Abstract
Activation-induced cytosine deaminase (AID) is essential for both somatic hypermutation (SHM) and class switch recombination (CSR), two processes involved in antibody diversification. Previously, various groups showed both in vitro and in vivo that AID initiates SHM and CSR by deaminating cytosines in DNA in a transcription-dependent manner. Although in vivo both DNA strands are equally targeted by AID, many in vitro and bacterial experiments found that AID almost exclusively targets the nontemplate strand of a transcribed substrate. Here, we report the detection of antisense transcripts in assembled Ig heavy chain (IgH) variable region exons and their immediate downstream region, as well as in switch regions, sequences that, respectively, are targets for SHM and CSR in vivo. In contrast, we did not detect antisense transcripts from the Cμ constant region exons, which lie between the IgH variable region exons and downstream S regions and which are not normally an AID target. Expression of the antisense variable region/flanking region and the S-region transcripts were found in all lymphocytes that transcribe these sequences in the sense direction. Steady-state levels of antisense transcripts appeared very low, and start sites potentially appeared heterogeneous. We discuss the potential implications of antisense IgH locus transcription for AID targeting or other processes.
Keywords: class switch recombination, somatic hypermutation, activation-induced cytosine deaminase, bidirectional transcription
B cells generate a highly diverse repertoire of immunoglobulins (Igs) by assembling a variable region exon from an array of variable (V), diversity (D), and joining (J) gene segments upstream of the Ig constant region exons. This process is called V(D)J recombination and takes place in the early stages of B cell development (1). Upon antigen encounter, Ig genes can undergo further modifications in peripheral lymphocytes. Somatic hypermutation (SHM) introduces point mutations at a very high rate in Ig variable region exons, allowing for the generation of antibodies with increased affinity for their antigen (2). Class switch recombination (CSR) promotes replacement of the μ-constant region (Cμ) exons with one of a set of downstream constant region exons (Cγ3, Cγ1, Cγ2b, Cγ2a, Cε, and Cα) that encode different effector functions (3). This replacement is achieved by breakage and joining of specialized switch (S) regions that precede every constant region that undergoes CSR. Breaks in the donor Sμ region are joined to breaks in a downstream acceptor S region via classical or alternative end joining, resulting in juxtaposition of the downstream CH exons to the expressed VHDJH exon (4).
The main function of S regions appears to be the generation of double-strand breaks (DSBs) by serving as a target for activation-induced cytosine deaminase (AID) (5–8). AID is essential for both SHM and CSR (9) and is believed to function by deaminating cytosine (C) to uracil (U) in DNA (10). The mismatched uracils resulting from AID activity are processed by base excision repair (BER) and/or mismatch repair (MMR) pathways to ultimately generate DSBs that are the intermediates in CSR (11). SHM also is initiated by cytosine deamination by AID, leading to transition mutations upon replication if the uracils are unrepaired. If the uracil gets excised, however, in the course of BER, uracil-DNA glycosylase generates an abasic site that can give rise to transition and transversion mutations upon replication. Moreover, the U·G mismatches can get repaired by error-prone MMR that predominantly mutates A·T base pairs close to the U·G mismatch (2, 12).
Targeting experiments of the murine IgH locus showed that the active transcription of the Ig variable region exon and S regions is necessary for both SHM (13) and CSR (14, 15), respectively. In addition, various studies showed that AID is an ssDNA-specific cytidine deaminase that accesses dsDNA in a transcription-dependent manner (16–20). Transcribed mammalian S regions form R loops, a DNA–RNA hybrid structure that typically forms when S regions are transcribed in the sense direction (21, 22), allowing AID to act mainly on the displaced nontemplate strand in vitro (17, 19, 20, 23). Transcribed SHM targets do not form R loops. However, in vitro studies suggest that AID may obtain access to transcribed variable region gene sequences in the context of a complex with the ssDNA-binding protein replication protein A (RPA) (24). Again, however, in such studies, AID only targets the nontemplate strand. Likewise, AID expressed in bacteria can generate SHM, but again primarily on the nontemplate DNA strand (20). Various findings also suggest that AID may access S regions via a non-R-loop mechanism that may relate to the mechanism by which it accesses variable regions during SHM (3, 25). Despite findings that AID targets mainly the nontemplate strand in most in vitro and bacterial experiments, both the template and nontemplate strands are targeted equally by AID during normal SHM and CSR in vivo (11, 26, 27).
