Abstract
We have designed MI-219 as a potent, highly selective and orally active small-molecule inhibitor of the MDM2–p53 interaction. MI-219 binds to human MDM2 with a Ki value of 5 nM and is 10,000-fold selective for MDM2 over MDMX. It disrupts the MDM2–p53 interaction and activates the p53 pathway in cells with wild-type p53, which leads to induction of cell cycle arrest in all cells and selective apoptosis in tumor cells. MI-219 stimulates rapid but transient p53 activation in established tumor xenograft tissues, resulting in inhibition of cell proliferation, induction of apoptosis, and complete tumor growth inhibition. MI-219 activates p53 in normal tissues with minimal p53 accumulation and is not toxic to animals. MI-219 warrants clinical investigation as a new agent for cancer treatment.
Keywords: cancer therapy, MDM2–p53 protein–protein interaction, selective toxicity to tumors, small-molecule inhibitor
The tumor suppressor p53 plays a central role in the regulation of cell cycle, apoptosis, DNA repair, and senescence (1–4). Because of the prominent role played by p53 in suppressing oncogenesis (5), it is not surprising that p53 function is impaired in all human cancers. Several distinct approaches have been pursued to restore p53 function as a new cancer therapeutic strategy (6–9). Three recent studies, using unique genetic mouse models, have demonstrated that the restoration of p53 leads universally to a rapid and robust regression of established sarcomas, lymphomas, and liver tumors (10–14). These studies provide strong evidence that established tumors remain persistently vulnerable to p53 tumor-suppressor function and that restoration of p53 function is therefore a powerful cancer therapeutic strategy (13).
In ≈50% of human cancers, the gene encoding p53 is either deleted or mutated, rendering the p53 protein inactive (5, 15). In the remaining cancers, p53 retains its wild-type status but its function is effectively inhibited by its primary cellular inhibitor, the human MDM2 oncoprotein (mouse double minute 2, also termed HDM2 in humans) (5, 16, 17). One attractive pharmacological approach to p53 reactivation is to use a small molecule to block the MDM2–p53 interaction (6–8, 18). The discovery of the Nutlins provided the important proof of the concept for this approach (7). Nutlins were shown to bind to MDM2, block the MDM2–p53 interaction, and activate wild-type p53 (7, 19–21). Nutlin-3a exhibits strong anti-tumor activity in multiple xenograft mouse models of human cancer (7, 19). The discovery of the Nutlins has fueled enthusiasm for the development of small-molecule MDM2 inhibitors as a new class of anticancer therapy (6, 8, 22, 23).
One critical question in the development of MDM2 inhibitors for cancer treatment is their potential toxicity to normal tissues. This concern was heightened by a recent genetic study, which showed that p53 activation in the absence of the MDM2 gene causes severe toxicity to radiosensitive normal adult mouse tissues, leading to rapid animal death (24). Previous studies on Nutlin-3 reported that therapeutically effective doses do not cause animal weight loss or other gross abnormalities (7, 19), but whether an MDM2 inhibitor would activate p53 and cause toxicity in normal tissues remains unaddressed.
For an unambiguous evaluation of pharmacological activation of p53 by blocking the MDM2–p53 interaction, it is essential first to create an ideal small molecule with the following properties: (i) a high binding affinity to MDM2, (ii) excellent specificity for MDM2 over other proteins, (iii) potent cellular activity and specificity, and (iv) a highly desirable pharmacokinetic (PK) profile in animals for in vivo evaluation. Employing a de novo structure-based approach, we have now designed MI-219 as an ideal MDM2 inhibitor. Using MI-219, we have carried out critical evaluation of the pharmacological activation of p53 by targeting the MDM2–p53 interaction as a potential cancer therapeutic strategy. Our present study provides compelling evidence that activation of p53 by a potent and specific MDM2 inhibitor is a promising cancer therapeutic strategy and that MI-219 warrants clinical investigation for cancer treatment.
Results and Discussion
Rational Design of MI-219 as a Potent, Specific, and Orally Available Small-Molecule Inhibitor of the MDM2–p53 Interaction.
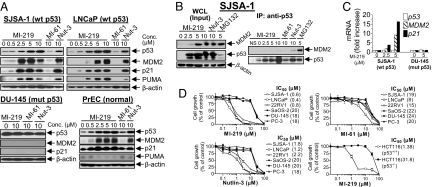
Employing a structure-based approach and based on the crystal structure of the MDM2–p53 complex (25), we have designed spiro-oxindoles (Fig. 1A) as a new class of inhibitors of the MDM2–p53 interaction (26, 27), as exemplified by MI-63. MI-63 binds to MDM2 with high affinity, activates the p53 pathway, and selectively inhibits cell growth in cancer cell lines with wild-type p53 (27). Furthermore, MI-63 effectively induces apoptosis ex vivo in chronic lymphocyte leukemia patient samples with functional p53 (42). However MI-63 has a poor PK profile and is unsuitable for in vivo evaluation. Extensive modifications of MI-63 have now yielded MI-219 as a potent and selective MDM2 inhibitor with a desirable PK profile [Fig. 1 A and B and supporting information (SI) Methods]. Computational modeling predicted that MI-219 mimics the key binding residues in p53 (Phe-19, Leu-22, Trp-23, and Leu-26) (Fig. 1C and SI Fig. 6) and achieves optimal interactions with MDM2. Indeed, MI-219 binds to MDM2 with a Ki value of 5 nM, and is >1,000-fold more potent than the p53 peptide (Fig. 1B). In this assay, Nutlin-3 has a Ki value of 36 nM.
