into distinct physiological responses mediated, in large measure, by repression of genes encoding orexigenic neuropeptides such as neuropeptide Y (NPY), Agouti-related protein (AgRP), orexin, and melanin-concentrating hormone (MCH), together with activation of genes encoding anorexigenic neuropeptides such as α-melanocyte-stimulating hormone (α-MSH) and cocaine- and amphetamine-regulated transcript (CART) (24, 25). To determine whether the impaired compensatory refeeding behavior observed in adult Ubb−/− mice is associated with impaired hypothalamic neurohormonal signaling, we measured the responses of the genes encoding, NPY, AgRP, and proopiomelanocortin (POMC), the precursor of α-MSH (Fig. 3C). In young mice fed ad libitum (Fig. 3C Upper), the levels of these neuropeptide mRNAs were similar, irrespective of genotype. Upon fasting, the levels of these mRNAs in both young and adult littermate controls exhibited stereotypical responses, with increased expression of genes encoding orexigenic neuropeptides, NPY and AgRP, and decreased expression of the gene encoding the anorexigenic neuropeptide precursor POMC. In young Ubb−/− mice, whereas NPY and POMC mRNA levels, respectively, increased and decreased in response to food deprivation, AgRP mRNA levels failed to respond to fasting (Fig. 3C, Upper). By contrast, in adult Ubb−/− mice (Fig. 3C Lower), the response of all three mRNA levels to fasting was severely attenuated. Moreover, the basal levels of AgRP mRNA were significantly reduced, even in fed Ubb−/− mice. Thus, loss of Ubb results in an early-onset defect in AgRP signaling and a profound, late-onset disruption in the responsiveness of orexigenic and anorexigenic neuropeptide genes to feeding status.
Adult-Onset Degeneration of Hypothalamic Neurons in Ubb−/− Mice.
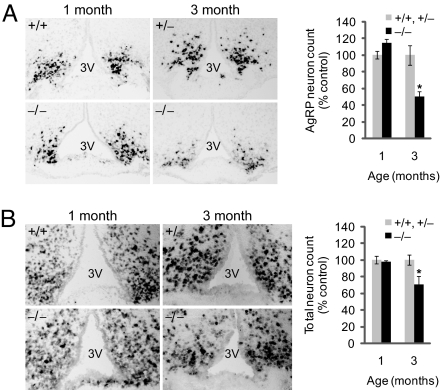
The dramatic impairment in AgRP transcripts in adult Ubb−/− mice could be attributable either to silencing of AgRP gene transcription or to a loss of AgRP neurons. To discriminate between these two possibilities, we examined brains of adult Ubb−/− mice for evidence of neurodegeneration. The spatial distribution of AgRP mRNA expression in the arcuate nucleus, assessed by in situ hybridization, revealed a significant, nearly 50% decrease in Ubb−/− mice at 3 months of age; this difference was not evident in 1-month-old mice (Fig. 4A). Strikingly, total neuron counts in the arcuate nucleus of Ubb−/− mice, assessed by in situ hybridization with a panneuronal probe, neuron-specific enolase (NSE), were significantly reduced by ≈30% in 3-month-old Ubb−/− mice compared with controls, whereas no difference was evident at 1 month of age (Fig. 4B). These findings suggest that adult-onset obesity in mice lacking Ubb is strongly correlated with the selective degeneration of neurons that control energy balance in the arcuate nucleus and imply that Ubb must play a unique role among ubiquitin genes in the survival of these neurons.
Fig. 4.
Degeneration of hypothalamic neurons in adult Ubb−/− mice. In situ hybridization of AgRP (A) and NSE (B) mRNA from Ubb−/− mice (n = 3 or 4) and littermate controls (n = 3 or 4). Images are representative for each age and group. For quantification, stereological counts were obtained from eight serial sections generated from each brain. 3V, third ventricle. The data are expressed as means ± SEM from the indicated number of mice. ∗, P < 0.05.
Contribution of Ubb to the Maintenance of Ub Levels in Hypothalamic Neurons.