Despite intense ongoing research, it is still unknown how the highly mutagenic activity of AID gets targeted mainly to Ig loci and not to other highly transcribed genes. Moreover, it is not known how AID effects are targeted to the Ig variable region exon and switch regions while the intervening IgH intronic enhancer (Eμ) and CH exons are spared. Furthermore, a full explanation is still missing as to why in vitro and in bacteria AID gets almost exclusively targeted to the nontemplate strand (17, 19, 20, 23), whereas in vivo AID appears to act equally on the template and the nontemplate strand (11, 26, 27). As for the latter question, several nonmutually exclusive possibilities have been suggested. In the in vitro experiments, there could be additional factor(s) missing that would allow AID to target both the template and nontemplate strands in vivo. In addition, the different RNA polymerases (viral T7 and T3 polymerases in the in vitro experiments vs. RNA polymerase II in cells undergoing SHM in vivo) might result in different access of AID to the sense and antisense strands. In the latter context, it was reported that AID can target both DNA strands in the context of transcription with Escherichia coli RNA polymerase (28). Another possibility is that differences in chromatin structure or supercoiled DNA in vivo could allow AID to access both strands (29). Finally, it is conceivable that antisense transcription through variable regions and S regions could target AID to both strands (30).
To further evaluate the potential role of antisense transcription as a mechanism of AID targeting, we have assayed, for the generation of antisense transcripts of IgH variable region exons, the Cμ constant region exons and S regions in various types of B-lineage cells.
Results
Sense and Antisense Transcription Within IgH Variable Region Exons.
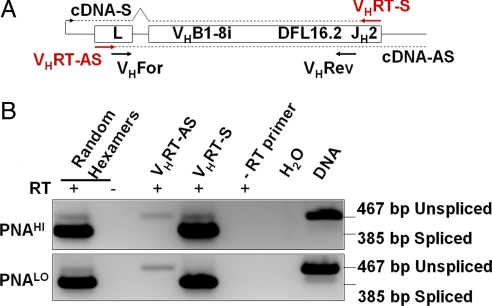
We first assayed for the existence of antisense transcripts within the rearranged VHDJH exons, a known target region for SHM. To this end, we used VB1–8 VHDJH knockin mice (31), which carry an assembled IgH variable region exon in their germ line. These mice offer the advantage for these studies that all B cells produce transcripts of an identical sequence, which facilitates transcript detection by RT-PCR. Naïve (B220+, PNALO) and activated (B220+, PNAHI) B cells were sorted from Peyer's patches of VB1–8 knockin mice, and RNA was prepared. Reverse transcription was conducted with random hexamers or with strand-specific primers to help distinguish between sense and antisense transcripts. PCR amplification of the cDNAs was designed in a way that the forward primer anneals in the leader exon, whereas the reverse primer anneals at JH2 within the knocked-in VHDJH exon (Fig. 1A). This design allows spliced sense transcipts and antisense transcripts, which would be unspliced, to be distinguished by size differences of the PCR products. Moreover, this design ensures that we only score transcripts from the assembled VB1–8 knock in and not potential sense and antisense transcripts from upstream unrearranged VH gene segments (32) that are similar in sequence. In these assays, the sense primer detected a major 385-bp band corresponding to processed sense transcripts and a very low-level 467-bp band that might reflect unprocessed precursors (Fig. 1B, VHRT-S lane). In contrast, the antisense primer detected only a band the size of unprocessed transcripts and no processed size transcripts (Fig. 1B, VHRT-AS lane) from VB1–8 in RNA from both naïve (Fig. 1B Lower) and activated (Fig. 1B Upper) B cells. These data strongly suggest that the knockin VB1–8 VHDJH rearrangement is transcribed in the antisense direction.
Fig. 1.
Bidirectional transcription within a rearranged VH gene. (A) RT-PCR design for the heavy chain knockin gene (31). Sense (VHRT-S)- and antisense (VHRT-AS)-specific reverse transcription primers are depicted in red, and PCR primers (VHFor and VHRev) are depicted in black and were designed within the leader (L) and JH2, respectively. Sense (cDNA-S)- and antisense (cDNA-AS)-specific cDNA is shown as dashed lines. (B) RT-PCR of ex vivo-sorted naïve (PNALO) (Lower) and activated (PNAHI) (Upper) B cells from Peyer's patches. Above the lanes, the used reverse transcription primers are indicated with (+) or without (−) reverse transcriptase (RT).
