Abstract
Understanding the prevalence and polymorphism of antibiotic resistance genes in soil bacteria and their potential to be transferred horizontally is required to evaluate the likelihood and ecological (and possibly clinical) consequences of the transfer of these genes from transgenic plants to soil bacteria. In this study, we combined culture-dependent and -independent approaches to study the prevalence and diversity of bla genes in soil bacteria and the potential impact that a 10-successive-year culture of the transgenic Bt176 corn, which has a blaTEM marker gene, could have had on the soil bacterial community. The bla gene encoding resistance to ampicillin belongs to the beta-lactam antibiotic family, which is widely used in medicine but is readily compromised by bacterial antibiotic resistance. Our results indicate that soil bacteria are naturally resistant to a broad spectrum of beta-lactam antibiotics, including the third cephalosporin generation, which has a slightly stronger discriminating effect on soil isolates than other cephalosporins. These high resistance levels for a wide range of antibiotics are partly due to the polymorphism of bla genes, which occur frequently among soil bacteria. The blaTEM116 gene of the transgenic corn Bt176 investigated here is among those frequently found, thus reducing any risk of introducing a new bacterial resistance trait from the transgenic material. In addition, no significant differences were observed in bacterial antibiotic-resistance levels between transgenic and nontransgenic corn fields, although the bacterial populations were different.
Keywords: antibiotic resistance, GMO, HGT
Because of their efficiency against susceptible Gram-positive and proteobacteria (1), their specificity, and a general absence of toxicity for humans, beta-lactam antibiotics, including penicillin, ampicillin, cephalosporins, and monobactams, are the largest antibiotic family used in medicine. However, the use of these antibiotics is readily compromised by bacterial antibiotic resistance (2, 3), partly because of the production of beta-lactamases. The most common beta-lactamases are the TEM beta-lactamases that are encoded by the blaTEM1 gene and its descendants. TEM1 beta-lactamase hydrolyzes only penicillins and some older cephalosporins, but descendants of TEM1 have evolved the ability to hydrolyze modern cephalosporins and monobactams (4). Indeed, the widespread use of beta-lactams in the past few decades has led to the evolution of a new generation of beta-lactamases, which have extended substrate spectra [i.e., extended spectrum beta-lactamases (ESBLs)]. ESBLs are generally plasmid-mediated enzymes that confer resistance to oxyimino-beta-lactams, such as cefotaxime, ceftazidime, and aztreonam (5). Numerous questions remain about the role that natural environments play in the maintenance and dispersion of antibiotic resistance genes and about the frequency with which these genes are exchanged among indigenous bacteria and whether they can spread from these commensal strains to clinical isolates. After many years of contradictory data about the actual production of antibiotics by indigenous microorganisms in soil (6), several reports were published confirming that antibiotics are produced in soils at sufficiently high concentrations to inhibit bacterial growth in the vicinity of the producers (7–10). These results suggest that in situ conditions are selective enough to drive the development of antibiotic resistance mechanisms as a necessary survival strategy to guard against antibiotic-producing strains. Moreover, the use of antibiotics in therapeutic treatments or as growth promoters and field cultivation of some genetically modified plants (GMPs) are suspected to increase the risk of antibiotic resistance gene dissemination. Several commercial GMPs contain antibiotic resistance genes that are still under the control of bacterial promoters as remnants of the bacterial vectors used to construct the GMPs. These former bacterial genes could be transferred more easily than other plant genes to soil bacteria because of a high degree of homology facilitating recombination in potential bacterial recipients (11). This might be the case for the insect-protected Bt176 corn that confers protection against attack by Lepidopteran insects because of the production of a Bt toxin. A bla gene under a bacterial promoter control that encodes a TEM116 beta-lactamase conferring resistance to ampicillin was cointroduced into corn during the Bt176 event.
The possible impact of GMPs in terms of gene transfer to bacteria and modification of the soil microbial community with antibiotic resistance genes cannot be evaluated without considering the initial ecology of these genes in this community. Indeed, the impact level would strongly depend upon the natural prevalence of these genes in soil bacteria and their polymorphism that reflects their actual evolutionary level susceptible of providing the community with the ability to hydrolyze a more or less wide range of antibiotic molecules.
We addressed the questions of antibiotic resistance gene evolution and GMPs impact by investigating the diversity of bla genes in soil bacteria from fields cultured for as many as 10 successive years with transgenic (Bt176 event) and traditional corn lines and from a prairie soil in the same area. The study was based on a combination of culture-dependent and -independent approaches that aimed to determine the actual state of evolution of bla genes in soils. Then, we addressed the potential impact of the transgenic plant on the prevalence of bla genes and the emergence of new resistant strains. In addition, the modifications of the bacterial community structure due to a potential (trans)gene transfer to bacteria and/or a direct effect of the GMPs was monitored by these methodological approaches and by the use of a 16S rRNA microarray (phylochip).
Results and Discussion
The study was conducted on soils sampled in Bazièges (Station Inter Institut Association Générale des Producteurs de MaïsAGPM-Technique, Haute-Garonne, France), where transgenic Bt176 corn (T1 and T2 soil samples) and traditional corn (C1 and C2 soil samples) have been cultured in separate fields for 10 successive years. Soil samples from a prairie (P1 and P2 soil samples), recovered in the same area, were also analyzed. The insect-protected Bt176 corn was obtained by microprojectile bombardment into immature embryos of inbred corn line CG00526 (Zea mays L.), using two different plasmids, including pCIB4431 and pCIB3064, both containing a copy of the blaTEM116 gene that confers resistance to ampicillin and carbenicillin under the control of a bacterial promoter.
