Abstract
Bacterial cells avoid lysis in response to hypoosmotic shock through the opening of the mechanosensitive channel MscL. Upon channel opening, MscL is thought to expand in the plane of the membrane and form a large pore with an estimated diameter of 3–4 nm. Here, we set out to analyze the closed and open structure of cell-free MscL. To this end, we characterized the function and structure of wild-type MscL and a mutant form of the protein (G22N MscL) that spontaneously adopts an open substate. Patch-clamp analysis of MscL that had been reconstituted into liposomes revealed that wild-type MscL was activated only by mechanical stimuli, whereas G22N MscL displayed spontaneous opening to the open substate. In accord with these results, Ca2+ influx into G22N MscL-containing liposomes occurred in the absence of mechanical stimulation. The electrophoretic migration of chemically cross-linked G22N MscL was slower than that of cross-linked wild-type MscL, suggesting that G22N MscL is in an expanded form. Finally, electron microscopy using low-angle rotary shadowing revealed the presence of a pore at the center of G22N MscL. No pore could be detected in wild-type MscL. However, wild-type MscL possessed a protrusion at one end, which was absent in G22N MscL. The deletion of carboxyl-terminal 27 residues resulted in the loss of protrusion and proper multimerization. The structures of wild-type and G22N MscL reveal that the opening of MscL is accompanied by the dissociation of a carboxyl-terminal protrusion and pore formation.
Keywords: electon microscopy, patch clamp, liposome, low-angle rotary shadowing, bacterial ion channel
Ion channels are a class of membrane proteins that form pores allowing for the flow of ions down their electrochemical gradient. Ion flux through these channels gives rise to a variety of physiological activities including neural excitation, stimulus perception, volume regulation, ion transport, and muscle contraction. Precise regulation of the channel pore is essential to control these activities. In fact, most channels are regulated by factors such as voltage, ligand binding, temperature, and membrane stretch. However, little is known about how channels rearrange to form pores, and this remains a fundamental question in understanding the mechanisms underlying ion channel gating.
MscL, a mechanosensitive channel ubiquitous among prokaryotes, undergoes a drastic conformational change on gating. MscL has a conductance of ≈3 nS (1) and forms a pore as large as 3–4 nm in diameter in the open state (2). In contrast, the pore is constricted to ≈0.2 nm in the closed state (3). The large pore in the open state is likely to be formed by tilting and sliding of the transmembrane helices (4–6). Structural modeling and molecular dynamics simulations also support the notion that a dynamic in-plane expansion occurs on gating (7–11). Indeed, a depression forms at the center of MscL when it is adsorbed on a loose alkylsilane monolayer, as seen by atomic force microscopy (12).
MscL is a homopentamer, with each subunit being composed of two transmembrane helices known as TM1 and TM2 (1, 3). TM1 lines the channel pore and forms a constriction near the cytoplasmic end of the pore. The hydrophobic nature of this constricted area probably stabilizes the closed state of MscL because introduction of a hydrophilic motif into this region destabilizes the closed state and reduces the gating threshold (13). An extreme example is a mutant channel formed by hydrophilic substitution of Gly-22 by asparagine (G22N MscL), which yields a channel that spontaneously adopts an open substate, even in the absence of membrane stretch. Similarly, spontaneous activity is also observed on incubation of G22C MscL with 2-(trimethylammonium)ethyl methane thiosulfonate (MTSET), a hydrophilic molecule that binds to cysteine (14). Based on these findings, Gly-22 and the nearby residues have been proposed to comprise a hydrophobic “lock” that is released by membrane tension (13, 15, 16).
Although full opening of MscL occurs through several open substates, most of the in-plane expansion of MscL occurs while the pore is still closed (17). Thus, transitions between the open substates and the fully open state occur without a significant increase in the cross-sectional area. In support of this view, an analysis of G22N MscL revealed that G22N MscL is already in an expanded form in the open substate (18).