Fig. 1.
MI-219 binds potently and selectively to MDM2 and mimics key p53 residues. (A) Chemical structures of MDM2 inhibitors and control compounds. (B) Competitive binding to recombinant human MDM2 and MDMX proteins. (C) Predicted binding model of MI-219 in complex with MDM2 and superpositioned to the MDM2–p53 crystal structure. MI-219 is shown in ball and stick representation, and the key residues are shown as sticks. Surface representation of MDM2 is shown with carbons in gray, nitrogen in blue, oxygen in red, and sulfur in yellow. For clarity, hydrogen atoms are not included. The figure was generated with the program Pymol.
Although MDMX is a closely related homolog of MDM2 (28, 29), MI-219 binds to MDMX with an IC50 value of >100 μM (estimated Ki = 55.7 μM) and is >10,000 times less potent in binding than to MDM2 (Fig. 1B). The p53 peptide binds to MDM2 and MDMX with similar affinities (Fig. 1B), and Nutlin-3 binds to MDMX with a Ki value of 9.4 μM (Fig. 1B), similar to that reported in refs. 21 and 30. Although Bcl-2 and Bcl-xL also have hydrophobic binding grooves that interact with amphiphilic α-helix domains of their binding partners, similar to the MDM2-p53 interaction, MI-219 showed no or minimal binding at 100 μM (SI Fig. 7A). Thus, MI-219 binds to MDM2 with a high affinity and selectivity.
MI-61 was designed as an inactive analogue and binds to MDM2 with a >2,000-times weaker affinity than MI-219 (Fig. 1 A and B).
MI-219 Blocks MDM2–p53 Interaction and Activates the p53 Pathway.
A potent and specific inhibitor of the MDM2–p53 interaction should activate the p53 pathway in cells with wild-type p53 (2, 31, 32). Indeed, MI-219 dose-dependently induced p53 accumulation and up-regulation of MDM2, p21, and PUMA, three p53-target gene products, in the SJSA-1 (osteosarcoma), LNCaP, and 22Rv1 (prostate cancer) cell lines with wild-type p53 (Fig. 2A and SI Fig. 8A). Similar results were obtained for Nutlin-3. The inactive analogue MI-61 had no effect. Furthermore, MI-219 and Nutlin-3 had no effect on the levels of p53, MDM2, and p21 proteins in cell lines with mutated/deleted p53 (Fig. 2A and SI Fig. 8A), indicating that up-regulation of MDM2 and p21 by MI-219 is strictly p53-dependent.
Fig. 2.
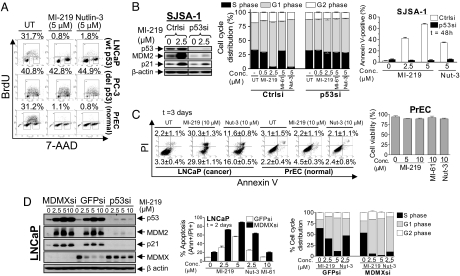
MI-219 blocks intracellular MDM2–p53 interaction, activates the p53 pathway, and inhibits cell growth. (A) Cells were treated for 24 h with different compounds. Whole-cell lysates (WCL) were analyzed by Western blotting. (B) SJSA-1 cells were treated for 15 h with MDM2 inhibitors or MG132. WCL were analyzed by immunoblotting, and coimmunoprecipitation was performed. (C) Cells were treated for 15 h with MI-219, and a Taqman real-time PCR assay was performed. (D) Cell growth inhibitory activity was evaluated in a WST cell growth assay. Data are shown as mean IC50 values. Error bars represent SEM.
MI-219 was evaluated for its effect on seven primary human normal cells or cell lines [primary normal prostate epithelial cells (PrEC); human mammary epithelial cells (HMEC); human renal epithelial cells (HREC); WI-38, CCD-18Co, and foreskin fibroblasts; and MCF-12F normal-like cell line]. MI-219 induced functional activation of p53 in these normal cells, as indicated by up-regulation of p53, MDM2, and/or p21, whereas MI-61 had no effect (Fig. 2A and SI Fig. 9A). Thus, these data indicate that MI-219 activates the p53 pathway in both normal and cancer cells with wild-type p53. However, both MI-219 and Nutlin-3 failed to up-regulate PUMA, a potent mediator of p53-dependent apoptosis, in normal PrEC (Fig. 2A).
MI-219 was evaluated for its ability to block the intracellular MDM2–p53 interaction. Coimmunoprecipitation showed that although MI-219 caused up-regulation of p53 and MDM2 in SJSA-1 (Fig. 2B) and 22Rv1 (SI Fig. 8B) cancer cell lines, little or no MDM2 protein coimmunoprecipitated with p53. Similar results were obtained for Nutlin-3. MG132, a proteasome inhibitor known to cause p53 and MDM2 accumulation by blocking protein degradation (33), indeed increased the levels of both proteins but failed to block their interaction.