Total and Beta-Lactam-Resistant Cultivable Bacteria in Soils.
Soil suspensions plated on appropriate media led to the recovery of cultivable bacteria and among them ampicillin-resistant isolates potentially carrying a bla gene. The total number of cultivable bacteria was not significantly different (0.36 < P < 0.62) between the three soils (1.7 × 104 in C1 to 2.5 × 105 in T2) [supporting information (SI) Table 1], but the level of resistant isolates was significantly different (P < 0.01) between cultivated and prairie soils. In the corn fields, the prevalence of cultivable ampicillin-resistant bacteria exhibited some heterogeneity that correlated not to the transgenic status of the plant but rather to field location. Percentages of resistant bacteria varied from 0.4% to 6.5% of total cultivable bacteria between samples T2 and T1, respectively, and from 5.5% to 8.0% in samples C2 and C1, respectively (SI Table 1). These results are consistent with other studies that found that the population levels of ampicillin-resistant bacteria in corn field soils did not exhibited significant difference between Bt176 transgenic and nontransgenic corns but did significantly differ with field location, sampling stage, and year of study (12). In prairie soil, prevalence of ampicillin-resistant bacteria is significantly higher, ranging from 54.4% to 69.6% in P2 and P1 samples, respectively (SI Table 1). This is a possible indication that bacterial communities not disturbed by agricultural practices may be more exposed to antibiotic-producing strains or in better contact for gene exchange and, as a consequence present a higher degree of antibiotic resistance.
A library of 576 ampicillin-resistant isolates (96 isolates per soil sample) was constructed. Beta-lactam resistance pattern of these isolates was determined by testing growth on media supplemented with antibiotics of the penicillin group, including ampicillin, carbenicillin, amoxicillin, penicillin G, and oxacillin, and the cephalosporin group, including cephalothin, cefamandole, and ceftriaxone, which are first-, second-, and third-generation cephalosporins, respectively.
Initially, we determined the resistance pattern of Escherichia coli DH5α and found that the presence of pCIB4431 plasmid, which was used to construct transgenic corn Bt176 and carried the blaTEM116 gene, conferred a robust resistance to penicillins, but would not provide the resistance to second- and third-generation cephalosporins. Then, the resistance pattern was established for all soil isolates, which exhibited a broad beta-lactam resistance spectrum (SI Fig. 2). This led to the observation that none of the 576 isolates tested exhibited the same resistance pattern as E. coli containing pCIB4431 because of their natural extended antibiotic resistance pattern. Almost all of the isolates from the three soils were resistant to penicillins and cephalosporin antibiotics belonging to the first and second generation, except sample T2, where bacteria were slightly less resistant to the second cephalosporin generation (94.6%). The only discriminating antibiotic belonged to the third cephalosporin generation, with resistance percentages ranging from 38.7% in sample P2 to 96.4% in sample C1. No marked difference was observed in bacterial antibiotic resistance level and pattern between transgenic and nontransgenic corn fields (SI Fig. 2 and SI Table 1). These data indicate that the growth of transgenic plants for >10 successive years on the same field has not increased the measurable level and spectrum of antibiotic resistance in the cultivable soil bacterial fraction, thus excluding any cumulative effect related to the constant release of transgenic plant DNA. The observed multiantibiotic resistance patterns of soil communities has been reported with bacteria resisting up to eight antibiotics belonging to various other antibiotic families (13, 14).
Diversity of blaTEM Genes in Bacterial Isolates and PCR-Amplified DNA from Soils.
Presence and putative diversity of the blaTEM genes in the 576 ampicillin and carbenicillin-resistant soil isolates was studied after genomic DNA was amplified by PCR. The expected 828-bp PCR product, which generates 295- and 553-bp RsaI cut fragments according to the published sequence (GenBank accession no. AY425988), were obtained for 505 of these 576 genomic DNA (87.7%), confirming that the use of the two antibiotics efficiently selected blaTEM gene harboring strains or that there is a natural prevalence of this gene among ampicillin–carbenicillin-resistant soil bacteria. For the 71 other sequences, each digestion pattern was sequenced and did not correspond to a bla sequence (results not shown).
For soil isolates, 80 PCR products were sequenced (15 from T1, 10 from T2, 16 from C1, 11 from C2, 12 from P1, and 16 from P2). Ten were identical to the blaTEM116 sequence of the transgenic corn (four T1, two C2, one P1, and three P2 sequences). This specific sequence was detected in bacteria isolated from the transgenic and nontransgenic corn fields and from the prairie soil. Some of these bacteria were resistant to ceftriaxone, indicating that these bacteria contained other antibiotic resistance mechanisms in addition to that encoded by the blaTEM116 gene or that its expression in soil bacteria increased the antibiotic resistance spectrum to third-generation cephalosporins. The 70 other PCR products had one nucleotide different from the blaTEM116 sequence (nucleotide 232, where G replaces A), which would substitute an isoleucin by a valin residue (neutral evolution) to provide a bla sequence used in cloning vectors (GenBank accession nos. X56095, L42764, AJ299425, and AJ431686) and encoding ampicillin resistance (15). Several PCR products yielded both A and G at position 232. Their cloning and sequencing revealed that some soil isolates carried at least two blaTEM genes. This agrees with data demonstrating that clinical isolates can produce as many as five distinct beta-lactamases (16). The observation of only two types of blaTEM gene sequences in soil isolates might demonstrate the restricted soil diversity compared with clinical environments or that the high ampicillin and carbenicillin concentration and the culture medium used strongly selected genes and/or bacteria. To test these hypotheses, we first conducted the analysis of 16S rDNA sequences of 16 of these isolates (4 strains from T soil, 6 strains from C soil, and 6 strains for P soil). The 16S rDNA sequences classified isolates as belonging to only two genera, 15 as Pseudomonas genus (99% similarity) and 1 as Stenotrophomonas sp. (99% similarity).