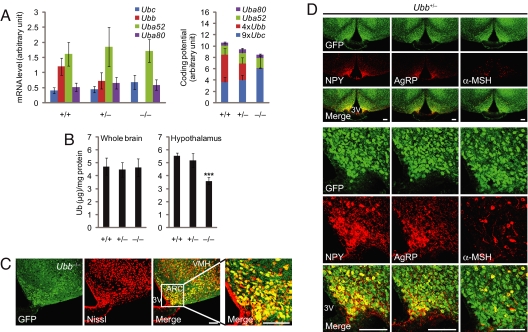
Quantitative real-time RT-PCR analysis of mRNA products of the four ubiquitin genes in hypothalamus indicated that Ubb expression levels are ≈3 times higher than that of the other polyubiquitin gene, Ubc, in wild-type mice. Thus, the slight increase in Ubc expression in Ubb−/− mice could only partially compensate for the loss of Ubb (Fig. 5A). Indeed, in hypothalamus of wild-type mice, the Ub-coding potential of Ubb (i.e., the absolute level of transcript multiplied by the number of ubiquitins per transcript) is clearly dominant. Whereas total Ub levels did not differ significantly among different Ubb genotypes in whole brain, Ub content was selectively reduced by nearly 30% in the hypothalamus of Ubb−/− compared with wild-type mice (Fig. 5B), consistent with the predictions based on mRNA quantification (Fig. 5A).
Fig. 5.
Dominant contribution of Ubb to Ub content of hypothalamus. (A) Quantification of ubiquitin transcript levels in hypothalamus of 3- to 4-month-old mice. (Left) Indicated mRNA levels, measured by quantitative real-time RT-PCR (+/+, n = 5; +/−, n = 6; −/−, n = 4). (Right) Relative contribution of ubiquitin genes to total Ub-coding potential normalized to the number of Ub moieties encoded by each ubiquitin transcript. The data are expressed as means ± SEM from the indicated number of mice. (B) Total Ub content in whole brain (n = 6 per genotype) and hypothalamus (+/+, n = 6; +/−, n = 3; −/−, n = 4) from 5- to 6-month-old mice. The data are expressed as means ± SEM from the indicated number of mice. ∗∗∗, P < 0.001 vs. Ubb+/+ mice. (C) Neuronal pattern of Ubb expression in the arcuate nucleus. Representative confocal images of GFP-Puror and Nissl stain from 4-month-old Ubb−/− mice (n = 3) are shown. The far right image shows the arcuate nucleus at higher magnification. VMH, ventromedial hypothalamus; ARC, arcuate nucleus; 3V, third ventricle. (Scale bars, 100 μm.) (D) Colocalization of NPY, AgRP, and α-MSH immunoreactive neurons in the arcuate nucleus to Ubb-expressing neurons. Representative confocal images of GFP-Puror and NPY, AgRP, and α-MSH immunoreactivity from 4-month-old Ubb+/− mice (n = 4) are shown. 3V, third ventricle. (Scale bars, 100 μm.)
The spatial distribution of Ubb gene expression, determined by confocal microscopy of the GFP-puror fusion protein knocked in to the Ubb locus, revealed that Ubb was highly expressed in a subset of neurons within the arcuate nucleus (Fig. 5C). By contrast, fluorescence of GFP-puror knocked in to the Ubc locus (15) was distinctly less intense and more diffusely distributed in hypothalamus and cerebral cortex (SI Fig. 7), consistent with the conclusion that Ubb is the predominant polyubiquitin gene in these brain regions. Indeed, Ubb expression, as assessed by GFP fluorescence, corresponded precisely with the localization of the neuropeptides NPY, AgRP, and α-MSH, indicating that the neurons that are most vulnerable to ablation of Ubb are those that express the highest levels of this polyubiquitin gene (Fig. 5D).
Discussion
We report here that disruption of murine Ubb gives rise to a novel neurodegenerative syndrome characterized initially by dysfunction of neurons within the CNS and progressing, over the course of 4 months, to neuronal loss within the hypothalamus. These findings demonstrate that Ubb transcription is essential for neuronal function and survival.
Sufficient Ub Levels Are Essential for Neuronal Survival.