RT-PCR analysis showed that MI-219 increased the transcription of both MDM2 and p21 genes in SJSA-1 cells but had no effect on p53 transcription, and had no effect on any of these genes in DU-145 cells with mutant p53 (Fig. 2C). These data showed that MI-219 induces up-regulation of MDM2 and p21 proteins at the transcriptional level but causes accumulation of p53 at the posttranscriptional level.
MI-219 Potently and Selectively Inhibits Cell Growth in Cancer Cell Lines with Wild-Type p53.
Activation of the p53 pathway by MI-219 leads to cell cycle arrest and/or cell death induction and cell growth inhibition (34, 35). MI-219 potently inhibited cell growth in the SJSA-1, LNCaP, and 22Rv1 cancer cells with wild-type p53, having IC50 values ranging between 0.4 and 0.8 μM (Fig. 2D), but showed a 20- to 50-fold weaker activity in cancer cell lines lacking wild-type p53. Using HCT-116 p53+/+ and p53−/− isogenic cell lines [a kind gift from B. Vogelstein (Johns Hopkins University, Baltimore)], we confirmed that the cell growth inhibitory activity of MI-219 is p53-dependent (Fig. 2D). MI-61 only weakly inhibited cell growth in cancer cell lines with wild-type p53 and significantly displayed no selectivity over those lacking wild-type p53. In agreement with a previous report (7), Nutlin-3 inhibited cell growth in cancer cell lines with wild-type p53 and exhibited ≈10-fold selectivity over those without wild-type p53 (Fig. 2D).
MI-219 Induces Cell Cycle Arrest in both Cancer and Normal Cells.
Both MI-219 and Nutlin-3 effectively depleted S-phase cells and arrested LNCaP cells in G1 and G2 phases but had no effect on all cell lines lacking wild-type p53 (Fig. 3A and SI Fig. 10A). Transient transfection of SJSA-1 and LNCaP cells with p53 siRNA decreased the p53 basal levels; attenuated the up-regulation of p53, MDM2, and p21 by MI-219; and rendered MI-219 much less effective at induction of cell cycle arrest (Fig. 3B and SI Fig. 10B). MI-61 had little or no effect. These data indicate that the cell cycle arrest by MI-219 in cancer cells is p53-dependent. MI-219 also effectively halted cell cycle progression in all normal cells examined (Fig. 3A and SI Fig. 9B), and, therefore, it induces cell cycle arrest in both cancer and normal cells with wild-type p53.
Fig. 3.
MI-219 induces apoptosis selectively in cancer cells with wild-type p53, and downregulation of MDMX enhances MI-219 activity. (A) Flow cytometric cell cycle analysis of BrdU incorporation was performed after treatment of cells with the inhibitors for 24 h. Data shown are percentage of S-phase cells. (B) SJSA-1 cells, transfected with p53 siRNA, were treated with inhibitors for 24 h and analyzed by Western blotting. Cell cycle was analyzed by PI staining, and apoptosis was analyzed by Annexin V staining. (C) Cells were treated with or without inhibitors, followed by analyses of apoptosis (Annexin V staining) and cell viability (trypan blue dye exclusion). (D) LNCaP cells, transfected with MDMX, p53, or GFP siRNA were treated with inhibitors for 24 h and analyzed by Western blotting. Cell cycle was analyzed by PI staining, and apoptosis was analyzed by Annexin V staining. Data are shown as mean ± SEM.
MI-219 Induces Apoptosis in Cancer but Not in Normal Cells.
Because p53 is also a central regulator of apoptosis (2, 3, 31, 32), we examined whether activation of p53 by MI-219 leads to apoptosis in cancer and normal cells. MI-219 effectively induced cell death in three cancer cell lines with wild-type p53 in a dose-dependent manner but had little effect on cancer cells lacking wild-type p53 (SI Fig. 11A). Cell death induced by MI-219 was apoptotic, as confirmed by Annexin V staining (Fig. 3 B and C and SI Fig. 11B). Nutlin-3 also induced apoptotic cell death in cancer cells with wild-type p53, albeit less potently than MI-219 (Fig. 3 B and C and SI Fig. 11B). MI-61 showed no significant effect on any cancer cell line. Induction of apoptosis by MI-219 was strictly p53-dependent, because silencing p53 via siRNA abolished its effect (Fig. 3B and SI Figs. 10B and 11B). Furthermore, apoptosis induction in SJSA-1 cells by MI-219 is preceded by cleavage of caspase 3 and PARP (SI Fig. 12A) and is caspase-dependent because a pan caspase inhibitor ZVAD almost abrogated apoptosis induction (SI Fig. 12B).
In contrast to cancer cell lines with wild-type p53, all normal cells or cell lines were resistant to induction of apoptosis and cell death by MI-219 and Nutlin-3 (Fig. 3C and SI Fig. 9 C and D). Thus, MI-219 effectively and selectively induces apoptosis in cancer cells expressing wild-type p53 but has minimal effect on normal cells.