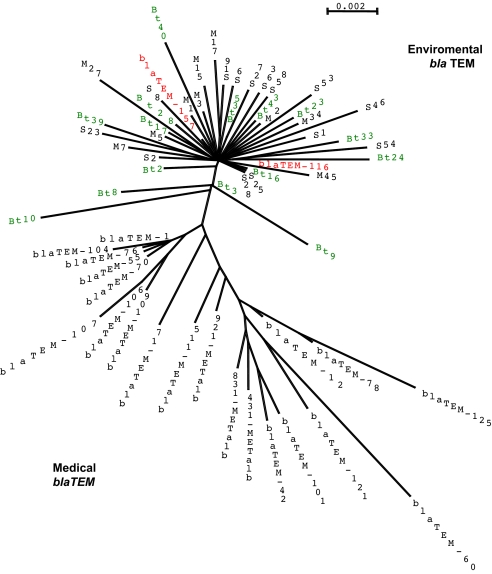
Because cultivable bacteria represented <1% of total bacteria, diversity of blaTEM genes in soil bacterial community was further investigated by a culture-independent strategy. Bacterial metagenomic DNA was extracted from soil samples and submitted to PCR, using primers bla1 and bla2 (SI Table 2). PCR products were cloned, and 178 inserts were selected on the basis of RsaI digestion pattern were sequenced (GenBank accession nos. EF514028–EF514205). This included 54, 56, and 68 PCR products from transgenic corn (named Bt1 to Bt54), nontransgenic corn (named M1 to M56), and prairie (named S1 to S68) soils, respectively. A group of 10 DNA inserts that originated from transgenic (five Bt sequences) and prairie (five S sequences) soil DNA exhibited the blaTEM116 gene sequence. A khi2 test, performed to compare the distribution of the blaTEM116 gene in the three soils demonstrated the absence of a transgenic plant effect (P < 0.10), thus confirming previous isolate-based data about its natural prevalence in environmental bacteria. In addition, this blaTEM116 gene was detected in DNA from another prairie soil (La Batie-Divisin, France) located 400 km away from the transgenic field, thus eliminating any possibility of contamination by the Bt176 plant DNA and reinforcing the potentially ubiquitous occurrence of this gene in prairie and agricultural soils. This result would justify a posteriori the choice of the blaTEM116 gene in the transgenic plant construction strategy, because it might have been already common in environmental bacteria and, thus, would limit the impact of a putative transfer from the transgenic plant to microbial biota. In addition to the blaTEM116 sequence, 153 different blaTEM nucleotide sequences were found in the PCR-amplified bacterial DNA, indicating a particularly high polymorphism level of this family of genes and confirming the interest in using a direct approach to avoid biases related to cultivation and selection methods. Polymorphism in the blaTEM genes was already noticed for clinical isolates with >150 variants (17). A phylogenetic relationship was established by using environmental and National Center for Biotechnology Information database sequences aligned between nucleotides 28 and 845 of blaTEM116 gene, corresponding to nucleotides 237 to 1054 of blaTEM1 gene, using the amino acid residue numeration as established in ref. 18. All of the blaTEM genes originating from soils grouped in a single cluster with only four sequences (Bt3, Bt8, Bt9, and Bt10) at the interface with sequences derived from medical facilities (Fig. 1) because of the presence of a G at position 458 (SI Fig. 3). Considering the low number of mismatches between blaTEM genes, the low bootstrap values, and the absence of a clear ancestor, the best representation was an unrooted phylogenic tree that demonstrates the separation between clinical and environmental strains except for the blaTEM116 and blaTEM157 genes, which might have arisen from environmental origins. An alignment analysis clarified the differences between medical and environmental blaTEM sequences. Some bases are preferentially represented in medical genes and others are preferentially represented in environmental sequences, such as positions 442, 458, and 759 in blaTEM1 sequence (SI Fig. 3). Substitution of a G by a A at position 458 and substitution of a C by a T at position 759 are particularly specific of soil sequences, because no medical bla other than blaTEM116 and blaTEM157 possessed these substitutions. Such results cannot be due to PCR biases related to bla1 and bla2 primers that were designed to amplify all blaTEM and most bla genes but would confirm that beta-lactamases can easily evolve through point mutations. Furthermore, strains characterized by intermediate mutation frequencies have significantly higher levels of antibiotic resistance than strains with low and high mutation frequencies (19). Correlation between point mutation and polymorphism could explain why uncultured soil bacteria are a reservoir of antibiotic resistance genes (20).
Fig. 1.
Phylogenic relationship of blaTEM sequences isolated from medical origin and amplified from a transgenic corn field (Bt), a traditional corn field (M), and a prairie soil (S). Green shading identifies blaTEM sequences from the transgenic corn field. Black shading identifies blaTEM sequences from other origins. Red shading identifies the two blaTEM medical sequences that might originate from the environment.
