Abstract
TDP-43 (for TAR DNA binding protein) is a highly conserved heterogeneous nuclear ribonucleoprotein (hnRNP) involved in specific pre-mRNA splicing and transcription events. TDP-43 recently has been identified as the main component of cytoplasmic inclusions in frontotemporal lobar degeneration (FTLD) and amyotrophic lateral sclerosis (ALS), two neurodegenerative disorders. The cellular role of this protein remains to be identified. Here, we show that loss of TDP-43 results in dysmorphic nuclear shape, misregulation of the cell cycle, and apoptosis. Removal of TDP-43 in human cells significantly increases cyclin-dependent kinase 6 (Cdk6) protein and transcript levels. The control of Cdk6 expression mediated by TDP-43 involves GT repeats in the target gene sequence. Cdk6 up-regulation in TDP-43-depleted cells is accompanied by an increase in phosphorylation of two of its major targets, the retinoblastoma protein pRb and pRb-related protein pRb2/p130. TDP-43 silencing also is followed by changes in the expression levels of several factors that control cell proliferation. Morphological nuclear defects and increased apoptosis upon TDP-43 loss are mediated via the pRb pathway because pRb-negative cells (Saos-2) do not undergo programmed cell death or nuclear shape deformation upon TDP-43 removal. Our results identify a regulatory target of TDP-43 and show that TDP-43 depletion has important consequences in essential metabolic processes in human cells.
TDP-43 belongs to the family of heterogeneous nuclear ribonucleoproteins (hnRNPs) and binds single-stranded RNA via its N-terminal RNA recognition motif (1). Members of the hnRNP family serve multiple roles in the generation and processing of RNA, including transcription, splicing, transport, and stability. TDP-43 inhibits exon recognition during splicing upon recruitment to the 3′ splice site of the cystic fibrosis transmembrane conductance regulator (CFTR) and apolipoprotein AII transcripts via a sequence of GU repeats (2–5). The binding affinity of the recombinant human, worm, and fly homologues for this target sequence is remarkably high, measured in the low nanomolar range (6). TDP-43 also has been implicated in the transcription regulation of HIV and the spermatid-specific gene SP-10 through promoter association (7, 8). More recently, TDP-43 was identified as the main ubiquitinated component of cytoplasmic inclusions in neurodegenerative diseases, specifically frontotemporal lobar degeneration (FTLD) and amyotrophic lateral sclerosis (ALS) (9, 10). Abnormal aggregation of TDP-43 in the cytoplasm now is thought to define a class of frontotemporal dementias termed TDP-43 proteinopathies. Mislocalization and the consequent loss of TDP-43 function in neuronal cells may represent a common event in FTLD pathogenesis. Despite the increasing awareness of processes involving TDP-43, the cellular role of the protein still is poorly defined.
We depleted TDP-43 by RNA interference (RNAi) to identify TDP-43-regulated transcripts. Our results point to cyclin-dependent kinase 6 (Cdk6) as a unique target of TDP-43 regulation and suggest that TDP-43 inhibits Cdk6 expression through recruitment to the GU-rich transcript. Simultaneously, we found that TDP-43 silencing alters cell cycle distribution and induces apoptosis.
Results
TDP-43 Down-Regulation Alters the Expression of pRb-Related Factors.
TDP-43 was depleted from HeLa cells by RNAi routinely achieving >90% silencing as measured by Western blot, 48 h after small interfering RNA (siRNA) transfection (3, 5). RNA microarray analysis was performed on TDP-43 depleted and control treated cells. The data obtained indicated altered levels of several cell proliferation factors in TDP-43-silenced cells. Table 1 lists 16 of these proteins whose functions have been associated with retinoblastoma protein (pRb) activity. The tumor suppressor pRb is essential for the control of cell cycle progression, cellular differentiation, and maintenance of genome integrity. Inactivation of pRb occurs through its gradual phosphorylation by Cdks during the G1 phase of the cell division cycle resulting in the activation of transcription factors that promote cell proliferation and enable transition on to the S phase (see ref. 11 for review). Our RNA microarray analyses showed altered levels of transcripts coding for proteins whose functions are related to the control of cell cycle progression (Cdk6, POLD4, cyclin B1, Cdk2, UBE2C, and SKP2). In addition, some of these factors are known to either directly interact with pRb (e.g., HDAC1, RBBP4, and CRI1), or act in response to pRb modulation (e.g., E2F8 and NAP1L1) (12–22).