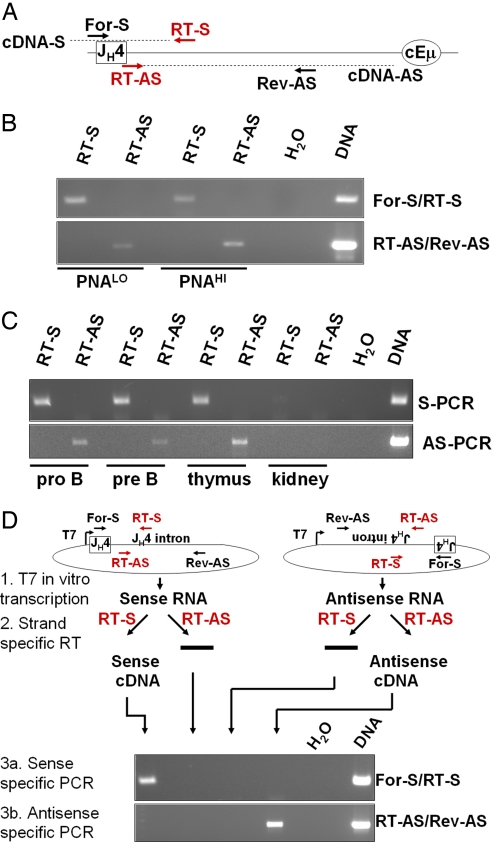
To unequivocally confirm antisense transcription in the region of the V(D)J exon and to extend these assays to wild-type mice, we designed a strand-specific RT-PCR assay to distinguish, in a very clean way, between sense and antisense transcripts through the SHM target region just downstream of the JH segments. In this assay, all primers annealed within or downstream of JH4, a SHM target region that is identical in every B cell of a population carrying nonclonally rearranged VHDJH genes. In the reverse transcription reaction, strand-specific reverse transcription primers (Fig. 2A, RT-S and RT-AS) generate sense (cDNA-S)- and antisense (cDNA-AS)-specific cDNA, respectively. Both generated cDNAs are then used as template for two different PCRs: one sense-specific (using primers For-S/RT-S) and one antisense-specific PCR (primers RT-AS/Rev-AS). The two PCRs amplify overlapping products. As a consequence, the sense-specific PCR (Fig. 2B Upper) will only amplify a signal from sense-specific cDNA (RT-S lanes), whereas antisense-specific cDNA (RT-AS lanes) will not result in a product because the RT reaction was primed in between this set of PCR primers. Similarly, the antisense-specific PCR (Fig. 2B Lower) will generate a signal from antisense-specific cDNA (RT-AS lanes) but not from sense-specific cDNA (RT-S lanes). This design allows for clear differentiation between sense and antisense transcripts, and it controls for DNA contamination and nonspecific priming of the reverse transcription reaction, both of which would result in a signal in reactions that are supposed to stay blank (Fig. 2B Upper, RT-AS lanes; Lower, RT-S lanes). The specificity of this assay was unequivocally confirmed by applying the described RT-PCR assay on in vitro-transcribed RNA from a linearized plasmid that contains the JH4 intron in either sense or antisense orientation. Even with the large amounts of RNA used in these test reactions, the sense and antisense transcripts were detected with complete specificity (Fig. 2D).
Fig. 2.
Bidirectional transcription within the JH–CH intron. (A) RT-PCR design downstream of JH4. Sense (RT-S)- and antisense (RT-AS)-specific reverse transcription primers are depicted in red; all primers are used for either sense (For-S/RT-S)- or antisense (RT-AS/Rev-AS)-specific PCRs. JH4 and the Eμ enhancer core are depicted as a rectangle and an oval, respectively. Sense (cDNA-S)- and antisense (cDNA-AS)-specific cDNA is shown as dashed lines. (B) RT-PCR of ex vivo-sorted naïve (PNALO) and activated (PNAHI) B cells. Above the lanes, the used reverse transcription primers are indicated with (+) or without (−) reverse transcriptase. (Upper) Sense-specific PCR. (Lower) Antisense-specific PCR. (C) Expression pattern of bidirectional transcripts in the JH–CH intron. Experimental design is as described in A. RT-PCR of ex vivo-sorted pro-B and pre-B cells, thymus, and kidney. (Upper) Sense-specific PCR. (Lower) Antisense-specific PCR. (D) In vitro-specificity control of strand-specific RT-PCR assay. JH4 and the JH4 intron were cloned into pGEM T-easy (Promega) in both orientations. In vitro transcription with T7 RNA polymerase results in either sense or antisense transcripts (1). Strand-specific reverse transcription reactions (primers depicted in red) generate either sense- or antisense-specific cDNA (2), and sense (3a)-, and antisense (3b)-specific PCRs produce only a product when the according cDNA is used as a template.