BLA Mutation Analyses.
To complete the previous study on bla diversity, we studied the potential activity of the partial BLA sequences retrieved from soils by comparing them to known beta-lactamases. The TEM1 beta-lactamase, a plasmid-borne enzyme (GenBank accession no. AF309824), was the first enzyme described in 1963 and is the most commonly detected in clinical bacteria (17, 21). Point mutations in plasmid bla genes expand the resistance spectrum of beta-lactamases even to the third cephalosporin generation, and most of the ESBLs from clinical isolates are mutants of TEM1 with one to four amino acid substitutions (5, 21).
In the present study, translation into a single protein was possible for only 139 of the 178 soil DNA PCR-amplified sequences. Other sequences contained internal stop codons or exhibited evidence that insertion or deletion events had modified the reading frame, and, therefore, these other sequences could be pseudogenes. A unique amino acid sequence was detected for 112 of the 178 analyzed fragments. Among the 139 sequences able to be translated, 35 sequences were 100% similar to BLA-TEM116 (15 Bt, 7 M, and 13 S sequences).
Comparisons to TEM1 were done by using the amino acid residues numeration as established in ref. 18 and according to www.lahey.org/studies. The environmental sequences included 120 different amino acid substitution positions (SI Table 3) with up to four amino acid residues modified. Some of them corresponded to sequence patterns detected in clinical environment (SI Table 4), such as mutations F16L, L21F, A42V, L51P, M69V, E104K, S130G, R164H, N175I, M182T, R204Q, G218E, E240K, N276D, and A280V. In addition, two specific mutations present in TEM116 and TEM157 and not in other medical sequences (V84I and A184V) were also detected in soil sequences. Mutation V84I was generally observed in soil sequences but was not present in Bt3, Bt8, Bt9, and Bt10 sequences. These sequences were at the interface between soil and clinical sequences in the phylogenetic tree (Fig. 1). Mutation A184V was detected in all soil sequences except in clone M27, presenting instead mutation A184I, and can be thus considered as the signature of BLA-TEM sequences in soil bacteria. Considering these data, BLA-TEM116 and BLA-TEM157 could originate from soil.
Amino acid substitutions at positions located in close proximity to the active-site cavity, including positions 104, 164, 237, 238, and 239, are expected to open up novel enzyme-substrate interactions (22). Indeed, incorporation of a lysine at position 104, a serine at position 164 or 238, or the doublet threonine-lysine at positions 237 and 239 conferred an increased substrate range (23, 24). These substitutions were detected in five TEM beta-lactamases, e.g., TEM12 and TEM17 (5, 24–26). Accordingly, mutations E104K and R164H detected in clones Bt17 and Bt10, respectively, should produce an ESBL phenotype. Because serine 70 is a key catalytic residue, its substitution by another amino acid is expected to have a strong effect on enzyme efficiency, including resistance loss to beta-lactams (27). Clone M34 exhibits mutation S70G, which was never observed in clinical isolates. However, the PCR-based selection of blaTEM genes among soil DNA did not exclude those that no longer confer the resistance phenotype or pseudogenes.
These data indicate that the numbers of possible mutational combination pathways that increase or maintain blaTEM enzyme activity without decreasing its stability are not unlimited. For instance, resistance to cefotaxime conferred by a blaTEM1 gene to an E. coli hypermutator strain submitted to directed evolution was only observed for the E104K/M182T/G238S mutation combination (28), and DNA shuffling approaches revealed that antibiotic resistance level increased for alleles containing A18V, A42G, G92S, and R241H mutations (29). All but one of these blaTEM mutations, which increase enzyme activity (E104K, M182T, A18P, A42V, and G92S) or substrate range (R241H), were detected in PCR-amplified soil DNA and in several clinically referenced bacteria. Thus, soil bacteria apparently have access to a wide range of adaptive alleles including some that guard against novel and synthetic antimicrobial molecules.
Bacterial Diversity in Soils.
Bacterial diversity in the different soils used was studied to determine whether the similarity observed between bla genes in the three soils was due to the presence of similar microbial structures. A microarray approach was developed to analyze and compare microbial community composition of soils recovered from a prairie and traditional and transgenic corn fields. For this purpose, a 16S rRNA-based taxonomic microarray composed of 575 probes targeting bacterial diversity (30) at various taxonomic levels from phyla to species was used. Principal component analysis (PCA) and k-means clustering of total bacterial community hybridization results (SI Fig. 4) showed significant differences between the three soils. However, the major difference was between the soils from corn fields (traditional and transgenic) and the prairie soil (43% of the total variability).
Changes in microbial communities due to the Bt trait have been shown to be smaller than changes due to soil type, plant growth stage, different (non-Bt) corn cultivar, and different crops (such as grass) (31). Other studies demonstrated that bacterial diversity as measured by metabolic profiling and molecular analysis of 16S rRNAs in nontransgenic and transgenic corn rhizospheres was not significantly different (32, 33). Indeed, rhizospheric bacterial diversity is affected more by soil texture or growing season than by cultivation of transgenic varieties (32, 34). In contrast, genetically modified potatoes expressing antibacterial compounds were observed to induce different microbial activity rates and structures (35). However, in that study, the impact of the genetic modification cannot be correlated to the transgenic trait, but rather to the plant genotype as the traditional corn was not the isogenic variety of the Bt corn.
In conclusion, the similarity observed between bla genes in the three soils was not correlated to the microbial structures, because they were significantly different.