MDMX Attenuates the Activity of MI-219.
Both MDM2 and MDMX inhibit p53 function through direct binding. Because MI-219 binds to MDMX with a very weak affinity, it may not be able to fully activate p53 in cells expressing high levels of MDMX protein (21). However, MI-219 induced significant MDMX degradation in LNCaP cells expressing high levels of MDMX (Fig. 3D). Silencing p53 with siRNA attenuated MDM2 up-regulation and markedly decreased MDMX degradation induced by MI-219 (Fig. 3D), consistent with a report that MDMX degradation induced by Nutlin-3 is mediated by MDM2 (36). Transfection with MDMX siRNA, compared with GFP siRNA, enhanced apoptosis induction and cell cycle arrest by MI-219 in LNCaP cells (Fig. 3D and SI Fig. 13), indicating that MDMX attenuates p53 activation by MI-219. These data indicate that, although it does not bind to MDMX, MI-219 can induce MDMX degradation, which may contribute to its antitumor activity in cells with high levels of MDMX.
MI-219 Activates p53, Inhibits Cell Proliferation, and Induces Apoptosis in Xenograft Tumors.
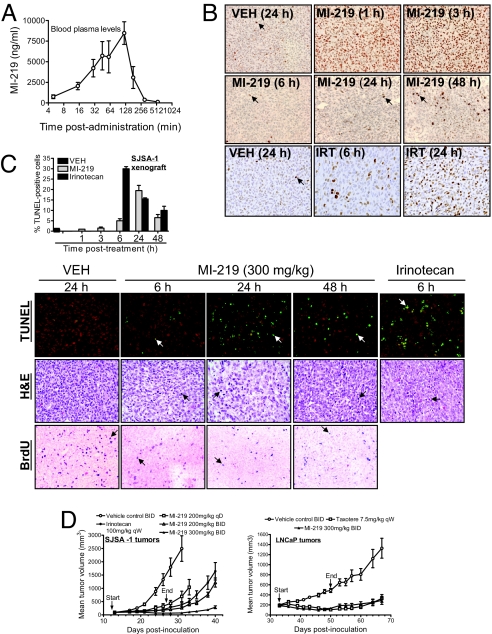
Because MI-219 achieved an excellent oral bioavailability in PK studies (Fig. 4A and SI Table 1), we investigated in vivo activation of p53 by MI-219, using mouse xenograft models of human cancer. Immunohistochemical (IHC) analysis showed that a single oral dose of MI-219 induced strong accumulation of p53 in SJSA-1 tumor xenograft tissues at 1- and 3-h time points, but p53 levels were barely detectable at 6 h and thereafter (Fig. 4B). Western blot data confirmed these findings and further showed that p53 accumulation induced by MI-219 was accompanied by up-regulation of MDM2 and p21 proteins in tumor tissues (SI Fig. 14A). Activation of p53 in tumor tissues is positively correlated with plasma concentrations of MI-219 in PK studies (Fig. 4A). When the concentrations of MI-219 decreased, the level of p53 activation in tumor tissues rapidly diminished, indicating that activation of p53 induced by MI-219 is transient. Irinotecan (IRT) also caused p53 accumulation in tumor tissues but had little effect on the levels of MDM2 and p21 proteins (Fig. 4B and SI Fig. 14A). MI-219 also induced p53 accumulation in LNCaP xenograft tumor tissues (SI Fig. 14A).
Fig. 4.
MI-219 activates p53 in vivo, inhibits cell proliferation, induces apoptosis in tumor tissues, and achieves robust in vivo anti-tumor activity. (A) A single oral dose of MI-219 (50 mg/kg) was given to mice, and MI-219 concentrations in blood plasma were analyzed by LC/MS/MS. Error bars represent SD of data from three animals. (B) Mice (two to three mice per group), bearing established SJSA-1 xenograft tumors, were treated with a single oral dose of MI-219, irinotecan (IRT), or vehicle, and p53 in tumor tissues was analyzed by IHC staining. Arrows indicate a small fraction of p53-positive cells. A wide-spread p53 staining is observed at 1 and 3 h of treatment with MI-219. (C) SJSA-1 xenograft tumor tissues, harvested from mice treated with a single oral dose of MI-219, were examined for cell proliferation by IHC staining of BrdU and apoptosis by TUNEL and H&E staining. Arrows indicate the positivity of the BrdU and TUNEL staining. H&E shows apoptotic cells with nuclear condensation/fragmentation. To score apoptosis, at least 1,000 cells were analyzed for TUNEL staining. (D) Nude mice (8–10 per group), bearing s.c. SJSA-1 and LNCaP xenograft tumors, were administered with MI-219 (p.o.), irinotecan (i.p.), Taxotere (i.v.), or vehicle control (p.o.). Data are represented as mean tumor volumes ± SEM.
A single oral dose of MI-219 inhibited cell proliferation and induced apoptosis in tumor tissues in a time-dependent manner and achieved maximum effects at 24 h (Fig. 4C and SI Fig. 14B). Thus, rapid p53 activation by MI-219 precedes its maximum biological effect. IRT induced maximum apoptosis at the 6-h time point. Induction of apoptosis by MI-219 and IRT in tumor tissues was further supported by IHC staining of cleaved PARP and H&E staining (Fig. 4C and SI Fig. 14C).