Table 1.
pRb pathway group microarray transcripts
| Transcripts | Fold change |
|---|---|
| Cyclin-dependent kinase 6 (Cdk6) | +10 |
| Nucleosome assembly protein 1-related protein (NAP1-L1) | +4 |
| Cyclin-dependent kinase 2 delta T (d-HSCdk2) | +3 |
| Cdk adapter protein (CKS1B) | +3 |
| Cyclin G1 | +3 |
| Polo-like kinase 1 (PLK1) | +2 |
| Cyclin B1 | −2 |
| CREBBP/EP300 inhibitory protein 1 (CRI1) | −3 |
| Polymerase delta 4 (PolD4) | −3 |
| Topoisomerase I (Top1) | −3 |
| Ribonucleotide reductase M2 (RRM2) | −4 |
| Retinoblastoma binding protein 4 (RBBP-4) | −4 |
| Ubiquitin-conjugating enzyme E2-C (UBE2C) | −4 |
| S-phase kinase-associated protein (SKP2) | −5 |
| Histone deacetylase 1 (HDAC1) | −6 |
| Elongation factor E2F8 | −12 |
Shown are transcripts identified by RNA microarray analysis whose levels either increased or decreased in the absence of TDP-43 compared to control-treated cells.
TDP-43 Inhibits Cdk6 Expression.
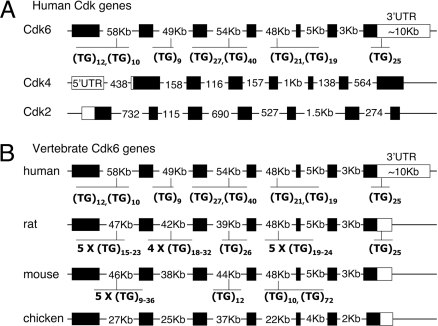
Among the transcripts whose levels were altered upon TDP-43 loss, the levels of Cdk6 showed the highest increase in the absence of TDP-43, specifically 10-fold. Cdk6 belongs to the family of Cdks and along with Cdk4 regulates the early G1 phase transition during cell growth (23, 24). In addition to pRb, Cdk6 substrates include pRb2/p130, histone H1, and Bcl-2 (25–27). The appropriate regulation of Cdk6 activity determines differentiation of many cell types and a variety of tumors have been associated with elevated levels of Cdk6 expression (28–33). Careful inspection of human Cdk6 revealed a peculiar gene structure with particularly long introns and a predicted 3′ untranslated region (UTR) spanning nearly 11 Kb (Fig. 1A). Remarkably, its pre-mRNA contains multiple (GU)n repeats, the TDP-43 target sequence. The repeat tracts are present once or twice in all of the larger introns and the 3′ UTR contains a (GU)25 sequence 180 nt upstream of the first predicted polyadenylation site. In contrast, introns in other human Cdk genes, e.g., Cdk4 and Cdk2, are significantly shorter and lack (GT)n (Fig. 1A). We observed that the repeats are conserved in primates and rat and mouse genes. The abundance of the TDP-43 target sequence within the Cdk6 gene urged us to confirm the up-regulation of Cdk6 expression upon loss of TDP-43 in human cells. We quantified Cdk6 protein and mRNA levels by immunoblotting and quantitative PCR amplification, respectively (Fig. 2). In agreement with the microarray results, we obtained a significant increase in Cdk6 protein and transcript in response to TDP-43 loss. The use of a different dsRNA oligonucleotide for RNAi (T1) showed a similar increase in Cdk6 protein levels [supporting information (SI) Fig. 5A]. Moreover, Cdk6 up-regulation upon RNAi treatment was studied in cells transiently expressing a FLAG-tagged TDP-43 mutant resistant to siRNA depletion (TDP-43siR). Cdk6 protein levels after depletion of TDP-43 in three independent experiments were 35 to 45% lower in the presence of TDP-43siR compared with control treated cells as seen by quantification of immunoblotting experiments, using tubulin as loading control (SI Fig. 6). Taken together these results strongly suggest that Cdk6 levels are specifically regulated by TDP-43.
Fig. 1.
GT repeats are unique to Cdk6 and are conserved in different mammals. Schematic representation of different Cdk genes, including coding exons (■) UTRs (□) and introns. (A) Human Cdk6 and the closely related Cdk4 and Cdk2 are shown. Highlighted in human Cdk6 are the GT repeat sequences. Two stretches ranging from 10 to 40 repeats are found in introns 1, 3, and 4. The repeats located at the 3′ UTR of the gene are 400 nt downstream of the stop codon and 180 nt upstream of the first polyadenylation site. In contrast to Cdk6, Cdk4, and Cdk2 have significantly shorter introns and 3′ UTR and lack GT repeat sequences. (B) TG repeats are present in primates, mouse, and rat Cdk6 genes. The length of the introns is more or less conserved, i.e., introns 1, 2, 3, and 4 are >35-Kb long, whereas introns 5 and 6 range from 1 to 5 Kb. Although chicken Cdk6 gene structure is similar to that of other vertebrates analyzed, the sequence lacks GT repeats entirely.