We immunized wild-type 129sv mice with sheep RBCs (SRBCs) and sorted naïve (B220+, PNALO) and activated (B220+, PNAHI) B cells from spleens. Applying the strand-specific RT-PCR assay described above, we detected sense and antisense transcripts in both naïve and activated B cells (Fig. 2B), thus confirming the results obtained from the VB1–8 knockin mice (Fig. 1B). Transcripts in the sense orientation were verified by signals from the sense-specific PCR (Fig. 2B Upper) exclusively when RNA was primed with the sense-specific reverse transcription primer (Fig. 2B, RT-S lanes). Antisense transcripts were substantiated through products from the antisense-specific PCR (Fig. 2B Lower) only when the reverse transcription reaction was primed with antisense-specific primers (Fig. 2B, RT-AS lanes). Control lanes (Fig. 2B Upper, RT-AS lanes; Lower, RT-S lanes) stayed blank as expected. When we assayed transcripts within the VDJH segment of VB1–8 knockin mice (Fig. 1), we observed a rather faint antisense-specific band (467 bp, VHRT-AS lane), whereas the sense-specific reverse transcription reaction resulted in a major band representing the spliced mRNA (385 bp) and a faint band representing unspliced pre-mRNA (467 bp, VHRT-S lane). In contrast, when we investigate transcripts within the JH4 intron in wild-type mice (Fig. 2 B and C), we find sense- and antisense-specific bands of comparable intensities. One obvious explanation for this observed difference in sense/antisense transcript ratios is that the reverse transcription primers used for the assay described in Fig. 2 anneal within the JH4 intron (see Fig. 2A), which gets spliced out in the mature mRNA. Therefore, the sense-specific PCR can only detect a signal from the minor unspliced pre-mRNA fraction.
Antisense Transcripts Have Heterogeneous Start Sites.
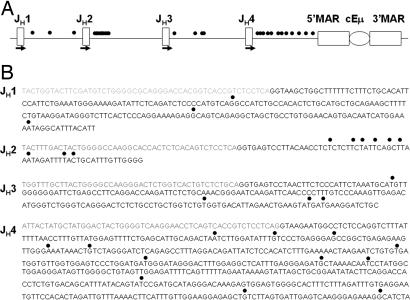
To identify a potential promoter of the antisense transcript in the JH locus, we carried out 5′RACE on total RNA of activated B cells (B220+, PNAHI) from spleens of SRBCs immunized wild-type 129sv mice. Several 5′RACE experiments were performed; gene-specific primers were designed within JH1, JH2, JH3, and JH4 (Fig. 3A). The 5′RACE products were cloned and sequenced. Potential heterogeneous start sites were identified downstream of every JH element (Fig. 3 A and B). The 5′RACE experiments were performed twice in the regions downstream of JH1, JH2, and JH3, and the region downstream of JH4 was tested five times. All experiments resulted in independent heterogeneous start sites, indicating the lack of a strong single promoter. To further elucidate these possibilities, we performed an RNase protection assay to confirm the 5′RACE results. However, we were not able to detect a signal from the antisense transcript. In contrast, sense transcripts could be readily detected (data not shown). These results argue that cellular levels of antisense transcripts are very low because of either low transcription rates or high instability of the antisense transcript. After identifying potential transcriptional start sites downstream of JH4, we tested the region between JH4 and Eμ for potential promoter activity in the antisense direction in a dual luciferase assay. The luciferase assay was performed in Ed20 pro-B and NS1 cells (TIB-18; American Type Culture Collection), which both express sense and antisense transcripts (data not shown) from the region downstream of JH4. Constructs containing different stretches of the JH4 intron and the Eμ enhancer were tested in the sense and antisense orientations [supporting information (SI) Fig. 5A]. Although we detected promoter activity in the sense direction, correlating with Iμ start sites (33), we did not detect luciferase signals above background in the antisense orientation (SI Fig. 5B), which might suggest that open chromatin can be sufficient to promote low-level antisense transcription from random start sites or that true start sites are located elsewhere.
Fig. 3.