MscL has a cytoplasmic carboxyl-terminal segment of ≈40 residues (1, 19). Removal of the 27 carboxyl residues does not remove channel activity; however, it does result in frequent open substates (19, 20). The crystal structure of Mycobacterium tuberculosis MscL (TbMscL) revealed that the carboxyl-terminal residues form an α helix that assembles into a pentameric bundle (3). Molecular dynamics simulation indicates that this bundle, which was obtained at very low pH, is not stable under physiological conditions (21). The bundle appears to be formed in the closed state but in a manner different from that in the crystal structure, as shown through cross-linking of the helices (20). Given the drastic in-plane enlargement of the transmembrane domain on channel gating, the bundle may be required to maintain the organization of the transmembrane domain in the open state as well as in the closed state (20). Alternatively, the bundle may be disassembled in the open state because of the large scale of the expansion (7).
In this study, we investigated the effect of G22N mutation on the function and structure of MscL. Because the expression of G22N MscL impedes cell growth, we synthesized MscL with a cell-free translation system (22). Channel activities were assessed by patch-clamp measurement and Ca2+ permeability measurement by using a fluorescent probe. The structure of single MscL molecules was analyzed by electron microscopy (EM) by using low-angle rotary shadowing. Our results demonstrate that the opening of MscL is accompanied by the dissociation of a protrusion and pore formation.
Results and Discussion
Biochemical Characterization of Cell-Free Translated MscL.
On translation of wild-type MscL in the absence of detergent, only a small proportion of MscL was present in the soluble fraction. However, a larger amount of MscL was in soluble form when translation was performed in the presence of the nonionic detergent, Brij-58 (1–6 mM). Nickel-nitrilotriacetic acid (NiNTA) column purification yielded almost pure preparations of MscL (Fig. 1A).
Fig. 1.
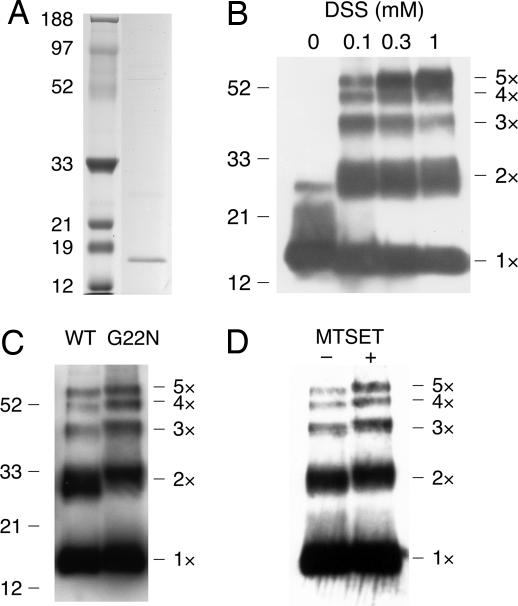
Electrophoresis of cell-free translated MscL. (A) Wild-type MscL (right column) and molecular marker (left column) separated by SDS/PAGE. Protein was visualized with Coomassie brilliant blue. (B) Wild-type MscL cross-linked with 0–1 mM DSS. (C) Wild-type and G22N MscL cross-linked with 0.1 mM DSS. (D) G22C MscL cross-linked with 0.1 mM DSS in the presence or absence of MTSET. (B–D) Western blots using anti-MscL antibody.
Purified MscL was cross-linked with disuccinimidyl suberate (DSS) and subjected to SDS-gel electrophoresis to characterize the assembly of MscL subunits. In the absence of DSS, MscL monomers were primarily detected. In contrast, the addition of DSS generated four additional bands representing dimeric, trimeric, tetrameric, and pentameric forms of the protein (Fig. 1B). Consistent with these results, cross-linking of MscL from intact Escherichia coli cells generates a similar electrophoretic ladder (3, 23). As increasing concentrations of DSS were added, the amount of pentamer increased, whereas amounts of the monomer decreased, indicating that the pentamer is the most abundant form of translated MscL.