Abnormal accumulation of Ub conjugates within cytoplasmic or nuclear inclusion bodies in affected neurons is a diagnostic feature of nearly all adult-onset neurodegenerative diseases, long suggesting that disruption of Ub homeostasis may be a common factor in the pathogenesis of these otherwise diverse disorders (21, 22). The obvious possibility that sequestration of Ub into intracellular inclusion bodies may lead to depletion of Ub availability has not received serious attention because it is widely assumed that the cellular supplies of this highly abundant protein are never limiting. However, the extent to which Ub levels are in excess of demand under basal or stressed conditions in any mammalian cell or tissue is not known. The finding that modest reduction in the level of free Ub in brain is linked to synaptic dysfunction and neuronal degeneration associated with loss-of-function mutations in DUBs in the axJ mouse (20) and the gad mouse (6), respectively, suggests that adequate neuronal Ub supply appears to be maintained by a surprisingly precarious homeostasis. It remains to be determined whether the phenotypes of these DUB mutants are simply attributable to reduced availability of free Ub or are associated with other manifestations of altered Ub homeostasis that could arise from defects in the activity or specificity of DUBs (26). However, because the sole known function of Ubb is to provide Ub molecules (which are chemically identical to the products of the other ubiquitin genes), the simplest interpretation of our data is that reduced Ub availability causes a progressive syndrome beginning with impaired neuronal function and resulting in neuronal death.
The primary translation product of Ubb is a tetraubiquitin polyprotein that is assumed to be efficiently processed into individual Ub moieties during or shortly after its synthesis (27, 28). Although unlikely, we cannot, at present, exclude the possibility that this short-lived Ub polyprotein could serve some unknown, but essential, function in neurons before proteolytic processing. Considering that Ub levels were reduced by ≈30% in the hypothalamus, the most affected region of the brains of Ubb−/− mice, and that the contribution of Ubb to total Ub was likely to be higher in the subset of neurons within the arcuate nucleus than the regional average, it is likely that Ub levels are even more substantially decreased in specific neuronal subpopulations. Indeed, we find a striking correspondence between the vulnerable neurons within the arcuate nucleus and those that express the highest levels of GFP-puror.
Despite the critical role played by Ub in a broad spectrum of cellular processes including cell cycle regulation, DNA repair, transcriptional regulation, stress response, protein quality control, signal transduction, endocytosis, antigen processing, and apoptosis (1, 2), surprisingly little is known about the mechanisms that control ubiquitin gene expression in metazoan organisms. The data reported here indicate that a deeper understanding of the homeostatic mechanisms that control neuronal ubiquitin gene expression to maintain Ub levels may help to illuminate the pathogenic mechanisms that underlie degenerative diseases and perhaps may suggest new therapeutic avenues in their treatment.
Hypothalamic Phenotype of the Ubb Knockout.
Although Ubb−/− mice are smaller than wild-type or heterozygous littermates at birth and fail to exhibit a growth surge after weaning, they do eventually achieve and maintain stable body weights comparable with those of Ubb+/+ or Ubb+/− mice. This is reminiscent of the phenotype of mice lacking melanocortin-3 receptors (Mc3r), which exhibit normal food intake but store fat more efficiently (29, 30). Like Ubb−/−, Mc3r−/− mice exhibit decreased linear growth and mild late-onset obesity without hyperphagia. Mice lacking Ubb resemble Mc3r-deficient mice in that both models display modestly increased postweaning body fat content with a compensating reduction in lean body mass, such that their body weights remain within the normal range. Both knockout strains exhibit normal levels of circulating glucose and insulin but elevated leptin. The similarity between the metabolic phenotypes of Ubb−/− and Mc3r−/− mice suggest that Ub may serve a particularly important function in Mc3r signaling or, perhaps, even for the survival of Mc3r neurons.
The obesity of Ubb−/− mice differs from the phenotypes of other murine obesity models, particularly those that disrupt the proximal signals in leptin circuitry. For example, disruption of the genes encoding leptin, leptin receptor (Ob-R) (31), or STAT3, which transduces signals directly downstream of Ob-R (32), result in severe early-onset obesity characterized by constitutive hyperphagia, greatly increased body weight, and a ≈5-fold increase in adiposity. Likewise, morbid obesity results from impairment of melanocortin signaling by disruption of the gene encoding POMC (33) or overexpression of the melanocortin receptor antagonist AgRP (34, 35).