Start sites of antisense transcripts within the JH region. Potential start sites were identified downstream of all JH elements. (A) Dots indicate antisense transcript start sites, and arrows indicate primers used for 5′RACE experiments. cEμ, core IgH intronic enhancer; MAR, matrix attachment region. (B) Detailed listing of start sites downstream of all JH elements.
Correlation of Sense and Antisense Transcript Expression Patterns.
Detection of sense and antisense transcripts in a rearranged VH gene and in the JH locus might provide a mechanism for how AID can target both strands during SHM. However, we detected sense and antisense transcripts in both naïve and activated B cells, although SHM does not occur in naïve B cells. Thus, the antisense transcripts are not expressed exclusively in cells undergoing SHM (Figs. 1B and 2B). To assess the expression pattern of the antisense variable region transcripts in cells other than naïve and activated B cells, we prepared RNA from various tissues of wild-type 129sv mice and examined the samples for the presence of sense and antisense transcripts. We prepared RNA from the thymus, the kidney, and sorted pro-B (B220int, CD43+) and pre-B (B220+, CD43lo) cells from bone marrow. We applied the RT-PCR assay for general VHDJH antisense transcripts (see Fig. 2A) and detected both sense (Fig. 2C Upper, RT-S lanes) and antisense (Fig. 2C Lower, RT-AS lanes) transcripts in pro-B cells, pre-B cells, and the thymus, but not in the kidney, which was used as a negative control. Sense transcripts of the JH locus in the early stages of B cell development (34) and in thymocytes (35) have been described previously. Our data show that antisense transcripts in the JH4 intron appear to be expressed whenever sense transcripts are present in all of the tissues that we examined. Moreover, the IgH intronic enhancer core (cEμ) is not necessary for sense or antisense transcript expression in the JH locus because we detected both types of transcripts in splenic B lymphocytes from Eμ KO mice (SI Fig. 6) (36).
Antisense Transcripts in Switch Regions.
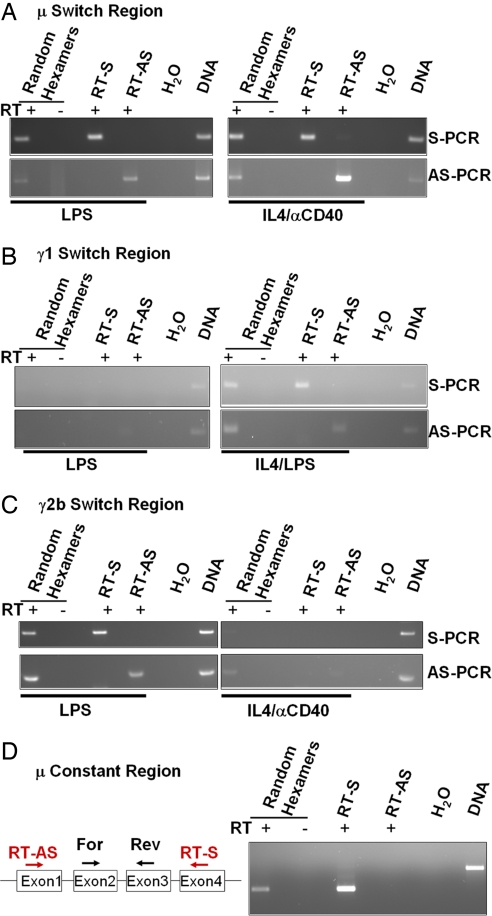
It was shown that AID equally targets both DNA strands of S regions in vivo (11, 26, 27). However, in the context of in vitro transcription experiments, mainly the nontemplate strand is a target for AID (17, 19, 20, 23). Therefore, we also tested S regions for the expression of antisense transcripts in addition to sense transcripts. Splenocytes from wild-type 129sv mice were MACS-sorted to enrich for CD43− B cells and cultured. RNA was prepared after 4 days of stimulation with LPS or with IL4/αCD40. Cultured B cells were stimulated with LPS to induce switching to Cγ2b and Cγ3 and were stimulated with LPS/IL4 or IL4/αCD40 to induce switching to Cγ1 and Cε (3). A similar RT-PCR assay to that described in Fig. 2A was applied to distinguish between sense and antisense transcripts. Sμ was transcribed in sense (Fig. 4A Upper, RT-S lanes) and antisense (Fig. 4A Lower, RT-AS lanes) orientation when stimulated with either LPS or IL4/αCD40, consistent with the fact that DSBs in Sμ are necessary to switch to any of the downstream constant regions. Sense and antisense transcripts within Sγ1 were readily detected when stimulated with IL4/αCD40, but not upon stimulation with LPS (Fig. 4B). Finally, Sγ2b was transcribed in both orientations when stimulated with LPS, but much less upon stimulation with IL4/αCD40 (Fig. 4C). When we assay transcripts within the different S regions via the RT-PCR assay (Fig. 4 A–C), primers anneal within the S regions, which get spliced out in the mature sense transcripts. Therefore, the sense-specific PCR only detects a signal from the unprocessed pre-mRNA fraction that could underestimate the total level of sense transcripts relative to antisense transcripts, assuming the latter do not get processed. Notably, we also assayed the Cμ exons, which are not a target of AID, and only detected transcripts in the sense orientation, but not in the antisense orientation (Fig. 4D) (32) in splenic B cells stimulated with LPS, LPS/IL4, or αCD40/IL4. An in vitro assay employing SP6 sense and T7 antisense transcripts for the Cμ exons confirmed the specificity and sensitivity of the assay to prove that both sense or antisense Cμ transcripts could be detected if present (SI Fig. 7).