Hypothesis for a Transgenic Contamination.
The continuously increasing antibiotic resistance in pathogenic (and maybe commensal) bacteria is one of the most striking demonstrations that human activity might accelerate evolutionary changes in other species, especially in highly adaptable organisms, such as bacteria. Ampicillin is one of the numerous examples of an antibiotic, which has rapidly lost its efficacy. Ampicillin was deployed in 1961, and resistance was first observed 15 years later (36). Antibiotic resistance genes detected on numerous plasmids today were apparently lacking from bacterial replicons isolated in the preantibiotic era (37, 38). These dramatic changes confirm the fundamental role of horizontal gene transfer (HGT) in the dispersion of these genes between bacteria. However, several fundamental questions remain about whether HGT occurs in hospitals or in the environment and the role of commensal bacteria, including those from soils as donors or recipients of DNA, thus justifying studies to evaluate the possible impact that transgenes could have as templates for further gene evolution.
Transgene transfer from plants to bacteria has been detected under greenhouse conditions with specifically selected donor and recipient organisms (39). Despite these experiments that simulated the environment, plant bacteria HGT events remain undetected under field conditions. These limitations justified carrying out studies on soil samples originating from fields cultivated with the Bt176 corn plants for 10 successive years. This period could increase the probability that the gene would be transferred to soil bacteria and that the soil bacterial community would demonstrate some effect. In addition, this plant was one of the first transgenic plants to have been developed and will certainly remain as one of the rare GMPs used in which a resistance gene has been cloned with its own promoter. The prokaryotic origin of the blaTEM116 gene and presence of a bacterial promoter are two factors susceptible to facilitate its transfer to bacteria and to improve the fitness of the recombinants that would be able to express this gene.
The potential that the diversity of the blaTEM gene in metagenomic DNA could have evolved from the transgene was evaluated by comparing the divergence of soil blaTEM sequences with blaTEM116 gene. To further test this hypothesis, synonymous (silent, dS) and nonsynonymous (amino acid altering, dN) nucleotide substitutions were estimated (40). dN/dS ratios <1, >1, or equal to 1 correspond to negative, positive, or neutral evolutions (related to selective pressure), respectively. A coding sequence is considered to be positively selected if the nonsynonymous substitution rate is significantly higher than the synonymous substitution rate. Among the 153 different metagenomic sequences tested, only five sequences (3.3%) were positively selected (SI Fig. 5). Two sequences arose from the transgenic corn soil (Bt10 and Bt24), and the three others came from the prairie soil (S46, S54, and S63). These genes could have evolved through environmental pressure leading to more adapted sequences. Negatively selected sequences include 36.6% of the cloned sequences leading to less adapted genes, which might not even be functional. Their activity could not be tested, because PCR primers used were designed inside the bla gene. However, this relatively high percentage might illustrate the general lack of selection pressure in these soils. Finally, there are 60.1% of sequences with the dN/dS ratio equal to 1, indicating that these genes are in the same evolution state as the transgenic sequence without any apparent additional selective pressure and that these genes were either well adapted to their environment or perhaps unnecessary for bacterial survival. Thus, the environmental selective pressure cannot be shown to influence the diversity of the blaTEM genes.
Conclusion
After the first cultivation of GMPs in the fields, questions have soon arisen about the risk of transfer of the transgenes to soil bacteria with possible consequences on the bacterial community structure and on the spread of antibiotic resistance to soil and clinical strains.
The successful transfer of transgene-borne antibiotic resistance genes to bacteria might be unavoidable according to a plethora of scientific data. This includes the long-term DNA persistence in soil, the heterogeneous soil structure favoring contact between DNA and bacteria, the prokaryotic origin of the plant transgene sequences that represent a specific risk for a facilitated integration in a bacterial genome by HGT as demonstrated under laboratory (41), and greenhouse conditions (39). In addition, in the Bt176 event that we investigated here, the bacterial promoter was introduced concomitantly with the antibiotic resistance gene that would facilitate its expression in a potential recipient. Finally, these GMPs were cultured in the same field for 10 successive years, making this GMP-field combination particularly suitable to address gene transfer questions and impact on soil bacteria.
However, the detection of such events remains very difficult, and, in this study, we, like others before, did not detect any cellular or molecular evidence that the blaTEM116 gene from the Bt176 transgenic plant was transferred to bacteria. If such transfer events ever happened (although undetectable), they apparently remain without consequences on the soil bacterial community structure. In addition, the use of the sensitive microarray-based hybridization technique failed to detect any significant changes in soil bacteria that could be specifically related to the presence of the transgenic plants or to the expression of the transgene, including cry genes.
These results are probably partly due to the low frequency at which these transfer events happen and at a limited efficiency of the investigation protocols. However, they are also and mainly because these genes are already present in soil. Bacteria that would have acquired a blaTEM116 gene from the plant would not have a specific selective advantage relative to other resistant bacteria. Our results indicate that indigenous bacteria are already involved in evolutionary processes by point mutations and HGT as evidenced by the polymorphism of the bla gene in the various soils tested. This could indicate that soil is certainly a reservoir in which all bacteria, including clinical pathogens, can acquire the genetic determinants that could permit them to adapt rapidly to present and future antibiotics. These results confirm the interest of considering the evolutionary potential of bacteria when evaluating the impact of GMPs. Our data are sufficiently informative to conclude that the risk that antibiotic-resistant genes in GMPs can pose to commensal and clinical bacteria should be considered as almost null. This risk has to be neglected not because these genes cannot be transferred but because the plethora of genes already present in soil bacteria and the constant evolution to which they are subjected limit the impact that a newly acquired, yet identical, gene from a plant can have.