Depletion of Ub from hypothalamic neurons is likely to interfere with the integrated signaling network that controls energy balance and feeding in complex ways that reflect the balance between the maintenance of Ub supply and the demands of the most Ub-intensive pathways in the subsets of neurons in which the contribution of Ubb to Ub supply is greatest. In this respect, the phenotype of Ubb−/− mice is more akin to neuron ablation than to individual gene knockouts. However, loss of Ubb led to degeneration of ≈30% of total neurons from the arcuate nucleus, implying that this gene is required for the survival of more than just AgRP neurons. Indeed, recent data indicate that multiple neuronal signaling and survival pathways are disrupted throughout the hypothalamus of Ubb−/− mice (K.-Y.R. and R.R.K., unpublished data). Thus, the disruption of body weight homeostasis in Ubb−/− mice is likely the result of impaired function of multiple hypothalamic networks. Deconvolution of the relative contributions of these different circuits to the overall phenotype will require targeted, cell-type specific gene knockout approaches.
Injection of neonatal mice with monosodium glutamate (MSG) produces hypothalamic lesions that selectively destroy the arcuate nucleus (36). Like Ubb−/− mice, adult mice with MSG-induced lesions are obese but not hyperphagic and have reduced snout–anus length (36). By contrast, selective ablation of AgRP neurons does not result in obesity (37). The finding of high GFP-puror expression in several types of neurons in Ubb+/− mice, together with the demonstration of ≈30% reduction in the number of total neurons in Ubb−/− mice (assessed by NSE in situ hybridization), suggests that Ubb is particularly important for the survival and function of neurons within the arcuate nucleus. These findings imply that either the relative contribution of Ubb to the maintenance of basal Ub levels or the ability of other ubiquitin genes to compensate for the loss of Ubb is greatest in the arcuate nucleus. Our observations do not exclude the possibility that Ubb−/− mice may exhibit other neurological phenotypes resulting from the contribution of this gene to the maintenance of Ub homeostasis and neuronal function in other parts of the brain. Nevertheless, these data reveal an essential role for Ubb in the maintenance of neuronal Ub levels and establish that decreased Ub availability is sufficient to cause neuronal dysfunction and death. These findings suggest that a deeper investigation of the role of Ub depletion in the pathogenesis of both sporadic and hereditary neurodegenerative diseases is needed.
Materials and Methods
Mouse Studies.
All mice were kept in plastic cages with ad libitum access to food and water, with 12-h light cycle (7 a.m. to 7 p.m.). All procedures followed National Institutes of Health guidelines with the approval of Stanford University Administrative Panel on Laboratory Animal Care. For the fasting/refeeding study, mice were individually housed for 1 week and daily food intake was measured at 4 p.m. to 5 p.m. for 7 consecutive days. For fasting, food was removed at 4 p.m. to 5 p.m. for 48 h and mice only had ad libitum access to water. Refeeding was also started at 4 p.m. to 5 p.m., and food intake was measured at 24 and 48 h later. Body fat content was determined by PIXImus DEXA. Mice were anesthetized by i.p. injection of 2.5% avertin (0.011–0.014 ml/g body weight). After body composition measurement, mice were always monitored for wake up and returning to normal behavior.
Quantitative Real-Time RT-PCR.
Real-time RT-PCR was performed as described previously (15). Additional details can be found in SI Materials and Methods.
In Situ Hybridization.
In situ hybridization was performed essentially as described previously (38–40). See SI Materials and Methods for details.
Confocal Microscopy.