MI-219 Shows Robust in Vivo Antitumor Activity.
We next evaluated the anti-tumor activity of MI-219 as an oral agent employing SJSA-1 and LNCaP xenograft mouse models. MI-219 was highly effective in the inhibition of tumor growth in both models (Fig. 4D and SI Fig. 15). At 200 mg/kg once a day (qD) for 14 days, MI-219 inhibited tumor growth by 75% in SJSA-1 xenografts compared with the vehicle-treated group (P = 0.0011, t test) (Fig. 4D). Because p53 activation by MI-219 is transient, we explored whether more frequent dosing of MI-219 would achieve an even stronger antitumor activity. Indeed, MI-219 at 200 mg/kg twice a day (BID) for 14 days inhibited tumor growth by 86% compared with the vehicle control (P = 0.0004, t test) and is more effective than the qD dosing (P = 0.0163, ANOVA) (Fig. 4D and SI Fig. 15). MI-219 at 300 mg/kg BID for 14 days completely inhibited tumor growth, and the tumor volume was decreased from 95 ± 13 mm3 at the start of the treatment to 67 ± 18 mm3 after the treatment, whereas the mean tumor volume in the vehicle-treated group grew from 95 ± 21 mm3 to 1,328 ± 633 mm3 in the same period. MI-219 at 300 mg/kg BID was much more effective than IRT at its maximum tolerated dose (P < 0.0001, ANOVA).
Furthermore, MI-219 was also very effective in the inhibition of LNCaP tumor growth (Fig. 4D). Treatment with MI-219 for 17 days reduced the tumor size from 184 ± 69 mm3 to 134 ± 67 mm3, whereas the vehicle-treated tumors grew from 200 ± 67 mm3 to 487 ± 128 mm3. No significant weight loss or other signs of toxicity were observed for mice treated with MI-219 at any dose schedule (SI Fig. 16). These in vivo data thus showed that MI-219 achieves strong antitumor activity at nontoxic dose schedules. The in vivo antitumor activity of MI-219 is p53-dependent because MI-219 failed to achieve significant antitumor activity (P > 0.05, ANOVA) in the MDA-MB-231 (2LMP) xenografts expressing mutated p53 (SI Fig. 17).
MI-219 Is Not Toxic to Normal Tissues.
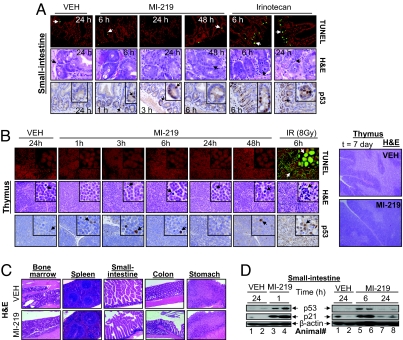
We next examined the toxicity of MI-219 on normal tissues, particularly radio-sensitive tissues, such as small-intestine crypts and thymus, which are known to be sensitive to p53-induced apoptosis (37, 38). TUNEL and H&E analyses showed that treatment of nude (Fig. 5A) and BALB/c (SI Fig. 18A) mice by a single dose of MI-219 did not induce apoptosis at any time point examined in the small-intestinal crypts. In contrast, a single dose of IRT or ionizing radiation (IR) induced clear apoptosis in the crypts (Fig. 5A and SI Fig. 18A). In addition, MI-219 caused no apoptosis in thymus in BALB/c mice, whereas IR induced profound tissue damage (Fig. 5B). These data were further confirmed by using C57BL/6 mice (SI Fig. 19).
Fig. 5.
MI-219 does not induce tissue damage in normal mouse tissues. (A) Apoptosis in normal small-intestine crypts from mice (three mice per group), treated with a single dose of MI-219 or irinotecan, was examined by TUNEL and H&E staining. p53 accumulation was analyzed by IHC. (B) BALB/c mice (three mice per group) were treated with MI-219 or IR. TUNEL, H&E, and p53 IHC staining were performed in thymus. In addition, H&E staining of thymus was also performed after 7 days of treatment with MI-219. (C) Six nude mice were treated with MI-219 (300 mg/kg BID) for 14 days, and H&E staining of normal mouse tissues was performed. (D) Western blot analysis of p53 activation in small intestine.
To investigate whether repeated and long-term dosing of MI-219 is toxic to animals, nude mice were treated with MI-219 at 300 mg/kg [BID and per os (p.o.)] for a total of 14 days, a highly efficacious dose schedule for its antitumor activity. Histopathology revealed that MI-219 did not cause damage to either radio-sensitive or -resistant tissues, such as those from bone marrow, spleen, small-intestine, and colon (39) (Fig. 5C and SI Fig. 20A). Furthermore, no tissue damage was found after the treatment was stopped for 2 weeks (data not shown). In addition, treatment of BALB/c mice with MI-219 at 300 mg/kg (BID and p.o.) for 7 days did not cause damage to thymus (Fig. 5B), small intestine (SI Fig. 20B), or any other tissues examined (data not shown). Hence, MI-219 is not toxic to normal mouse tissues.