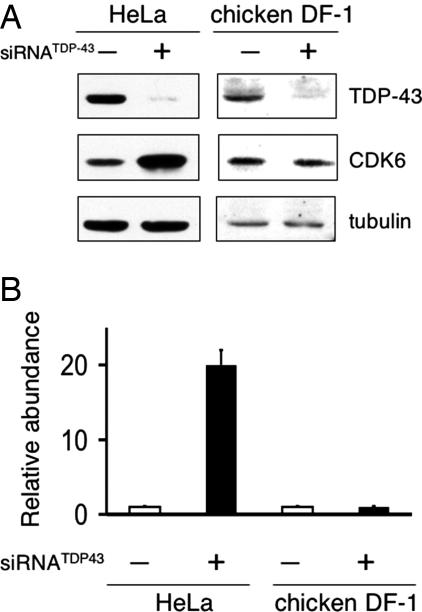
Fig. 2.
TDP-43 depletion causes up-regulation of Cdk6 in human but not in chicken cells. (A) Immunoblot analysis of HeLa and chicken embryo fibroblast (DF-1) cell extracts. Human and chicken cells were transfected with control or human/chicken TDP-43-targeted siRNA to check for changes in Cdk6 expression levels. Equal loading was confirmed by tubulin detection. Other human cell lines tested showed a similar increase in Cdk6 upon TDP-43 depletion. (B) Real-time RT-PCR analysis of Cdk6 cDNA in TDP-43 depleted (■) and control-treated (□) HeLa and DF-1 cells. Values shown are the average of three independent experiments. Error bars indicate SD.
To address whether TDP-43 regulates Cdk6 expression via the GT tracts, we took advantage of the fact that chicken Cdk6 lacks GT repeats completely, despite its otherwise highly similar exon and intron structure. Although depletion of TDP-43 leads to Cdk6 up-regulation in human cells, loss of TDP-43 has no effect on Cdk6 protein levels in chicken cells (DF-1) (Fig. 2B). This human specific increase in Cdk6 was confirmed with real time PCR quantification of Cdk6 mRNA derived from RNAi treated HeLa and DF-1 cells. The analysis showed a 20-fold increase in Cdk6 mRNA upon TDP-43 depletion from human cells compared with control transfected cells (Fig. 2C). However, TDP-43-depleted and control treated DF-1 chicken cells showed no variation. These data suggest that human Cdk6 expression is modulated by TDP-43 recruitment to (GU)n repeats in the pre-mRNA.
TDP-43 Depletion Increases pRb and pRb2/p130 Phosphorylation.
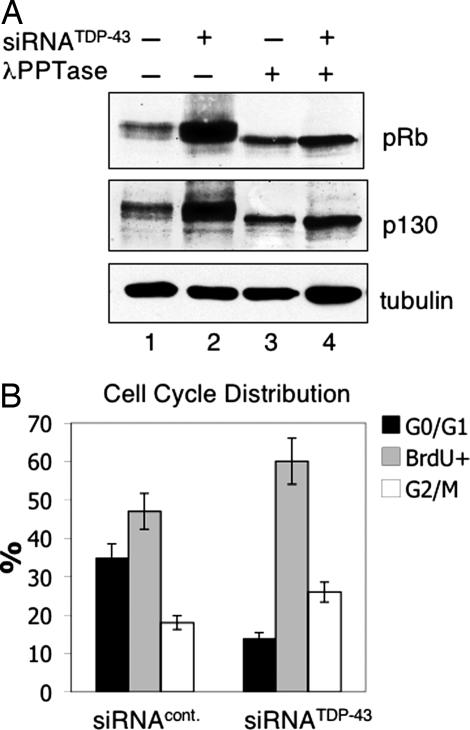
We next tested whether Cdk6 misregulation in the absence of TDP-43 extends to aberrant pRb phosphorylation. The phosphorylation levels of pRb and of the pRb-related protein pRb2/p130 were determined after TDP-43 silencing. To avoid artifacts deriving from disruptions of the pRb pathway caused by papilloma virus functional genes present in HeLa cells (34), we have carried out our experiments in human osteosarcoma U2OS cells. Immunoblot analysis showed that TDP-43 silencing resulted in a 7 and 6-fold increase in the phosphorylation of pRb and pRb2/p130, respectively (Fig. 3A). A modest increase in pRb and pRb2/p130 protein levels (2- and 1.5-fold) was observed in phosphatase treated samples from TDP-43-depleted cells (lanes 3 and 4 of Fig. 3A). These results indicate that the higher intensity of the pRb signals (Fig. 3A, lanes 1 and 2) in the absence of TDP-43 were primarily attributable to increased levels of phosphorylation, corresponding to the increase in Cdk6 levels in these cells. Moreover, pRb or pRb2/p130 phosphorylation levels did not change after depletion of TDP-43 in chicken embryo fibroblasts, where Cdk6 levels are unresponsive to TDP-43 silencing (SI Fig. 7). These data strongly suggest that changes in pRb and pRb2/p130 posttranslational modification, observed in U2OS after TDP-43 removal, are mediated by the up-regulation of Cdk6 in these cells.