Translation of G22N MscL yielded quantities of protein comparable with that seen with wild-type MscL. The migration of monomers was indistinguishable between wild-type and G22N MscL. However, the bands representing the four multimers migrated more slowly than those of wild type (Fig. 1C). The increase in molecular weight resulting from the Gly-to-Asn mutation could not account for this large decrease in electrophoretic mobility. For example, in the case of the dimer, the increase in weight was calculated to be 114 Da, which is far less than the ≈2.1-kDa increase in the apparent molecular mass determined by electrophoretic migration. A similar decrease in the migration rate was observed on the incubation of G22C MscL with MTSET (Fig. 1D). Again, the slow migration could not be completely attributed to the attachment of the MTSET, which increases the molecular mass by only 120 Da/subunit. Slowed electrophoretic migration in the absence of any significant increase in the molecular mass would suggest that MscL retains its secondary structure as a result of the cross-link. Thus, G22N MscL and MTSET-bound G22C MscL may be present in a more expanded form than wild-type MscL. Similar changes in electrophoretic migration caused by alterations in protein conformation have been reported for Ca2+-binding proteins such as calmodulin (24).
MscL Channel Activity and Ca2+ Permeability.
To test whether the translated MscL is functional, MscL was reconstituted into liposomes and analyzed by patch-clamp. Application of a negative pressure to an excised membrane through a patch pipette elicited channel opening with an amplitude of ≈90 pA (Fig. 2A; +20 mV pipette voltage). The pressure needed to activate wild-type MscL ranged from 40 to 80 mmHg. Application of pressure to liposomal membranes reconstituted with translated green fluorescent protein did not elicit any channel activity in the 10 membrane patches examined. Therefore, the channel activity observed with MscL cannot be attributed to any contaminating components of the translation system.
Fig. 2.
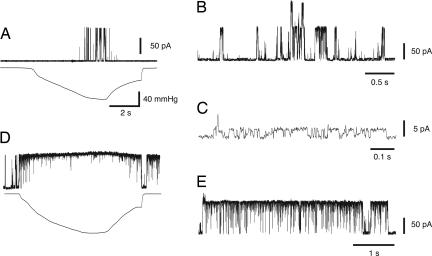
Electrophysiology of wild-type and G22N MscL. (A) Channel current (upper trace) of wild-type MscL in response to negative pressure (lower trace) applied to the membrane patch. (B) Spontaneous opening of G22N in the absence of pressure. (C) An expanded view of a portion of the trace in B, showing the open substate. (D) Response of G22N MscL to the application of negative pressure. (E) G22N MscL activity after pressure removal. The pipette potential was 20 mV positive to the bath in all experiments.
When G22N MscL was examined, channel opening was observed just after the formation of the patch, even without pressure (Fig. 2B). G22N MscL adopted an open substate most of the time (Fig. 2C), whereas the closed and fully open state occurred less frequently. G22N MscL completely entered in the fully open state at pressures <5 mmHg (Fig. 2D). Interestingly, G22N MscL remained predominantly in the fully open state after release of pressure (Fig. 2 D and E). This fully open state shifted to an open substate gradually 2–10 min after removal of pressure. These observations indicate that G22N MscL is most stable in the open substate when the membrane tension is absent, and the transition to the fully open state requires little tension. These findings are consistent with kinetic analyses showing that the transition from the smallest open substate to the fully open state is nearly tension-independent in wild-type MscL (17).
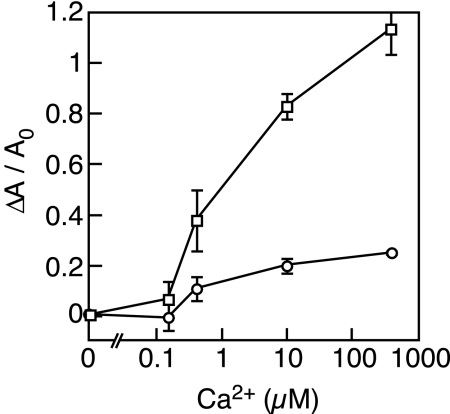
To ascertain whether the opening of G22N MscL occurs spontaneously in the absence of a patch pipette, the influx of Ca2+ into the liposome reconstituted with MscL was measured by using a fluorescent Ca2+ indicator. The addition of Ca2+ solution to G22N MscL-containing liposomes yielded a dose-dependent increase in the fluorescent signal (Fig. 3). On the other hand, the addition of increasing concentrations of Ca2+ to wild-type MscL-containing liposomes did not appreciably alter the fluorescent signal. Therefore, only G22N MscL-containing liposomes are freely permeable to Ca2+, consistent with the idea that G22N MscL is open in the absence of membrane stretch.