Ad libitum-fed mice were anesthetized and perfused transcardially with ice-cold PBS followed by 4% paraformaldehyde. Brains were postfixed in 4% paraformaldehyde at 4°C for 12 h, cryoprotected with 30% sucrose, and sectioned with microtome in the coronal plane. A series of free-floating sections (25 μm thick) were generated from 1.5 to 2.1 mm posterior to bregma and stored at −20°C in a cryoprotective medium (25% glycerol, 30% ethylene glycol, 45% PBS) until use. Ubb transcriptional activity was monitored by direct visualization of GFP fluorescence. Briefly, sections were washed with TBS and stained with TO-PRO-3 iodide (1:1,000; Molecular Probes) in 0.3% Triton X-100/Tris-buffered saline (TBST) for 30 min at room temperature for visualization of DNA. Sections were washed with TBST followed by TBS only and mounted on slides with ProLong Gold antifade reagent (Molecular Probes). To mark for neurons, Nissl staining was carried out by using the NeuroTrace red fluorescent Nissl staining kit (1:20; Molecular Probes) according to the protocol of the manufacturer. To stain the neuropeptide-containing neuronal cell bodies, mice were pretreated with i.c.v. injection of colchicine (Sigma) 48 h before euthanasia to block axonal transport. Deeply anesthetized mice received 1.5 to 2 μl of colchicine solution (15 μg/μl in 0.9% sterile saline, 0.75 μg/g body weight) over a 5-min period at 1 mm lateral and 0.3 mm posterior to bregma and 2 mm ventral to the surface of the skull. Free-floating sections were generated as described above, washed with TBS, permeabilized with TBST, and blocked with 3% normal goat serum in TBST for 1 h at room temperature. For NPY, AgRP, and α-MSH immunofluorescence, sections were incubated with anti-NPY (1:200; Immunostar), AgRP (1:500) (41), and α-MSH (1:200; Immunostar) polyclonal antibodies in blocking buffer at 4°C overnight, washed with TBST, and incubated with Alexa Fluor 555-conjugated goat anti-rabbit IgG (1:200; Molecular Probes) and TO-PRO-3 iodide (1:1,000) in blocking buffer for 1 h at room temperature. Sections were washed and mounted as described above. Confocal images were collected with a TCS SP2 laser scanning system (Leica) with sequential image recording.
Indirect Competitive ELISA.
Tissue lysates were treated with Usp2-cc (42, 43) and subjected to indirect competitive ELISA as described previously (7). Additional details can be found in SI Materials and Methods.
Statistical Analysis.
Two-tailed unpaired Student's t test with equal or unequal variance, which was determined by F test, was used to compare the data between two groups. P < 0.05 was considered to be statistically significant.
Supplementary Material
ACKNOWLEDGMENTS.
We thank our colleagues in the laboratory of R.R.K. for helpful discussions and comments; in particular, we are grateful to Catherine Gilchrist for help with screening the mouse genomic DNA library and Preeti Nayyar for help with body weight measurement. We thank Allison Xu, Nobuhiro Fujiki, and Seiji Nishino for technical assistance in animal studies; Melissa Kazantzis and Andreas Stahl for helpful discussions; and the Transgenic Research Center at Stanford University for DNA microinjection, ES cell manipulation, and generation of chimeric mice. This work was supported in part by a grant from the Fidelity Foundation and a grant from the National Institute of Aging.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800096105/DC1.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 3.D'Andrea A, Pellman D. Deubiquitinating enzymes: A new class of biological regulators. Crit Rev Biochem Mol Biol. 1998;33:337–352. doi: 10.1080/10409239891204251. [DOI] [PubMed] [Google Scholar]
- 4.Ohtani-Kaneko R, et al. Nerve growth factor (NGF) induces increase in multi-ubiquitin chains and concomitant decrease in free ubiquitin in nuclei of PC12h. Neurosci Res. 1996;26:349–355. doi: 10.1016/s0168-0102(96)01117-0. [DOI] [PubMed] [Google Scholar]
- 5.Takada K, Hibi N, Tsukada Y, Shibasaki T, Ohkawa K. Ability of ubiquitin radioimmunoassay to discriminate between monoubiquitin and multi-ubiquitin chains. Biochim Biophys Acta. 1996;1290:282–288. doi: 10.1016/0304-4165(96)00032-3. [DOI] [PubMed] [Google Scholar]