Fig. 4.
Bidirectional transcription in switch regions. (A–C) Strand-specific RT-PCR design similar as described in Fig. 2A, performed on in vitro-stimulated splenic B cells for Sμ (A), Sγ1 (B), and Sγ2b (C). Above the lanes, the used reverse transcription primers are indicated with (+) or without (−) reverse transcriptase. Below the lanes, respective B cell stimulation is indicated. (Upper) Sense-specific PCR. (Lower) Antisense-specific PCR. (D) Unidirectional transcription of Cμ. (Left) RT-PCR design of Cμ. Sense (RT-S)- and antisense (RT-AS)-specific reverse transcription primers are depicted in red, and PCR primers are depicted in black; the four exons of Cμ are depicted. (Right) Strand-specific RT-PCR assay performed on in vitro-stimulated splenic B cells for Cμ. Above the lanes, the used RT-primers are indicated with (+) or without (−) reverse transcriptase.
Discussion
In this article, we describe the presence of antisense transcripts, in addition to the known sense transcripts, in IgH variable region exons and IgH S regions, which are target regions for SHM and CSR. Transcription in the sense orientation through Ig variable regions and S regions has been shown to be necessary for SHM and CSR, respectively (13–15). Antisense transcripts in these regions of the IgH locus appear to be expressed whenever their counterparts in the sense orientation are expressed and are therefore not obviously subject to differential regulation. Antisense transcripts in the JH4 intron, a region that undergoes SHM, are expressed in different stages of B cell development and in thymocytes, but not in the kidney, just like sense transcripts in this region. Overall, the expression of antisense transcripts is not restricted only to cells that actually undergo SHM or CSR.
Based on 5′RACE experiments, the antisense transcripts through the VHDJH region appear to originate from heterogeneous start sites of a region without significant promoter activity. However, these putative start sites could not be readily detected by a sensitive RNase protection assay. One interpretation of the latter finding is that the transcripts are at too low a steady-state level to be detected by the RNase protection because of either low-level transcription or high turnover. Moreover, we cannot exclude the possibility that some of the apparent transcription initiation sites represent cDNA strong stops and that the true initiation site(s) is more distant. Antisense transcripts through S regions, the AID target regions for CSR, also can be readily detected in ex vivo B cells that are stimulated in culture to undergo CSR. Again the expression pattern of these antisense transcripts correlates with the expression of the corresponding sense transcripts. Notably, we find that the Cμ exons, which lie in between the VHDJH and S regions and are not targeted by AID, are only transcribed detectably in the sense direction. Although many other interpretations are possible, the finding of the lack of Cμ antisense transcripts, although Cμ lies in between the VHDJH and downstream CH exon, is consistent with the hypothesis that antisense transcripts contribute in some way to AID targeting in the context of SHM and CSR.
Antisense transcripts originating in various S regions have been described previously in the context of chromosomal translocations in murine and human B cell tumors and B cell tumor cell lines, resulting in hybrid c-Myc/Sγ2a (37), c-Myc/Sμ (38), or Pax5/Sμ (39) transcripts and thereby driving the expression of the oncogene. In two translocation studies involving Sμ, distinct antisense promoters have been described within the μ-switch region in human tumors and cell lines. These promoters could be the origin of the antisense transcript within Sμ that we describe in wild-type B cells in mice. Also, we cannot exclude the possibility that some fraction of the antisense transcripts that we described within the JH region originate from the promoters in Sμ, in addition to the potential start sites that we tentatively identified within the JH region by 5′RACE.