Fig. 3.
TDP-43 depletion promotes pRb and p130 phosphorylation and causes changes in cell cycle distribution. (A) Western blot analysis to detect pRb and pRb2/p130 levels in U2OS cell extracts from TDP-43 depleted (lanes 2 and 4) and control treated cells (lanes 1 and 3). Extracts were also treated with λ phosphatase (PPTase) (lanes 3 and 4) to compare unphosphorylated pRb and pRb2/p130 protein levels. Tubulin was used as loading control. (B) Bars show the percentage of cells in G0/G1, S, and G2/M phases of the cell cycle in proliferating siRNATDP-43 and siRNA control-treated U2OS cells. Propidium- and BrdU-labeled cells were analyzed by FACS after RNAi treatment. Results show average values of three independent experiments, and error bars show SD.
We next investigated the impact of silencing TDP-43 and the consequent alteration of the pRb pathway on cell cycle distribution by Fluorescent-activated cell sorting (FACS) and BrdU labeling experiments. U2OS cells showed an alteration of the cell cycle pattern after removal of TDP-43 (Fig. 3B) resulting in a 60% decrease of cells in G0/G1 accompanied by a corresponding increase in S and G2/M cells. The disruption of cell cycle distribution caused by the loss of TDP-43 is consistent with alterations in pRb phosphorylation and misregulation of factors that control cell growth.
Loss of TDP-43 Affects Nuclear Membrane Stability and Increases Apoptosis.
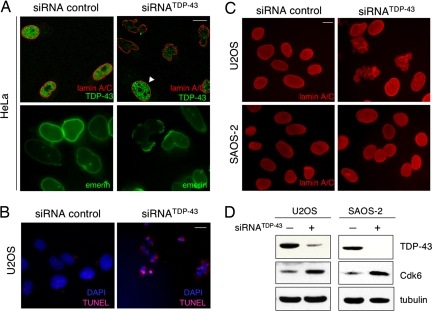
In addition to these changes we observed that TDP-43 depletion caused increased cell death and aberrant nuclear morphology throughout our experiments. Visualization of the nuclear lamina using antibodies against integral nuclear envelope proteins lamin A/C and emerin showed membrane blebbling and a significant disruption of the usual smooth and round nuclear shape (Fig. 4A). Similar results were observed upon depletion of TDP-43 using a different siRNA oligonucleotide sequence, T1 (SI Fig. 5B). Such nuclear membrane disruption was observed in nearly 50% of TDP-43 depleted cells and its severity correlated with the extent of TDP-43 depletion. The lack of TDP-43 led to an uneven distribution of the nuclear envelope protein emerin along the membrane, with discrete regions of either protein accumulation or lack of protein (Fig. 4A Lower). TDP-43 silencing did not lead to changes, as judged by immunoblotting, in the quantity or processing of the nuclear envelope proteins lamin A/C, lamin B, emerin, and Lap2β (data not shown).
Fig. 4.
Loss of TDP-43 results in nuclear membrane deformation and apoptosis. Immunofluorescence microscopy of siRNATDP-43 compared with control-treated cells is shown. (Scale bar, 10 μm.) (A) HeLa cells were stained with antibodies against lamin A/C and TDP-43 (red and green, Upper). Arrowhead points to a cell that escaped siRNA silencing. (Lower) Emerin-labeled nuclear membrane. Similar nuclear shape deformation was observed upon detection of lamin B and Lap2β. (B) U2OS cells were assayed for DAPI and TUNEL staining to detect apoptosis. (Scale bar, 10 μm.) (C) The nuclei of U2OS and Saos-2 cells were visualized by lamin A/C detection after TDP-43 depletion. (D) Down-regulation of TDP-43 in U2OS and Saos-2 cells and the consequent Cdk6 increase were verified by immunoblotting. Tubulin was used as loading control.