Fig. 3.
Ca2+ influx into liposomes containing wild-type MscL (circles) or G22N MscL (squares). Varying concentrations of Ca2+ were added to suspensions of liposomes that were loaded with the Ca2+ indicator, Fluo-4 dextran. Each point represents the mean and the standard deviation (n = 3) of the increase in the fluorescence (ΔA) normalized to the initial intensity (A0).
Electron Microscopic Analysis of MscL.
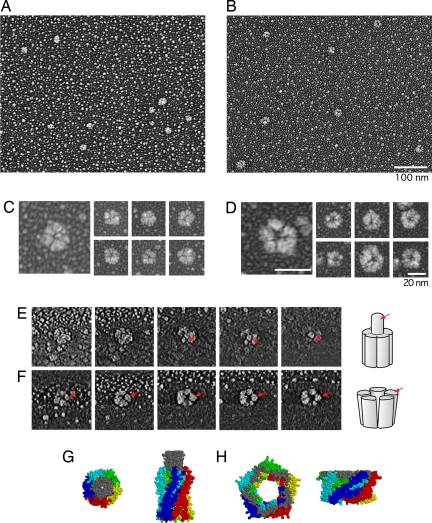
Purified MscL was examined by low-angle rotary shadowing. This method was selected because the specimen has a higher contrast than in negative staining and is able to withstand the high-dose electron beams necessary for three-dimensional tomography. As shown in Fig. 4 A and B, both wild-type and G22N MscL molecules appeared as round particles with a ≈20 nm diameter. Based on the area of the imaged particle of wild-type MscL (285 ± 81 nm2, n = 120), the diameter was estimated to be 18.9 ± 2.8 nm, assuming the shape to be a perfect circle. The large variation in size is probably because of differences in the thickness of the platinum coating and the tilt of the specimen on the mica surface. We examined the particles from four independently obtained translation products and obtained consistent results. Under higher magnification, wild-type MscL appeared to be pentagonal in shape and was frequently divided by fine clefts (Fig. 4C). These EM observations are consistent with the pentameric structure of MscL (3, 23). Generation of the three-dimensional structure through tomography and analysis of cross-sections revealed the presence of a protrusion on the upper part of the particle (Fig. 4E, arrow). The protrusion was directed almost perpendicular to the mica surface.
Fig. 4.
Electron microscopic analysis of MscL by using low-angle rotary shadowing. (A–D) Wild-type MscL (A and C) and G22N MscL (B and D) at low magnification (A and B) and high magnification (C and D). (E and F) Cross-sections of wild-type MscL (E) and G22N MscL (F) reconstructed by three-dimensional tomography. The leftmost images show the section closest to the bottom (mica surface). Arrows in E indicate the protrusion in wild-type MscL, and those in F indicate a G22N MscL subparticle. Schematic drawings are shown on the right. (G and H) Structure of MscL in the open (G) and the closed (H) state according to modeling by Sukharev et al. (7). Cytoplasmic (Left) and side (Right) views are shown. The carboxyl-terminal domain of the five subunits is displayed in gray, whereas the remainder of each subunit is shown in a different color.
In contrast to wild-type MscL, G22N MscL possessed a pore at its center (Fig. 4D). Moreover, the pentagonal shape was more apparent in G22N MscL than in wild-type MscL. The diameter of G22N MscL was estimated to be 19.5 ± 2.2 nm (based on an area of 302 ± 67 nm2). Subtraction of the thickness of the platinum coating (≈6 nm) from the diameters of wild-type and G22N MscL yields diameters of ≈7 – 8 nm. These values are in good agreement with the diameter of the TbMscL crystal structure (5 nm) (3) and with electron paramagnetic resonance data suggesting that the diameter of MscL increases to 7 nm on opening (5). Tomography confirmed the presence of a pore at the center of G22N MscL (Fig. 4F). Clefts radiating from the pore were also apparent. Although most of the G22N MscL (95%) had both pore and clefts, the protrusion seen in wild-type MscL was missing.