- 6.Osaka H, et al. Ubiquitin carboxy-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neuron. Hum Mol Genet. 2003;12:1945–1958. doi: 10.1093/hmg/ddg211. [DOI] [PubMed] [Google Scholar]
- 7.Ryu KY, Baker RT, Kopito RR. Ubiquitin-specific protease 2 as a tool for quantification of total ubiquitin levels in biological specimens. Anal Biochem. 2006;353:153–155. doi: 10.1016/j.ab.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 8.Hanna J, Leggett DS, Finley D. Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol Cell Biol. 2003;23:9251–9261. doi: 10.1128/MCB.23.24.9251-9261.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lund PK, et al. Nucleotide sequence analysis of a cDNA encoding human ubiquitin reveals that ubiquitin is synthesized as a precursor. J Biol Chem. 1985;260:7609–7613. [PubMed] [Google Scholar]
- 10.Wiborg O, et al. The human ubiquitin multigene family: some genes contain multiple directly repeated ubiquitin coding sequences. EMBO J. 1985;4:755–759. doi: 10.1002/j.1460-2075.1985.tb03693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker RT, Board PG. The human ubiquitin gene family: structure of a gene and pseudogenes from the Ub B subfamily. Nucleic Acids Res. 1987;15:443–463. doi: 10.1093/nar/15.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finley D, Bartel B, Varshavsky A. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature. 1989;338:394–401. doi: 10.1038/338394a0. [DOI] [PubMed] [Google Scholar]
- 13.Redman KL, Rechsteiner M. Identification of the long ubiquitin extension as ribosomal protein S27a. Nature. 1989;338:438–440. doi: 10.1038/338438a0. [DOI] [PubMed] [Google Scholar]
- 14.Baker RT, Board PG. The human ubiquitin-52 amino acid fusion protein gene shares several structural features with mammalian ribosomal protein genes. Nucleic Acids Res. 1991;19:1035–1040. doi: 10.1093/nar/19.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryu KY, et al. The mouse polyubiquitin gene UbC is essential for fetal liver development, cell-cycle progression and stress tolerance. EMBO J. 2007;26:2693–2706. doi: 10.1038/sj.emboj.7601722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryu KY, et al. The mouse polyubiquitin gene Ubb is essential for meiotic progression. Mol Cell Biol. 2008;28:1136–1146. doi: 10.1128/MCB.01566-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watts RJ, Hoopfer ED, Luo L. Axon pruning during Drosophila metamorphosis: Evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron. 2003;38:871–885. doi: 10.1016/s0896-6273(03)00295-2. [DOI] [PubMed] [Google Scholar]
- 18.Patrick GN. Synapse formation and plasticity: Recent insights from the perspective of the ubiquitin proteasome system. Curr Opin Neurobiol. 2006;16:90–94. doi: 10.1016/j.conb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 19.Mizuno Y, Hattori N, Mori H, Suzuki T, Tanaka K. Parkin and Parkinson's disease. Curr Opin Neurol. 2001;14:477–482. doi: 10.1097/00019052-200108000-00008. [DOI] [PubMed] [Google Scholar]
- 20.Anderson C, et al. Loss of Usp14 results in reduced levels of ubiquitin in ataxia mice. J Neurochem. 2005;95:724–731. doi: 10.1111/j.1471-4159.2005.03409.x. [DOI] [PubMed] [Google Scholar]
- 21.Lowe J, et al. Ubiquitin is a common factor in intermediate filament inclusion bodies of diverse type in man, including those of Parkinson's disease, Pick's disease, and Alzheimer's disease, as well as Rosenthal fibres in cerebellar astrocytomas, cytoplasmic bodies in muscle, and mallory bodies in alcoholic liver disease. J Pathol. 1988;155:9–15. doi: 10.1002/path.1711550105. [DOI] [PubMed] [Google Scholar]
- 22.Perry G, Friedman R, Shaw G, Chau V. Ubiquitin is detected in neurofibrillary tangles and senile plaque neurites of Alzheimer disease brains. Proc Natl Acad Sci USA. 1987;84:3033–3036. doi: 10.1073/pnas.84.9.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennett EJ, et al. Global changes to the ubiquitin system in Huntington's disease. Nature. 2007;448:704–708. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- 24.Friedman JM, Halaas JL. Leptin and the regulation of body weight in mammals. Nature. 1998;395:763–770. doi: 10.1038/27376. [DOI] [PubMed] [Google Scholar]