Antisense transcripts from the JH region and within the rearranged IgH variable region exon have been described in human lymphoma cell lines (30, 40). Two recent studies also describe antisense transcripts in the DHJH region of bone marrow pro-B cells. The authors suggest that antisense transcription increases DHJH accessibility (41) or decreases DHJH accessibility by RNA interference-mediated gene silencing (42), respectively. An increasing number of studies show that natural antisense transcripts (NATs), in addition to sense transcripts, are expressed in a significant portion of the murine (43) and human (44) genomes. Although several interesting studies were able to identify the function for few of these antisense transcripts (45), the role for most of them remains elusive. Although we initiated our study in the context of considering a role for antisense transcription in SHM and CSR, our findings, which are consistent with such a possibility, do not prove it or rule out other potential functions. Other suggested functions for NATs throughout the genome range from RNAi-related mechanisms, such as silencing and heterochromatin formation induced by the sense–antisense RNA pairs, to a role in RNA editing, DNA methylation, and monoallelic silencing in mammalian X chromosome inactivation and genomic imprinting (46). Such functions also might play a role in IgH expression or diversification mechanisms.
Materials and Methods
Oligonucleotides.
For sequences of primers and annealing temperatures, see Table 1.
Table 1.
Oligonucleotides and annealing temperatures
| Name | Sequence | Temp., °C |
|---|---|---|
| Reverse transcription primers | ||
| HC ki VHRT-S | TGAGGAGACTGTGAGAGTGGTGCC | 56.0 |
| HC ki VHRT-AS | CATGGGATGGAGCTGTATCA | 56.0 |
| 3′JH4 RT-S | CAAATATCCAAGATTAGTCTGCAA | 59.5 |
| 3′JH4 RT-AS | AATGGCCTCTCCAGGTCTT | 58.6 |
| Sμ RT-S | CTACTCCAGAGTAGCTCATTTCAGAT | 56.0 |
| Sμ RT-AS | CGCTAAACTGAGGTGATTACTCTG | 53.0 |
| Sγ1 RT-S | ACCTGTCCTAGCTCCCCAAT | 60.7 |
| Sγ1 RT-AS | TCAAGAGCTCAGGACACAAGAC | 55.5 |
| Sγ2b RT-S | CCCTCCACATGCCGCTCTCCCCAGGTATC | 60.3 |
| Sγ2b RT-AS | AGGAACCAACTATGAAAGACCTGAGTTAGG | 60.3 |
| Cμ RT-S | CGTGGCCTACAACACAGGTAT | 55.5 |
| Cμ RT-AS | GAGTCAGTCCTTCCCAAATGTC | 55.5 |
| PCR primers | ||
| HC ki VHFor | ATGGGATGGAGCTGTATCATGCTC | 61.0 |
| HC ki VHRev | CTTGGCCCCAGTAGTCAAAGTAGC | 61.0 |
| 3′JH4 For-S | GCGAAACTCGAGCCACTATTGTGATTACTATGCTATGG | 54.0 |
| 3′JH4 RT-S | CAAATATCCAAGATTAGTCTGCAA | 54.0 |
| 3′JH4 RT-AS | AATGGCCTCTCCAGGTCTT | 61.0 |
| 3′JH4 Rev-AS | AAAGAAAGTGCCCCACTCCAC | 61.0 |
| Sμ For-S | TTGAGAGCCCTAGTAAGCG | 55.0 |
| Sμ Rev-S | TCATCTCGGTTAAGCCAGCCTAGTTTAG | 55.0 |
| Sμ RT-AS | CGCTAAACTGAGGTGATTACTCTG | 55.0 |
| Sμ Rev-AS | GCTCACCCCATCTCACC | 55.0 |
| Sγ1 For-S | TGCAAGCTGCTCTGAGGG | 63.5 |
| Sγ1 Rev-S | CTTGGATCACCACACTTCCA | 63.5 |
| Sγ1 For-AS | AGAGCTGAGGCTGGTAAGAGTAA | 63.5 |
| Sγ1 Rev-AS | TATAGCTGCTGTGCCTGGATC | 63.5 |
| Sγ2b For-S | AGTTCTAGCAGCTATGGGGAATCTTGGTCAG | 55.0 |
| Sγ2b RT-S | CCCTCCACATGCCGCTCTCCCCAGGTATC | 55.0 |
| Sγ2b RT-AS | AGGAACCAACTATGAAAGACCTGAGTTAGG | 55.0 |
| Sγ2b Rev-AS | TCTCTGCTCTCATCTCTCGAAGCTTTCCATG | 55.0 |
| Cμ For | TAAAGGATGGGAAGCTCGTG | 55.0 |