The electrophysiological and Ca2+ permeability analyses indicate that the opening of G22N MscL does not require mechanical stretch. G22N MscL has also been shown to open spontaneously when examined with spheroplasts (13, 18). Therefore, it is likely that the pore observed in the electron micrograph of G22N MscL represents the ion pathway of the open channel. The decreased electrophoretic migration rate of G22N MscL also supports the idea that this protein is in an expanded and open form. In contrast, an obvious pore was not detected in wild-type MscL. This observation is in line with the electrophysiological and Ca2+ leakage data, which showed that wild-type MscL is closed in the absence of stretch. The failure to observe any pore is probably not because of the tilt of the molecule because the pore could not be detected with tomography and because the projection, which we suggest to be the carboxyl-terminal domain (see below), is almost perpendicular to the mica plate. Therefore, it is likely that the particles of wild-type and G22N MscL correspond to the closed and open structure of MscL, respectively.
The clefts radiating from the pore of G22N MscL separate the protein into rod-shaped subparticles (Fig. 4F). Judging from subparticles that are traceable from the bottom to the top of the channel, the subparticles did not display any apparent tilting (Fig. 4F, arrow). This upright structure contrasts with the tilted transmembrane helices in the modeled MscL structure (7, 8) (Fig. 4H). Several experimental studies have provided evidence in support of tilting and sliding of the transmembrane helices on gating (4, 5). A possible explanation for this inconsistency is that the upright conformation is stabilized in the absence of membrane because the mismatch between the thickness of the transmembrane domain and the membrane does not occur. According to this hypothesis, the tilting and the sliding of the transmembrane helices do not result from the intrinsic nature of the protein. Instead, the tilting and the sliding of the transmembrane helices occur only when the membrane restricts the height of the transmembrane domain. An alternative possibility is that the transmembrane helices move in parallel during the transition from the closed state to the open substate, and they tilt during the transition toward the fully open state.
Effect of Deletion of the Carboxyl-Terminal Residues.
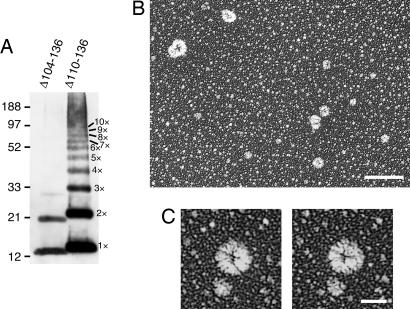
The protrusion observed at one end of the wild-type MscL molecule likely represents the bundle of the carboxyl-terminal helices, based on the crystal structure of M. tuberculosis MscL and the modeled structure of E. coli MscL (3, 7) (Fig. 4G). To test this idea, the mutants that lack the carboxyl-terminal residues were constructed. The residues 104–136 and 110–136 were deleted because the channel activity of these mutants had already been examined on spheroplasts (19, 20). When Δ104–136 MscL was expressed with a cell-free system and cross-linked chemically by DSS, monomer, dimer, and a faint amount of trimer were detected by electrophoresis (Fig. 5A). The absence of pentamer is consistent with the report that Δ104–136 MscL does not show channel activity on spheroplasts (19). On the other hand, Δ110–136 MscL, which shows channel activity on spheroplasts (19, 20), formed every multimer from dimer to octamer (Fig. 5A). A smear band was also present in the range of 100–200 kDa. The rotary-shadowed preparation of Δ110–136 MscL showed particles with various sizes (Fig. 5B), which probably represent diverse multimerization states. Protrusion was not present on the particles when examined with stereo pair images (Fig. 5C). This observation is consistent with the idea that the protrusion observed in wild-type MscL is composed of the carboxyl-terminal bundle. In addition, the low (Δ110–136) and high (Δ104–136) degrees of multimerization indicate that the carboxyl terminus determines the proper number of multimerization states in MscL.