- 25.Friedman JM. Modern science versus the stigma of obesity. Nat Med. 2004;10:563–569. doi: 10.1038/nm0604-563. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT., Jr The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson's disease susceptibility. Cell. 2002;111:209–218. doi: 10.1016/s0092-8674(02)01012-7. [DOI] [PubMed] [Google Scholar]
- 27.Finley D, Ozkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- 28.Ozkaynak E, Finley D, Solomon MJ, Varshavsky A. The yeast ubiquitin genes: A family of natural gene fusions. EMBO J. 1987;6:1429–1439. doi: 10.1002/j.1460-2075.1987.tb02384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butler AA, et al. A unique metabolic syndrome causes obesity in the melanocortin-3 receptor-deficient mouse. Endocrinology. 2000;141:3518–3521. doi: 10.1210/endo.141.9.7791. [DOI] [PubMed] [Google Scholar]
- 30.Chen AS, et al. Inactivation of the mouse melanocortin-3 receptor results in increased fat mass and reduced lean body mass. Nat Genet. 2000;26:97–102. doi: 10.1038/79254. [DOI] [PubMed] [Google Scholar]
- 31.Coleman DL. Obese and diabetes: two mutant genes causing diabetes-obesity syndromes in mice. Diabetologia. 1978;14:141–148. doi: 10.1007/BF00429772. [DOI] [PubMed] [Google Scholar]
- 32.Gao Q, et al. Disruption of neural signal transducer and activator of transcription 3 causes obesity, diabetes, infertility, and thermal dysregulation. Proc Natl Acad Sci USA. 2004;101:4661–4666. doi: 10.1073/pnas.0303992101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yaswen L, Diehl N, Brennan MB, Hochgeschwender U. Obesity in the mouse model of pro-opiomelanocortin deficiency responds to peripheral melanocortin. Nat Med. 1999;5:1066–1070. doi: 10.1038/12506. [DOI] [PubMed] [Google Scholar]
- 34.Ollmann MM, et al. Antagonism of central melanocortin receptors in vitro and in vivo by agouti-related protein. Science. 1997;278:135–138. doi: 10.1126/science.278.5335.135. [DOI] [PubMed] [Google Scholar]
- 35.Graham M, Shutter JR, Sarmiento U, Sarosi I, Stark KL. Overexpression of Agrt leads to obesity in transgenic mice. Nat Genet. 1997;17:273–274. doi: 10.1038/ng1197-273. [DOI] [PubMed] [Google Scholar]
- 36.Olney JW. Brain lesions, obesity, and other disturbances in mice treated with monosodium glutamate. Science. 1969;164:719–721. doi: 10.1126/science.164.3880.719. [DOI] [PubMed] [Google Scholar]
- 37.Xu AW, et al. Effects of hypothalamic neurodegeneration on energy balance. PLoS Biol. 2005;3:e415. doi: 10.1371/journal.pbio.0030415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, et al. The melanocortinergic pathway is rapidly recruited by acute emotional stress and contributes to stress-induced anorexia and anxiety-like behavior. Endocrinology. 2007;148:5531–5540. doi: 10.1210/en.2007-0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu XY, Barsh GS, Akil H, Watson SJ. Interaction between alpha-melanocyte-stimulating hormone and corticotropin-releasing hormone in the regulation of feeding and hypothalamo-pituitary-adrenal responses. J Neurosci. 2003;23:7863–7872. doi: 10.1523/JNEUROSCI.23-21-07863.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu XY, et al. Diurnal rhythm of agouti-related protein and its relation to corticosterone and food intake. Endocrinology. 2002;143:3905–3915. doi: 10.1210/en.2002-220150. [DOI] [PubMed] [Google Scholar]
- 41.Xu AW, et al. PI3K integrates the action of insulin and leptin on hypothalamic neurons. J Clin Invest. 2005;115:951–958. doi: 10.1172/JCI24301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Catanzariti AM, Soboleva TA, Jans DA, Board PG, Baker RT. An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Sci. 2004;13:1331–1339. doi: 10.1110/ps.04618904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker RT, et al. Using deubiquitylating enzymes as research tools. Methods Enzymol. 2005;398:540–554. doi: 10.1016/S0076-6879(05)98044-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.