Experimental Procedures
Bacteria, Media, and Plasmids.
Bacteria, media, and plasmids are described in SI Text.
Soil Sampling.
Soil was sampled (SI Text) by using geostatistical methods as described by Atteia et al. (42). The soil was sieved at 2 mm. Some soils characteristics are showed in SI Table 5. Trace elements in soil were analyzed by inductively coupled plasma-mass spectrometry (ICP-MS HP 4500; Agilent), using external calibration and internal standard correction (SI Table 6).
Isolation and Enumeration of Bacteria.
Recovery of bacteria was performed by using a Nycodenz gradient separation method (SI Text) as described in ref. 43. To enumerate total and resistant soil cultivable bacteria, appropriate dilutions of bacterial suspension were spread onto tryptic soil agar (TSA) 1/10 medium alone or supplemented with ampicillin and carbenicillin. Colonies were counted after a 2-day incubation at 28°C. Colonies grown on selective plates were used to constitute a library of 96 ampicillin and carbenicillin-resistant isolates for two samples of each of the three soils.
Antibiotics-Resistance Assays.
The following beta-lactam antibiotics were used in this study: ampicillin, carbenicillin, amoxicillin, penicillin G, oxacillin, cephalothin, cefamandole, and ceftriaxone. Details are provided in SI Text.
DNA Manipulations.
DNA extraction and cloning step are described in SI Text.
PCR Assays.
The different primer sets, the targeted genes, and the sizes of the generated PCR products are described in SI Table 2. Detection of blaTEM-like genes was performed by amplifying an ≈828-bp fragment with primers bla1 and bla2, whose sequences are complementary to a conserved portion of the blaTEM gene sequences. Amplifications of blaTEM genes from soil isolates were first performed in a final volume of 20 μl (SI Text). Samples presenting expected sizes were selected for a new PCR in a final volume of 100 μl for sequencing.
Amplifications of blaTEM sequences from metagenomic DNA (SI Text) were performed on 5 μl of extracted DNA in a final volume of 200 μl in presence of 1 μg of T4 gene 32 protein (Roche) and 1 unit of titanium Taq polymerase (Clontech). After purification on 1% (wt/vol) agarose gel, amplified DNA was cloned into vector TOPO-TA and transformed into E. coli TOP10. Cloned metagenomic blaTEM sequences were checked by PCR with primers M13F and M13R (provided in the cloning kit) on 2 μl of liquid culture of transformed E. coli strains with the Taq polymerase from Invitrogen in a final volume of 20 μl.
The 16S rDNA genes of ampicillin-resistant isolates and of metagenomic DNA were amplified by using the primers pA and pH′ (SI Table 2 and SI Text). Amplified DNA was then loaded on a 0.8% (wt/vol) agarose gel, and excised bands were purified by using the QIAquick Gel MiniElute Extraction kit (Qiagen).
Microarray Assays.
Fluorescence labeling of 16S rRNA PCR products from bacterial communities, manufacturing and processing of the 16S rRNA-based microarrays, hybridization, scanning, and data analysis of microarrays were performed as described in refs. 30 and 44. Comparisons of microarray data were carried out by PCA and k-means clustering, using the package ade4 and stats for R (www.r-project.org), respectively. PCA was performed by using the covariance matrix and k-means clustering the algorithm of Hartigan and Wong (45). For each soil sample, two hybridization replicates were done.
DNA Sequencing.
The nucleotide sequences of blaTEM and 16S rDNA were determined from soil isolates by direct sequencing of the PCR products after column purification performed by the sequencing society (Genoscreen, Lille, France or Genome Express, Meylan, France). The nucleotide sequence of blaTEM was also obtained from soil metagenomic DNA by sequencing cloned PCR products with primers M13F/M13R.
Sequence and Phylogenetic Analyses.
Sequences were submitted to the National Center for Biotechnology Information data bank and compared by using the BLASTN and BLASTX algorithm (46). On the basis of the results of the database analyses, bla sequences were aligned in between and with the bla sequence of plasmid pCIB4431, using ClustalX software, Version 1.64b. Phylogenetic reconstructions were generated with three different analytical methods, maximum parsimony, maximum likelihood, and neighbor-joining, using the Phylowin software package (47), and visualized with NjPLOT (48). The dN/dS ratio was calculated after the clustalX alignment (49) between DNA sequences with the SNAP software (40).
Chimeric 16S rRNA gene sequences were checked by using the chimera detection program Pintail version 0.33 (available at www.cardiff.ac.uk/biosi/research/biosoft/pintail/pintail.html). Data management and computation were performed with Microsoft Excel software.
Supplementary Material
ACKNOWLEDGMENTS.
We thank G. Grundmann for fruitful discussions with the microarray approach. This work was supported by the Programme National de Recherches sur les Organismes Génétiquement Modifiés from the Agence Nationale de la Recherche project Ploben Grant ANR-05-POGM-004-01 and Agence Française de Sécurité Sanitaire de l'Environnement et du Travail project AntiReSol Grant EST-2006/1/44.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EF514028–EF514205).
This article contains supporting information online at www.pnas.org/cgi/content/full/0800072105/DC1.