Fig. 5.
Effect of deletion of the carboxyl-terminal residues. (A) Electrophoresis of MscL that has a deletion of residues 104–136 (Δ104–136) and 110–136 (Δ110–136). The products of cell-free translation were cross-linked with 0.1% DSS and detected with Western blot by using anti-His antibody. (B and C) Electron microscopy of Δ110–136 MscL by using low-angle rotary shadowing. Shown are the particles observed at low magnification (B) and those shown as stereo pair images (C). (Scale bars: B, 100 nm; C, 20 nm.)
The disappearance of the protrusion in G22N MscL suggests that the bundle is disassembled in the open state. Although the disassembly may represent a physiological conformation, it is also possible that the bundle is dissociated during sample preparation. Even if this were the case, the absence of this protrusion only in G22N MscL suggests that the interaction among helices is weaker in the open state than in the closed state. Alternatively, the disassembly may be because of G22N mutation itself and unrelated to the channel gating. However, this idea is not very likely if we take into account the long linker between the transmembrane domain and the carboxyl-terminal helix.
The conjecture that the carboxyl-terminal helices form a bundle in the closed state and disassemble in the open state is in accord with the modeled MscL structure (Fig. 4 G and H) (7). Bundle formation in the closed state has been confirmed by cross-linking the helices (20). Anishkin et al. (20) reported that MscL retains the ability to open when the α helices are cross-linked. Note that this result does not exclude the possibility that helices disassemble under normal conditions. They also showed, by using molecular dynamics simulation, that the bundle does not disassemble when the linker to the transmembrane domain is stretched. Nevertheless, the interhelical interactions may be essentially sensitive to cellular conditions and will require further study through FRET or spin labeling.
In conclusion, we have provided the direct view of the closed structure of wild-type MscL and the open structure of G22N MscL. Our data reveal that the closed structure is characterized by a protrusion, whereas the open structure is characterized by a pore. Because MscL is one of the smallest channel proteins, future studies of other larger membrane proteins are expected to provide perspectives on conformational changes related to protein function.
Materials and Methods
Cell-Free Expression and Purification of MscL.
The linear template for cell-free expression was constructed by two-step PCR according to the manufacturer's instructions (Roche Diagnostics). To construct the PCR template, mscL was cloned into pB10c vector, which has multiple cloning sites in the direction opposite to pB10b (15). G22N mutation was introduced by a DpnI-based method (Quick Mutagenesis kit, Stratagene). The second round of PCR was performed to add regulatory regions to the 5′ end and to add linker (PGGGS) and 6×His tag to the 3′ end by overlap extension. The product was verified by bidirectional sequencing. MscL was expressed by using E. coli lysate in a rapid translation system (Roche Diagnostics). Template DNA and 6 mM Brij-58 was added to the reaction mixture and incubated for 6 h at 30° C. Insoluble product was removed by centrifugation. For NiNTA column purification, samples were supplemented with buffer containing a 50 mM final concentration of sodium phosphate, pH 8.0; 300 mM NaCl; 1% octyl glucoside; and 35 mM imidazole. The preparations were then applied to the NiNTA column (Qiagen) and washed three times with the same buffer. MscL was then eluted with buffer containing 250 mM imidazole. The eluate was desalted on a PD-10 column (Amersham Biosciences) by using buffer containing 20 mM potassium phosphate, pH 7.2, and 1% octyl glucoside and then concentrated with Amicon Ultra (Millipore).
Patch-Clamp Measurements of Liposome-Reconstituted MscL.
Purified MscL was reconstituted into liposome made from azolectin as described previously (25). Liposomes were collected by ultracentrifugation at 160,000 × g for 1 h and suspended in dehydration buffer [10 mM 4-morpholinopropanesulfonic acid (Mops), pH 7.2/5% ethyleneglycol]. The suspension was spotted onto a slide glass and dehydrated for 2 h at 4°C. The spot was rehydrated overnight at 4°C in rehydration buffer (250 mM NaCl/0.1 mM EDTA/5 mM Hepes, pH 7.2). Patch-clamp recording was performed on a blister membrane from rehydrated liposome in experimental solution (200 mM KCl/90 mM MgCl2/10 mM CaCl2/5 mM Hepes, pH 6.0) (25). The pipette contained the same solution and was held at 20 mV positive to the bath. Current was amplified by using an Axopatch 200B patch clamp amplifier (Axon Instruments).