To rule out the possibility that the lack of toxicity of MI-219 in mice could be due to low drug exposure in normal tissues, we performed tissue distribution analysis in nude mice bearing SJSA-1 xenografts. MI-219 was present at very high concentrations in all of the tissues except the brain (SI Table 2). The concentrations of MI-219 in SJSA-1 tumor tissues were very similar to those observed in spleen and plasma, but much lower than those in other tissues, such as lung, liver, and kidney. Therefore, the lack of toxicity of MI-219 to normal tissues is not due to low drug exposure.
MI-219 Functionally Activates p53 in Normal Mouse Tissues.
We next examined p53 activation in normal mouse tissues. IHC staining showed that although IR and IRT induced robust p53 accumulation in the small intestinal crypts and thymus, MI-219 caused minimal p53 accumulation in these tissues (Fig. 5 A and B and SI Fig. 18B).
To investigate whether p53 was still functionally activated, despite low accumulation, we examined p21 accumulation in small intestine, liver, stomach, and spleen. Western blot analysis showed that p21 is robustly up-regulated in small intestine (Fig. 5D) and in liver, stomach, and spleen (SI Fig. 21A). Because up-regulation of p21 by MI-219 and Nutlin-3 in human cells is strictly p53-dependent, these data suggest that, although p53 is not accumulated to a high level in normal tissues, it is functionally activated.
To exclude the possibility that p21 up-regulation by MI-219 in mouse tissues in vivo is p53 independent, MI-219 was evaluated in NIH 3T3 and B16 mouse cell lines with wild-type p53 (SI Fig. 21B). MI-61 and Nutlin-3 were used as the inactive and active control, respectively. MI-219 and Nutlin-3 induced p21 up-regulation in both mouse cell lines, whereas MI-61 had no effect. Furthermore, MI-219 and Nutlin-3 did not induce significant p53 accumulation either. These data indicate that MI-219 activates p53 in normal mouse tissues, despite low levels of p53 accumulation.
We also investigated the possible differences in the binding of MI-219 to mouse and human MDM2 by computational modeling. Comparison of the modeled three-dimensional (3D) structure of mouse MDM2 to the crystal structure of human MDM2 revealed that their p53 binding sites are very similar (SI Fig. 22A), with the exception that two leucine residues (i.e., Leu-54 and 57) in human MDM2 are replaced by isoleucine residues in mouse MDM2. Modeling further predicted that MI-219 binds to mouse and human MDM2 with essentially identical models and similar binding affinities (SI Fig. 22B and SI Table 3).
Activation of p53 by an MDM2 Inhibitor Is Different from Other Approaches.
Activation of p53 by a potent and specific MDM2 inhibitor is achieved by blocking the MDM2–p53 interaction, does not require posttranslational modifications, and takes place in the presence of MDM2. These important aspects of p53 activation by an MDM2 inhibitor may account for the lack of toxicity of MI-219 to normal tissues.
The important role of MDM2 in protecting normal tissue from damage upon p53 activation is supported by a genetic study (11, 40), which showed that mice with a 70% reduced level of MDM2 protein had increased p53 activity but were born viable and had only slightly reduced body weight (40). Importantly, genetic activation of p53 in an MDM2-null background is severely toxic to animals (24), whereas p53 activation in the presence of MDM2, using a different genetic approach, does not cause damage to normal tissues (11). In comparison, activation of p53 by MI-219 is always under the surveillance of MDM2 and, therefore, is never fully unleashed.
DNA damaging agents, such as IR and IRT, activate p53 by inducing its posttranslational modifications, such as extensive phosphorylation, therefore altering the protein stability. Indeed, IR and IRT induced robust accumulation of p53 in normal tissues, leading to tissue damage in p53 sensitive tissues, such as the small intestinal crypts and thymus. In comparison, MI-219 activated p53 in normal tissues with minimal p53 accumulation. Therefore, the higher levels of p53 accumulation induced by genotoxic agents (IR and IRT) than by MI-219 could account for their different toxicities.
Summary.
We have rationally designed MI-219 as an orally bioavailable and most potent and selective inhibitor of the MDM2–p53 interaction and have performed critical in vitro and in vivo evaluations of MI-219 as a potential new cancer therapy.
MI-219 disrupts the MDM2–p53 interaction and activates the p53 pathway in cells with wild-type p53. Activation of p53 by MI-219 leads to cell cycle arrest in both normal and tumor cells and selective apoptosis in the latter. It activates p53 in normal mouse tissues with minimal p53 accumulation and is not toxic to animals, indicating that normal cells and tissues are intrinsically resistant to activation of p53 by an MDM2 inhibitor. MI-219 stimulates rapid but transient p53 activation in xenograft tumor tissues, resulting in inhibition of cell proliferation, induction of apoptosis, and complete tumor growth inhibition consistent with the notion that established tumors remain persistently vulnerable to the p53 tumor-suppressor function (13). Our present study provides compelling evidence that pharmacological activation of the p53 by blocking the MDM2–p53 interaction is a promising cancer therapeutic strategy and MI-219 warrants clinical evaluation as a new cancer therapy.