We investigated whether the effect on cell survival was attributable to the activation of apoptosis or DNA damage. Loss of TDP-43 in HeLa cells resulted in increased double strand DNA breaks as indicated by the presence of γH2AX foci (35): ≈15% of TDP-43-depleted cells contained γH2AX foci compared with 1% of control treated cells (SI Fig. 8A). At the same time, U2OS showed an increase in TUNEL staining and nuclear fragmentation upon TDP-43 depletion (Fig. 4B). Typically, 30% of TDP-43-depleted cells were TUNEL positive in contrast to <1% of the control treated U2OS. U2OS cell extracts from TDP-43-depleted samples were also positive for poly(ADP-ribose) polymerase-1 (PARP-1) cleavage, consistent with the activation of programmed cell death (SI Fig. 8B). Remarkably, TDP-43 silencing in cells that lack functional pRb (Saos-2) did not cause defects in nuclear envelope shape or apoptotic cell death (Fig. 4C). Because Saos-2 cells are also p53 negative, it was necessary to rule out a role of p53 deficiency in the Saos-2 response to TDP-43 silencing. We depleted TDP-43 from MG-63 cells, an osteosarcoma cell line deficient in p53 but not defective in pRb, and observed increased apoptosis upon loss of TDP-43 in these cells, as seen with U2OS (SI Fig. 9). These results strongly suggest that the morphological defects and increased apoptosis upon TDP-43 loss are tied to disruption of the pRb pathway.
Discussion
We have shown here that the major component of cytoplasmic inclusions in frontotemporal dementias TDP-43 is involved in a key cellular pathway that has ramifications in essential metabolic processes. Loss of TDP-43 in human cells causes genomic instability and increased apoptosis as seen by γH2AX staining and TUNEL. These results underscore a critical role of TDP-43 for cell survival. Moreover, we identify the cyclin-D dependent kinase 6 as a target for TDP-43 control and provide a possible mechanism of regulation. TDP-43 strongly represses Cdk6 expression as seen from the large increase in protein and transcript levels upon TDP-43 depletion. We propose that TDP-43 recruitment to the GU-rich Cdk6 transcript reduces RNA levels. This mechanism is supported by the lack of change in Cdk6 levels upon TDP-43 depletion in chicken cells, which express a Cdk6 transcript devoid of GU repeats. The involvement of TDP-43 in the control of RNA processing has been seen in other systems where GU repeats are involved, i.e., CFTR and ApoAII alternative splicing (2–5). The elucidation of the precise step of RNA processing controlled by TDP-43 in the case of Cdk6 needs further investigation. The implications of such studies could be of great relevance because similar mechanisms of RNA processing may be at play to explain the newly discovered role of TDP-43 in neurodegenerative pathologies.
The hyperphosphorylation of pRb and pRb2/p130 caused by Cdk6 misregulation in TDP-43-silenced cells is likely to block pRb function and change the expression of proliferation associated factors as seen by our microarray analysis (Table 1). In chicken cells, TDP-43 loss leaves pRb and pRb2/p130 phosphorylation levels unchanged supporting a direct association between the gain in Cdk6 levels upon TDP-43 depletion in human cells and pRb phosphorylation. The observed decrease of cells in G0/G1 and the corresponding accumulation of cells in S and G2/M phases in U2OS TDP-43-depleted cells coincide with pRb hyperphosphorylation. More detailed studies will be required to determine whether these observations involve checkpoint activation and cycle arrest. The aberrant control of pRb function and cell proliferation may cause the observed activation of apoptosis in the absence of TDP-43. This interpretation is in line with findings that deregulation of the pRb pathway does not only affect proliferation, but also increases programmed cell death and alters DNA damage responses (36–38). Moreover, increased activity of Cdks also has been associated with apoptosis (39). Further investigation will be required to understand the onset of nuclear membrane instability upon loss of TDP-43. Interestingly, recent work showed that nuclear membrane dysfunction, particularly loss of emerin, is tied to misregulation of pRb pathway components (40, 41) suggesting that nuclear membrane abnormalities caused by TDP-43 loss are mediated by pRb. These conclusions are supported by the lack of apoptosis and nuclear membrane disruption after TDP-43 silencing in Saos-2 cells that lack a functional pRb pathway. At the same time, TDP-43 may be more directly involved in the activation of programmed cell death. New findings reveal that reduction of progranulin, characteristic in familial FTLD with ubiquitin-positive inclusions, causes a caspase-mediated cleavage of TDP-43 and results in mislocalization of TDP-43 to insoluble cytoplasmic fractions (42). Collectively, our results may help to shed light on the pathophysiological consequences of TDP-43 loss of function in neurodegenerative disorders.