MscL Cross-Linking Analysis.
To assess multimerization of MscL, purified MscL was cross-linked with DSS (Pierce) as detailed previously (25). In these experiments, MscL loaded onto PD-10 columns was eluted with reaction buffer (100 mM NaCl/30 mM sodium phosphate, pH 7.5/1% octyl glucoside). The preparation was incubated with DSS for 1 h at room temperature. The reaction was terminated by adding Tris to a final concentration of 25 mM. Electrophoresis was carried out by using 12% [bis(2-hydroxyethyl)imino]tris(hydroxymethyl)methane PAGE gel (Invitrogen). For Western blot analysis, protein was transferred onto PVDF membranes and probed with anti-MscL antibody (19) and donkey anti-rabbit Fab (Amersham Biosciences). Alternatively, mouse anti-His tag antibody and anti-mouse-IgG antibody (Amersham Biosciences) were used when the carboxyl-terminal residues were removed. The secondary antibody was detected by chemiluminescence (ECL Plus, Amersham Biosciences).
Measurement of Ca2+ Influx.
Ca2+ influx was measured by loading liposomes with Fluo-4 dextran (molecular weight of 10,000). The solution inside and outside liposome contained 100 mM KCl, 1 mM EGTA, and 30 mM Mops, pH 7.2. Ca2+ solution was added to the liposome suspension, and the fluorescence from Fluo-4 dextran was measured with a spectrofluorophotometer (excitation = 488 nm, emission = 530 nm; RF-5300PC, Shimazdu).
Electron Microscopy.
Single molecules of MscL were observed with electron microscopy by using methods described previously (26). Preparations of translated MscL were supplemented with glycerol (50%, final) and sprayed onto freshly cleaved mica. The mica was dried under vacuum for 10 min in a freeze-etching device FR 9000 (Hitachi). The sample was rotary-shadowed with platinum at an angle of 2.5° to the mica surface and then coated with carbon. The film of platinum and carbon was removed from the mica and placed on copper grids for observation by a transmission electron microscopy (1200 EX,JEOL). For tomography, the replica was tilted at angles between −70° and 70° at 1° steps in the transmission electron microscope (Tecnai G2 Polara, FEI). The specimen was cooled to −192°C and observed at an acceleration voltage of 300 kV. Images (2,048 × 2,048 pixels) were captured by using a CCD camera. Data were analyzed as described elsewhere (26). For obtaining stereo pair images, the grid was tilted at ±10° in a transmission microscope (H7600, Hitachi).
ACKNOWLEDGMENTS.