References
- 1.Thomson JM, Bonomo RA. The threat of antibiotic resistance in Gram-negative pathogenic bacteria: Beta-lactams in peril! Curr Opin Microbiol. 2005;8:518–524. doi: 10.1016/j.mib.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 2.Livermore DM. Are all beta-lactams created equal? Scand J Infect Dis Suppl. 1996;101:33–43. [PubMed] [Google Scholar]
- 3.Wilke MS, Lovering AL, Strynadka NC. beta-Lactam antibiotic resistance: a current structural perspective. Curr Opin Microbiol. 2005;8:525–533. doi: 10.1016/j.mib.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 4.Medeiros AA. Evolution and dissemination of beta-lactamases accelerated by generations of beta-lactam antibiotics. Clin Infect Dis. 1997;24(Suppl 1):S19–S45. doi: 10.1093/clinids/24.supplement_1.s19. [DOI] [PubMed] [Google Scholar]
- 5.Jacoby GA, Medeiros AA. More extended-spectrum beta-lactamases. Antimicrob. Agents. Antimicrob Agents Chemother. 1991;35:1697–1704. doi: 10.1128/aac.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottlieb D. The production and role of antibiotics in soil. J Antibiot (Tokyo) 1976;29:987–1000. doi: 10.7164/antibiotics.29.987. [DOI] [PubMed] [Google Scholar]
- 7.Li D-M, Alexander M. Factors affecting co-inoculation with antibiotic-producing bacteria to enhance rhizobial colonization and nodulation. Plant Soil. 1990;129:195–201. [Google Scholar]
- 8.Thomashow LS, Weller DM, Bonsall RF, Pierson LS. Production of the antibiotic phenazine-1-carboxylic acid by fluorescent Pseudomonas species in the rhizosphere of wheat. Appl Environ Microbiol. 1990;56:908–912. doi: 10.1128/aem.56.4.908-912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen LH, Ferrari B, Sorensen AH, Veal D, Sorensen SJ. Detection of oxytetracycline production by Streptomyces rimosus in soil microcosms by combining whole-cell biosensors and flow cytometry. Appl Environ Microbiol. 2001;67:239–244. doi: 10.1128/AEM.67.1.239-244.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anukool U, Gaze WH, Wellington EM. In situ monitoring of streptothricin production by Streptomyces rochei F20 in soil and rhizosphere. Appl Environ Microbiol. 2004;70:5222–5228. doi: 10.1128/AEM.70.9.5222-5228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldstein DA, et al. Human safety and genetically modified plants: A review of antibiotic resistance markers and future transformation selection technologies. J Appl Microbiol. 2005;99:7–23. doi: 10.1111/j.1365-2672.2005.02595.x. [DOI] [PubMed] [Google Scholar]
- 12.Badosa E, Moreno C, Montesinos E. Lack of detection of ampicillin resistance gene transfer from Bt176 transgenic corn to culturable bacteria under field conditions. FEMS Microbiol Ecol. 2004;48:169–178. doi: 10.1016/j.femsec.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Dcosta VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 14.Nwosu VC, Ladapo JA. Antibiotic resistance with particular reference to soil microorganisms. Curr Microbiol. 1999;39:249–253. [Google Scholar]
- 15.Smits TH, Seeger MA, Witholt B, Van Beilen JB. New alkane-responsive expression vectors for Escherichia coli and Pseudomonas. Plasmid. 2001;46:16–24. doi: 10.1006/plas.2001.1522. [DOI] [PubMed] [Google Scholar]
- 16.Bush K. New beta-lactamases in gram-negative bacteria: Diversity and impact on the selection of antimicrobial therapy. Clin Infect Dis. 2001;32:1085–1089. doi: 10.1086/319610. [DOI] [PubMed] [Google Scholar]
- 17.Jacoby GA. Beta-lactamase nomenclature. Antimicrob Agents Chemother. 2006;50:1123–1129. doi: 10.1128/AAC.50.4.1123-1129.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ambler RP, et al. A standard numbering scheme for the class A beta-lactamases. Biochem J. 1991;276:269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denamur E, et al. Intermediate Mutation Frequencies Favor Evolution of Multidrug Resistance in Escherichia coli. Genetics. 2005;171:825–827. doi: 10.1534/genetics.105.045526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riesenfeld CS, Goodman RM, Handelsman J. Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ Microbiol. 2004;6:981–989. doi: 10.1111/j.1462-2920.2004.00664.x. [DOI] [PubMed] [Google Scholar]
- 21.Bastarrachea F. On the origin of plasmid-borne, extended-spectrum, antibiotic resistance mutations in bacteria. J Theor Biol. 1998;190:379–387. doi: 10.1006/jtbi.1997.0563. [DOI] [PubMed] [Google Scholar]
- 22.Collatz E, Labia R, Gutmann L. Molecular evolution of ubiquitous beta-lactamases towards extended-spectrum enzymes active against newer beta-lactam antibiotics. Mol Microbiol. 1990;4:1615–1620. doi: 10.1111/j.1365-2958.1990.tb00537.x. [DOI] [PubMed] [Google Scholar]
- 23.Mabilat C, Courvalin P. Development of “oligotyping” for characterization and molecular epidemiology of TEM beta-lactamases in members of the family Enterobacteriaceae. Antimicrob Agents Chemother. 1990;34:2210–2216. doi: 10.1128/aac.34.11.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sougakoff W, Goussard S, Gerbaud G, Courvalin P. Plasmid-mediated resistance to third-generation cephalosporins caused by point mutations in TEM-type penicillinase genes. Rev Infect Dis. 1988;10:879–884. doi: 10.1093/clinids/10.4.879. [DOI] [PubMed] [Google Scholar]