Materials and Methods
Antibodies.
The following primary antibodies were used in the study: anti-p53 (FL-393 and CM5) for Western blot analysis, co-IP, and IHC; anti-MDM2 (SMP-14); anti-p53 (DO-1); anti-p21 (SX118); rabbit polyclonal anti-MDMX (Upstate Biosciences); and anti-PUMA (Ab-1) for Western blot analysis.
Chemical Synthesis, Computational Modeling, and FP-Based Binding Assays.
MI-219, MI-63, and MI-61 were synthesized by using our method published in refs. 26 and 27. Nutlin-3 was purchased from Cayman Chemical. The binding of small molecules to human MDM2 was predicted by using the GOLD program (26, 27), as described in SI Methods. Competitive binding assays were performed to determine binding to human MDM2, Bcl-2, and Bcl-xL (26, 27). The MDMX binding assay is described in SI Methods.
Cell Growth, Cell Cycle, Cell Death, and Apoptosis.
Cell growth inhibition activity was determined in a water-soluble tetrazolium (WST)-based assay (27). Cell cycle analysis was performed by flow cytometric analysis of DNA content after PI staining and BrdU incorporation, using the FITC BrdU kit (BD Biosciences). Cell death was measured by trypan blue staining, and apoptosis was determined by using an Annexin V-FLUOS staining kit (Roche Applied Science).
RNA Interference.
To down-regulate p53 and MDMX, cells were transiently transfected with p53 siRNA (41) or MDMX siRNA (Qiagen). GFP siRNA was used as a control. Transfections were performed by using Lipfectamine RNAiMAX (Invitrogen).
Intracellular MDM2–p53 Interaction.
Coimmunoprecipitation assay was performed to examine the ability of the inhibitors and proteasome inhibitor MG132 to dissociate MDM2–p53 complex formation, as described in ref. 30.
In Vivo Studies.
For anti-tumor efficacy studies, nude athymic mice bearing s.c. tumor xenografts were used. To determine p53 activation in vivo, tissues were harvested, and tissue lysates were analyzed by Western blot analysis and immunohistochemistry (IHC) as described in SI Methods. For histopathology analyses of tissues, paraffin-embedded tissues were stained with H&E. Cell proliferation was examined by IHC staining of BrdU, using a 5-Bromo-2′-deoxy-uridine labeling and detection kit II (Roche Applied Science). Apoptosis was examined by TUNEL staining, using an in situ cell death detection kit (Roche Applied Science). For whole-body γ radiation, mice were irradiated at 2 Gy/min. All animal experiments were performed under the guidelines of the University of Michigan Committee for Use and Care of Animals.
Statistical Analysis.
Statistical analyses were performed by two-way ANOVA and unpaired two-tailed t test, using Prism (version 4.0) from GraphPad. P < 0.05 was considered significant.
Supplementary Material
ACKNOWLEDGMENTS.
This work was supported by National Cancer Institute/National Institutes of Health Grant R01CA121279 (to S.W.); University of Michigan Prostate Cancer Specialized Program of Research Excellence P50CA069568 (S.W. and K.J.P.); the Leukemia and Lymphoma Society (S.W.); the Prostate Cancer Foundation (S.W.); Ascenta Therapeutics, Inc. (S.W.); and University of Michigan Cancer Center Grant P30CA046592.
Footnotes
Conflict of interest statement: Ascenta Therapeutics, Inc., has licensed the technologies related to the MDM2 inhibitors (MI-219, MI-63), in which the University of Michigan and S.W. own equity. S.W. serves as a consultant for Ascenta and is the principal investigator on a research contract from Ascenta to the University of Michigan. X.L., S.K., M.G., and D.Y. are employees of Ascenta.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708917105/DC1.