We thank Dr. T. Nomura for help in molecular biology and Dr. M. Murase for help in electrophysiology. This work was supported by Ministry of Education, Culture, Sports, Science, and Technology (Japan) Grants-in-Aid 16GS0308 (to M.S.) and 16370034 (to K.Y.) as well as a grant from the Japan Science and Technology Agency (to K.Y. and M.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Sukharev SI, Blount P, Martinac B, Blattner FR, Kung C. Nature. 1994;368:265–268. doi: 10.1038/368265a0. [DOI] [PubMed] [Google Scholar]
- 2.Cruickshank C, Minchin R, Le Dain A, Martinac B. Biophys J. 1997;73:1925–1931. doi: 10.1016/S0006-3495(97)78223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang G, Spencer RH, Lee AT, Barclay MT, Rees DC. Science. 1998;282:2220–2226. doi: 10.1126/science.282.5397.2220. [DOI] [PubMed] [Google Scholar]
- 4.Betanzos M, Chiang C-S, Guy R, Sukharev S. Nat Struc Biol. 2002;9:704–710. doi: 10.1038/nsb828. [DOI] [PubMed] [Google Scholar]
- 5.Perozo E, Cortes DM, Sompornpisut P, Kloda A, Martinac B. Nature. 2002;418:942–948. doi: 10.1038/nature00992. [DOI] [PubMed] [Google Scholar]
- 6.Perozo E, Kloda A, Cortes DM, Martinac B. Nat Struc Biol. 2002;9:696–703. doi: 10.1038/nsb827. [DOI] [PubMed] [Google Scholar]
- 7.Sukharev S, Durell SR, Guy HR. Biophys J. 2001;81:917–936. doi: 10.1016/S0006-3495(01)75751-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sukharev SI, Betanzos M, Chiang CS, Guy HR. Nature. 2001;409:720–724. doi: 10.1038/35055559. [DOI] [PubMed] [Google Scholar]
- 9.Kong Y, Shen Y, Warth TE, Ma J. Proc Natl Acad Sci USA. 2002;99:5999–6004. doi: 10.1073/pnas.092051099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gullingsrud J, Schulten K. Biophys J. 2003;85:2087–2099. doi: 10.1016/s0006-3495(03)74637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colombo G, Marrink SJ, Mark AE. Biophys J. 2003;84:2331–2337. doi: 10.1016/S0006-3495(03)75038-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ornatska M, Jones SE, Naik RR, Stone M, Tsukruk VV. J Am Chem Soc. 2003;125:12722–12723. doi: 10.1021/ja037686q. [DOI] [PubMed] [Google Scholar]
- 13.Yoshimura K, Batiza A, Schroeder M, Blount P, Kung C. Biophys J. 1999;77:1960–1972. doi: 10.1016/S0006-3495(99)77037-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshimura K, Batiza A, Kung C. Biophys J. 2001;80:2198–2206. doi: 10.1016/S0006-3495(01)76192-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ou X, Blount P, Hoffman R, Kung C. Proc Natl Acad Sci USA. 1998;95:11471–11475. doi: 10.1073/pnas.95.19.11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blount P, Moe P. Trends Microbiol. 1999;7:420–424. doi: 10.1016/s0966-842x(99)01594-2. [DOI] [PubMed] [Google Scholar]
- 17.Sukharev SI, Sigurdson WJ, Kung C, Sachs F. J Gen Physiol. 1999;113:525–540. doi: 10.1085/jgp.113.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anishkin A, Chiang C-S, Sukharev S. J Gen Physiol. 2005;125:155–170. doi: 10.1085/jgp.200409118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blount P, Sukharev SI, Schroeder MJ, Nagle SK, Kung C. Proc Natl Acad Sci USA. 1996;93:11652–11657. doi: 10.1073/pnas.93.21.11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anishkin A, Gendel V, Sharifi N, Chiang C-S, Shirinian L, Guy R, Sukharev S. J Gen Physiol. 2003;121:227–244. doi: 10.1085/jgp.20028768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elmore DE, Dougherty DA. Biophys J. 2001;81:1345–1359. doi: 10.1016/S0006-3495(01)75791-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berrier C, Park KH, Abes S, Bibonne A, Betton JM, Ghazi A. Biochemistry. 2004;43:12585–12591. doi: 10.1021/bi049049y. [DOI] [PubMed] [Google Scholar]
- 23.Sukharev SI, Schroeder MJ, McCaslin DR. J Memb Biol. 1999;171:183–193. doi: 10.1007/s002329900570. [DOI] [PubMed] [Google Scholar]
- 24.Cartaud A, Ozon R, Walsh MP, Haiech J, Demaille JG. J Biol Chem. 1980;255:9404–9408. [PubMed] [Google Scholar]
- 25.Blount P, Sukharev S, Moe P, Martinac B, Kung C. Methods Enzymol. 1999;294:458–482. doi: 10.1016/s0076-6879(99)94027-2. [DOI] [PubMed] [Google Scholar]
- 26.Matsui T, Hogetsu K, Usukura J, Sato T, Kumasaka T, Akao Y, Tanaka N. Genes Cells. 2006;11:439–452. doi: 10.1111/j.1365-2443.2006.00951.x. [DOI] [PubMed] [Google Scholar]