- 25.Collatz E, Tran Van Nhieu G, Billot-Klein D, Williamson R, Gutmann L. Substitution of serine for arginine in position 162 of tem-type beta-lactamases extends the substrate profile of mutant enzymes, tem-7 and tem-101, to ceftazidime and aztreonam. Gene. 1989;78:349–354. doi: 10.1016/0378-1119(89)90237-0. [DOI] [PubMed] [Google Scholar]
- 26.Sougakoff W, et al. Characterization of the plasmid genes blaT-4 and blaT-5 which encode the broad-spectrum beta-lactamases TEM-4 and TEM-5 in enterobacteriaceae. Gene. 1989;78:339–348. doi: 10.1016/0378-1119(89)90236-9. [DOI] [PubMed] [Google Scholar]
- 27.Herzberg O, Moult J. Bacterial resistance to beta-lactam antibiotics: crystal structure of beta-lactamase from Staphylococcus aureus PC1 at 2.5 Å resolution. Science. 1987;236:694–701. doi: 10.1126/science.3107125. [DOI] [PubMed] [Google Scholar]
- 28.Orencia MC, Yoon JS, Ness JE, Stemmer WP, Stevens RC. Predicting the emergence of antibiotic resistance by directed evolution and structural analysis. Nat Struct Biol. 2001;8:238–242. doi: 10.1038/84981. [DOI] [PubMed] [Google Scholar]
- 29.Stemmer WP. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 30.Sanguin H. Lyon, France: Université Lyon I; 2006. Development and validation of a 16S taxonomic microarray for characterizing and monitoring the rhizosphere bacterial community. PhD thesis. [Google Scholar]
- 31.Griffiths BS, et al. A comparison of soil microbial community structure, protozoa and nematodes in field plots of conventional and genetically modified maize expressing the Bacillus thuringiens is CryIAb toxin. Plant Soil. 2005;275:135–146. [Google Scholar]
- 32.Fang M, Kremer RJ, Motavalli PP, Davis G. Bacterial diversity in rhizospheres of nontransgenic and transgenic corn. Appl Environ Microbiol. 2005;71:4132–4136. doi: 10.1128/AEM.71.7.4132-4136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saxena D, Stotzky G. Bacillus thuringiensis (Bt) toxin released from root exudates and biomass of Bt corn has no apparent effect on earthworms, nematodes, protozoa, bacteria, and fungi in soil. Soil Biol Biochem. 2001;33:1225–1230. [Google Scholar]
- 34.Schmalenberger A, Tebbe CC. Genetic profiling of noncultivated bacteria from the rhizospheres of sugar beet (Beta vulgaris) reveal field and annual variability but no effect of a transgenic herbicide resistance. Can J Microbiol. 2003;49:1–8. doi: 10.1139/w02-111. [DOI] [PubMed] [Google Scholar]
- 35.Rasche F, et al. Rhizosphere bacteria affected by transgenic potatoes with antibacterial activities compared with the effects of soil, wild-type potatoes, vegetation stage and pathogen exposure. Fems Microbiol Ecol. 2006;56:219–235. doi: 10.1111/j.1574-6941.2005.00027.x. [DOI] [PubMed] [Google Scholar]
- 36.Palumbi SR. Humans as the world's greatest evolutionary force. Science. 2001;293:1786–1790. doi: 10.1126/science.293.5536.1786. [DOI] [PubMed] [Google Scholar]
- 37.Hughes VM, Datta N. Conjugative plasmids in bacteria of the “pre-antibiotic” era. Nature. 1983;302:725–726. doi: 10.1038/302725a0. [DOI] [PubMed] [Google Scholar]
- 38.Nwosu VC. Antibiotic resistance with particular reference to soil microorganisms. Res Microbiol. 2001;152:421–430. doi: 10.1016/s0923-2508(01)01215-3. [DOI] [PubMed] [Google Scholar]
- 39.Kay E, Vogel TM, Bertolla F, Nalin R, Simonet P. In situ transfer of antibiotic resistance genes from transgenic (transplastomic) tobacco plants to bacteria. Appl Environ Microbiol. 2002;68:3345–3351. doi: 10.1128/AEM.68.7.3345-3351.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 41.Gebhard F, Smalla K. Transformation of Acinetobacter sp. strain BD413 by transgenic sugar beet DNA. Appl Environ Microbiol. 1998;64:1550–1554. doi: 10.1128/aem.64.4.1550-1554.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atteia O, Dubois JP, Webster R. Geostatistical analysis of soil contamination in the Swiss Jura. Environ Pollut. 1994;86:315–327. doi: 10.1016/0269-7491(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 43.Bertrand H, et al. High molecular weight DNA recovery from soils prerequisite for biotechnological metagenomic library construction. J Microbiol Methods. 2005;62:1–11. doi: 10.1016/j.mimet.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 44.Bodrossy L, et al. Development and validation of a diagnostic microbial microarray for methanotrophs. Environ Microbiol. 2003;5:566–582. doi: 10.1046/j.1462-2920.2003.00450.x. [DOI] [PubMed] [Google Scholar]
- 45.Hartigan JA, Wong MA. A K-means clustering algorithm. Appl Statist. 1979;28:100–108. [Google Scholar]
- 46.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 47.Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: Two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 48.Perriere G, Gouy M. WWW-query: An on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- 49.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.