References
- 1.Levine AJ. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine AJ. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Vousden KH, Lu X. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 4.Fridman JS, Lowe SW. Oncogene. 2003;22:9030–9040. doi: 10.1038/sj.onc.1207116. [DOI] [PubMed] [Google Scholar]
- 5.Hainaut P, Hollstein M. Adv Cancer Res. 2000;77:81–137. doi: 10.1016/s0065-230x(08)60785-x. [DOI] [PubMed] [Google Scholar]
- 6.Chene P. Nat Rev Cancer. 2003;3:102–109. doi: 10.1038/nrc991. [DOI] [PubMed] [Google Scholar]
- 7.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 8.Vassilev LT. Trends Mol Med. 2007;13:23–31. doi: 10.1016/j.molmed.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Wiman KG. Cell Death Differ. 2006;13:921–926. doi: 10.1038/sj.cdd.4401921. [DOI] [PubMed] [Google Scholar]
- 10.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 12.Martins CP, Brown-Swigart L, Evan GI. Cell. 2006;127:1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Sharpless NE, DePinho RA. Nature. 2007;445:606–607. doi: 10.1038/nature05567. [DOI] [PubMed] [Google Scholar]
- 14.Kastan MB. Cell. 2007;128:837–840. doi: 10.1016/j.cell.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 15.Feki A, Irminger-Finger I. Crit Rev Oncol Hematol. 2004;52:103–116. doi: 10.1016/j.critrevonc.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 16.Freedman DA, Wu L, Levine AJ. Cell Mol Life Sci. 1999;55:96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fakharzadeh SS, Rosenblum-Vos L, Murphy M, Hoffman EK, George DL. Genomics. 1993;15:283–290. doi: 10.1006/geno.1993.1058. [DOI] [PubMed] [Google Scholar]
- 18.Vassilev LT. Cell Cycle. 2004;3:419–421. [PubMed] [Google Scholar]
- 19.Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, Hilton H, Zhao X, Vu BT, Qing W, Packman K, et al. Proc Natl Acad Sci USA. 2006;103:1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Efeyan A, Ortega-Molina A, Velasco-Miguel S, Herranz D, Vassilev LT, Serrano M. Cancer Res. 2007;67:7350–7357. doi: 10.1158/0008-5472.CAN-07-0200. [DOI] [PubMed] [Google Scholar]
- 21.Hu B, Gilkes DM, Farooqi B, Sebti SM, Chen J. J Biol Chem. 2006;281:33030–33035. doi: 10.1074/jbc.C600147200. [DOI] [PubMed] [Google Scholar]
- 22.Chen L, Yin H, Farooqi B, Sebti S, Hamilton AD, Chen J. Mol Cancer Ther. 2005;4:1019–1025. doi: 10.1158/1535-7163.MCT-04-0342. [DOI] [PubMed] [Google Scholar]
- 23.Kojima K, Konopleva M, Samudio IJ, Shikami M, Cabreira-Hansen M, McQueen T, Ruvolo V, Tsao T, Zeng Z, Vassilev LT, Andreeff M. Blood. 2005;106:3150–3159. doi: 10.1182/blood-2005-02-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ringshausen I, O'Shea CC, Finch AJ, Swigart LB, Evan GI. Cancer Cell. 2006;10:501–514. doi: 10.1016/j.ccr.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 25.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 26.Ding K, Lu Y, Nikolovska-Coleska Z, Qiu S, Ding Y, Gao W, Stuckey J, Krajewski K, Roller PP, Tomita Y, et al. J Am Chem Soc. 2005;127:10130–10131. doi: 10.1021/ja051147z. [DOI] [PubMed] [Google Scholar]
- 27.Ding K, Lu Y, Nikolovska-Coleska Z, Wang G, Qiu S, Shangary S, Gao W, Qin D, Stuckey J, Krajewski K, et al. J Med Chem. 2006;49:3432–3435. doi: 10.1021/jm051122a. [DOI] [PubMed] [Google Scholar]
- 28.Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M, van Ham RC, van der Houven van Oordt W, Hateboer G, van der Eb AJ, Jochemsen AG. EMBO J. 1996;15:5349–5357. [PMC free article] [PubMed] [Google Scholar]
- 29.Bottger V, Bottger A, Garcia-Echeverria C, Ramos YF, van der Eb AJ, Jochemsen AG, Lane DP. Oncogene. 1999;18:189–199. doi: 10.1038/sj.onc.1202281. [DOI] [PubMed] [Google Scholar]
- 30.Laurie NA, Donovan SL, Shih CS, Zhang J, Mills N, Fuller C, Teunisse A, Lam S, Ramos Y, Mohan A, et al. Nature. 2006;444:61–66. doi: 10.1038/nature05194. [DOI] [PubMed] [Google Scholar]
- 31.el-Deiry WS. Semin Cancer Biol. 1998;8:345–357. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- 32.Bouchet BP, de Fromentel CC, Puisieux A, Galmarini CM. Crit Rev Oncol Hematol. 2006;58:190–207. doi: 10.1016/j.critrevonc.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Maki CG, Huibregtse JM, Howley PM. Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- 34.el-Deiry WS, Harper JW, O'Connor PM, Velculescu VE, Canman CE, Jackman J, Pietenpol JA, Burrell M, Hill DE, Wang Y, et al. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 35.Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, Kinzler KW, Vogelstein B. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 36.Patton JT, Mayo LD, Singhi AD, Gudkov AV, Stark GR, Jackson MW. Cancer Res. 2006;66:3169–3176. doi: 10.1158/0008-5472.CAN-05-3832. [DOI] [PubMed] [Google Scholar]
- 37.Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 38.Potten CS, Wilson JW, Booth C. Stem Cells. 1997;15:82–93. doi: 10.1002/stem.150082. [DOI] [PubMed] [Google Scholar]
- 39.Rubin P, Casarett GW. Cancer. 1968;22:767–778. doi: 10.1002/1097-0142(196810)22:4<767::aid-cncr2820220412>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 40.Mendrysa SM, McElwee MK, Michalowski J, O'Leary KA, Young KM, Perry ME. Mol Cell Biol. 2003;23:462–472. doi: 10.1128/MCB.23.2.462-473.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brummelkamp TR, Bernards R, Agami R. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 42.Saddler C, Ouillette P, Kujawski L, Shangary S, Talpaz M, Kaminski M, Erba H, Shedden K, Wang S, Malek SN. Blood. 2008;111:1584–1593. doi: 10.1182/blood-2007-09-112698. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.