Materials and Methods
RNA Microarray.
Biotinylated cRNAs from TDP-43 and control siRNA treated HeLa were hybridized to Affymetrix GeneChip Human Genome U133A 2.0 arrays (Affymetrix). Values reported are the average of three independent experiments.
Vectors and Cloning.
FLAG-tagged TDP-43siR was constructed with the pFLAG-CMV-2 Expression Vector (Sigma E7398). Mutations in the protein were designed to change the nucleotide TDP-43 sequence to render the transcript resistant to siRNA oligonucleotide T2. These silent mutations do not alter the amino acid TDP-43 sequence. The mutant vector was generated by PCR mutagenesis using the following oligonucleotides: RNAi-forward, cttcctaattctaagcagtcccaggatgagcctttgagaagcagaaaag; RNAi-reverse, cttttctgcttctcaaaggctcatcctgggactgcttagaattaggaag.
Cell Culture and RNAi.
HeLa (ATCC, CCL-2, LGC Promochem, MI, Italy), DF-1 (ATCC CRL-12203, LGC Promochem, MI, Italy), U2OS and Saos-2 were grown in DMEM+GlutaMAX I (Invitrogen, 31966), 10% FBS (Euroclone). TDP-43 was silenced from human cells as described by using siRNA oligonucleotide T2 (3, 5), the same transfection protocol was used with chicken cells using the following sequence of double-stranded RNA: GCAAAGUCCCGAUGAGCCU (Dharmacon). A second dsRNA sequence (T1:GAUGAGAACGAUGAGCCCA) was also used to silence TDP-43. The luciferase siRNA, sicontrol#2, was used as control (Dharmacon).
Immunoblotting.
Cell pellets were resuspended in lysis buffer: 50 mM Tris·HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 1× protease inhibitors (Roche 1873580). When necessary, phosphatase inhibitor containing buffer was used (15 mM Hepes, pH 7.5, 0.25 M NaCl, 0.5% Nonidet P-40, 10% glycerol, 1× protease inhibitor, 25 mM NaF, 10 mM β-glycerolphosphate, 0.2 mM Na3VO4, 1 mM phenylmethylsulphonyl fluoride). Proteins were separated by SDS/PAGE and transferred to nitrocellulose (0.22 μm, Schleicher & Schuell). Membranes used to check protein phosphorylation were incubated in Western Blocking Reagent (Roche). Antibodies used were TDP-43 (2),tubulin (Sigma T5168), γH2AX (Upstate, 05–636), lamin A/C (Santa Cruz Biotechnology, sc-6215), emerin (Novocastra), pRb (BD Pharmingen, 554136), p130 (Santa Cruz Biotechnologies, sc-317), Cdk6 (Santa Cruz Biotechnologies, sc-177), and FLAG-tagged TDP-43 (TDP-43siR) (Sigma F1804).
RNA Isolation and Real-Time RT-PCR.
Cell RNA was isolated with TRI Reagent (Ambion) and treated with DNase I. After reverse transcription, real-time PCR was conducted on an ABI 7000 real-time PCR system (Applied Biosystems). Human Cdk6 was amplified with a probe spanning exons 5–6 (assay ID Hs01026372_m1, Applied Biosystems), and chicken Cdk6 was amplified with chickFW-TCAGATGTTGATCAGCTAGGAAAAA, chickRV-TCATTAGGCCAGTCCTCTTCTTCT, and the TaqMan probe CTTTGATGTAATTGGACTCC (Applied Biosystems). GADPH and 18S rRNA were used as endogenous controls.
Immunofluorescence Microscopy.
After fixation in 4% buffered paraformaldehyde in PBS for 15 min at room temperature, cells were permeabilized by using 0.5% Triton in PBS for 5 min on ice and blocked with 2% BSA/PBS for 15 min at room temperature. Immunolabeling using specific antibodies was carried out at room temperature for 1 h in 2% BSA/PBS. Cells were incubated with conjugated secondary antibodies at room temperature for 1 h in PBS. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI).
FACS Analyses and BrdU Labeling.
RNAi treated cells were pulsed by using 10 μM BrdU for 1h and fixed in ice-cold 70% ethanol. Cells were then stained with anti-BrdU-FITC (Becton Dickinson), followed by fluorescein goat anti-mouse (Jackson Laboratories), and propidium iodide. FACS analysis using CellQuest software (Becton Dickinson) was used to quantify BrdU-positive cells and determine the DNA content of the different samples.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Andrea D'Ambrogio and Emanuele Buratti [International Centre for Genetic Engineering and Biotechnology (ICGEB)] for useful discussions and for providing the TDP-43siR vector, María Elena López for help with tissue culture, Ramiro Mendoza for his help with FACS analyses, and Lawrence Banks (ICGEB) for useful discussions and U2OS and Saos-2 cells. This work was supported by National Science Foundation Postdoctoral Minority Research Fellowship 0414069 (to Y.M.A.); Telethon Grant GGP06147 (to F.E.B.); Eurasnet Grant LSHG-CT-2005-518238 (to F.E.B.); and the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800546105/DC1.
References
- 1.Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- 2.Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayala YM, Pagani F, Baralle FE. TDP43 depletion rescues aberrant CFTR exon 9 skipping. FEBS Lett. 2006;580:1339–1344. doi: 10.1016/j.febslet.2006.01.052. [DOI] [PubMed] [Google Scholar]
- 4.Buratti E, Brindisi A, Pagani F, Baralle FE. Nuclear factor TDP-43 binds to the polymorphic TG repeats in CFTR intron 8 and causes skipping of exon 9: A functional link with disease penetrance. Am J Hum Genet. 2004;74:1322–1325. doi: 10.1086/420978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mercado PA, Ayala YM, Romano M, Buratti E, Baralle FE. Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res. 2005;33:6000–6010. doi: 10.1093/nar/gki897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayala YM, et al. Human, Drosophila, and C.elegans TDP43: Nucleic acid binding properties and splicing regulatory function. J Mol Biol. 2005;348:575–588. doi: 10.1016/j.jmb.2005.02.038. [DOI] [PubMed] [Google Scholar]
- 7.Ou SH, Wu F, Harrich D, Garcia-Martinez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acharya KK, Govind CK, Shore AN, Stoler MH, Reddi PP. Cis-requirement for the maintenance of round spermatid-specific transcription. Dev Biol. 2006;295:781–790. doi: 10.1016/j.ydbio.2006.04.443. [DOI] [PubMed] [Google Scholar]
- 9.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 10.Davidson Y, et al. Ubiquitinated pathological lesions in frontotemporal lobar degeneration contain the TAR DNA-binding protein, TDP-43. Acta Neuropathol (Berl) 2007;113:521–533. doi: 10.1007/s00401-006-0189-y. [DOI] [PubMed] [Google Scholar]
- 11.Knudsen ES, Knudsen KE. Retinoblastoma tumor suppressor: Where cancer meets the cell cycle. Exp Biol Med (Maywood) 2006;231:1271–1281. doi: 10.1177/153537020623100713. [DOI] [PubMed] [Google Scholar]
- 12.Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu Rev Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 13.Moore JD, Kirk JA, Hunt T. Unmasking the S-phase-promoting potential of cyclin B1. Science. 2003;300:987–990. doi: 10.1126/science.1081418. [DOI] [PubMed] [Google Scholar]
- 14.Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem Sci. 2005;30:630–641. doi: 10.1016/j.tibs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432:588–595. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Kobayashi R, Galaktionov K, Beach D. p19Skp1 and p45Skp2 are essential elements of the cyclin A-CDK2 S phase kinase. Cell. 1995;82:915–925. doi: 10.1016/0092-8674(95)90271-6. [DOI] [PubMed] [Google Scholar]
- 17.Magnaghi-Jaulin L, et al. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 18.Qian YW, Lee EY. Dual retinoblastoma-binding proteins with properties related to a negative regulator of ras in yeast. J Biol Chem. 1995;270:25507–25513. doi: 10.1074/jbc.270.43.25507. [DOI] [PubMed] [Google Scholar]
- 19.MacLellan WR, Xiao G, Abdellatif M, Schneider MD. A novel Rb- and p300-binding protein inhibits transactivation by MyoD. Mol Cell Biol. 2000;20:8903–8915. doi: 10.1128/mcb.20.23.8903-8915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyake S, et al. Cells degrade a novel inhibitor of differentiation with E1A-like properties upon exiting the cell cycle. Mol Cell Biol. 2000;20:8889–8902. doi: 10.1128/mcb.20.23.8889-8902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christensen J, et al. Characterization of E2F8, a novel E2F-like cell-cycle regulated repressor of E2F-activated transcription. Nucleic Acids Res. 2005;33:5458–5470. doi: 10.1093/nar/gki855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simon HU, et al. Molecular characterization of hNRP, a cDNA encoding a human nucleosome-assembly-protein-I-related gene product involved in the induction of cell proliferation. Biochem J. 1994;297:389–397. doi: 10.1042/bj2970389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harbour JW, Dean DC. The Rb/E2F pathway: Expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 24.Ewen ME. Where the cell cycle and histones meet. Genes Dev. 2000;14:2265–2270. doi: 10.1101/gad.842100. [DOI] [PubMed] [Google Scholar]
- 25.Meyerson M, Harlow E. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol Cell Biol. 1994;14:2077–2086. doi: 10.1128/mcb.14.3.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ojala PM, Yamamoto K, Castanos-Velez E, Biberfeld P, Korsmeyer SJ, Makela TP. The apoptotic v-cyclin-CDK6 complex phosphorylates and inactivates Bcl-2. Nat Cell Biol. 2000;2:819–825. doi: 10.1038/35041064. [DOI] [PubMed] [Google Scholar]
- 27.Godden-Kent D, et al. The cyclin encoded by Kaposi's sarcoma-associated herpesvirus stimulates cdk6 to phosphorylate the retinoblastoma protein and histone H1. J Virol. 1997;71:4193–4198. doi: 10.1128/jvi.71.6.4193-4198.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ekholm SV, Reed SI. Regulation of G (1) cyclin-dependent kinases in the mammalian cell cycle. Curr Opin Cell Biol. 2000;12:676–684. doi: 10.1016/s0955-0674(00)00151-4. [DOI] [PubMed] [Google Scholar]
- 29.Grossel MJ, Hinds PW. Beyond the cell cycle: A new role for Cdk6 in differentiation. J Cell Biochem. 2006;97:485–493. doi: 10.1002/jcb.20712. [DOI] [PubMed] [Google Scholar]
- 30.Brito-Babapulle V, et al. Translocation t(2;7)(p12;q21–22) with dysregulation of the CDK6 gene mapping to 7q21–22 in a non-Hodgkin's lymphoma with leukemia. Haematologica. 2002;87:357–362. [PubMed] [Google Scholar]
- 31.Corcoran MM, et al. Dysregulation of cyclin dependent kinase 6 expression in splenic marginal zone lymphoma through chromosome 7q translocations. Oncogene. 1999;18:6271–6277. doi: 10.1038/sj.onc.1203033. [DOI] [PubMed] [Google Scholar]
- 32.Costello JF, et al. Cyclin-dependent kinase 6 (CDK6) amplification in human gliomas identified using two-dimensional separation of genomic DNA. Cancer Res. 1997;57:1250–1254. [PubMed] [Google Scholar]
- 33.Timmermann S, Hinds PW, Munger K. Elevated activity of cyclin-dependent kinase 6 in human squamous cell carcinoma lines. Cell Growth Differ. 1997;8:361–370. [PubMed] [Google Scholar]
- 34.Munger K, et al. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene. 2001;20:7888–7898. doi: 10.1038/sj.onc.1204860. [DOI] [PubMed] [Google Scholar]
- 35.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 36.Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24:2796–2809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- 37.Knudsen KE, et al. RB-dependent S-phase response to DNA damage. Mol Cell Biol. 2000;20:7751–7763. doi: 10.1128/mcb.20.20.7751-7763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Genovese C, Trani D, Caputi M, Claudio PP. Cell cycle control and beyond: emerging roles for the retinoblastoma gene family. Oncogene. 2006;25:5201–5209. doi: 10.1038/sj.onc.1209652. [DOI] [PubMed] [Google Scholar]
- 39.Meikrantz W, Schlegel R. Suppression of apoptosis by dominant negative mutants of cyclin-dependent protein kinases. J Biol Chem. 1996;271:10205–10209. doi: 10.1074/jbc.271.17.10205. [DOI] [PubMed] [Google Scholar]
- 40.Bakay M, et al. Nuclear envelope dystrophies show a transcriptional fingerprint suggesting disruption of Rb-MyoD pathways in muscle regeneration. Brain. 2006;129:996–1013. doi: 10.1093/brain/awl023. [DOI] [PubMed] [Google Scholar]
- 41.Melcon G, et al. Loss of emerin at the nuclear envelope disrupts the Rb1/E2F and MyoD pathways during muscle regeneration. Hum Mol Genet. 2006;15:637–651. doi: 10.1093/hmg/ddi479. [DOI] [PubMed] [Google Scholar]
- 42.Zhang YJ, et al. Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J Neurosci. 2007;27:10530–10534. doi: 10.1523/JNEUROSCI.3421-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.