| Cμ Rev | TCTTCTGTGGTGAAGGCAGA | 55.0 |
| 5′RACE primers | ||
| JH1 GSP1 | GTACTTCGATGTCTGGGGC | 53.0 |
| JH1 GSP2 | CGCAGGGACCACGGTCA | 55.0 |
| JH1 nested GSP | GTCACCGTCTCCTCAGGTAAGCT | 55.0 |
| JH2 GSP1 | ACTACTGGGGCCAAGGC | 53.0 |
| JH2 GSP2 | CAAGGCACCACTCTCACAGTCT | 55.0 |
| JH2 nested GSP | CACAGTCTCCTCAGGTGAGTCCTT | 55.0 |
| JH3 GSP1 | TGCTTACTGGGGCCAAG | 53.0 |
| JH3 GSP2 | CCAAGGGACTCTGGTCACTGTC | 55.0 |
| JH3 nested GSP | CACTGTCTCTGCAGGTGAGTCCT | 55.0 |
| JH4 GSP1 | GGACTACTGGGGTCAAGGA | 53.0 |
| JH4 GSP2 | ACCTCAGTCACCGTCTCCTCAG | 55.0 |
| JH4 nested GSP | AGAATGGCCTCTCCAGGTCTTTA | 55.0 |
RT-PCR Assays.
Total RNA from various tissues was prepared by using TRIzol Reagent (Invitrogen). RNA was treated with Turbo DNase (Ambion) and cleaned up with an RNeasy Mini Kit (Qiagen). Then, 300 ng to 1 μg of total RNA was reverse transcribed for 1 h with strand-specific primers at various temperatures (see below) or with random hexamers (Roche) at 50°C by using SuperscriptIII (Invitrogen). Subsequently, PCR was performed at various conditions: 30–39 cycles of 30 sec at 94°C, 30 sec at annealing temperature (see Table 1), and 20–30 sec at 72°C. PCR products were visualized on ethidium bromide gels, excised, gel-purified by using the QIAquick gel extraction kit (Qiagen), cloned into pGEM-T easy (Promega), and inserts were sequenced to confirm specific amplification.
B Cell Cultures.
Single-cell suspension from spleens was enriched for CD43− B cells by using a MACS separator (Miltenyi Biotec) according to the manufacturer's instructions. B cells were cultured for 4 days with 20 μg/ml LPS (Sigma–Aldrich) or 1 μg/ml αCD40 antibody (BD PharMingen) and 20 ng/ml IL-4 (R&D Systems).
Identification of Transcriptional Start Sites by 5′RACE.
Wild-type 129sv mice were immunized with SRBCs, activated B cells (B220+, PNAHI) were sorted from spleens, and total RNA was prepared by using TRIzol reagent (Invitrogen). RNA was treated with Turbo DNase (Ambion) and cleaned up with an RNeasy Mini Kit (Qiagen). Then, 1 μg of total RNA was used for reverse transcription, and rapid amplification of cDNA ends was performed with 5′RACE system version 2.0 (Invitrogen). The primers used are listed in Table 1. Products of the nested PCR were visualized on ethidium bromide gels and were directly cloned into pGEM-T Easy (Promega). Inserts were sequenced to identify the start sites of antisense transcripts.
Immunization.
Wild-type 129sv mice were injected i.p. with 108 SRBCs to isolate naïve (B220+, PNALO) and activated (B220+, PNAHI) B cells from spleens 12 days after immunization.
Flow Cytometry.
Single-cell suspensions from immunized spleens (2 × 107 cells per ml) were stained with FITC-labeled anti-mouse PNA (Vector Laboratories) and CyChrome (CyC)-labeled anti-mouse B220 (BD PharMingen) antibodies, and single-cell suspensions from bone marrow cells were stained with FITC-labeled anti-mouse CD43 (BD PharMingen) and CyC-labeled anti-mouse B220 antibodies. Cell sorting was performed on a FACSAria cell-sorting system (BD Biosciences).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Uttiya Basu, Meinrad Busslinger, Tirtha Chakraborty, Jayanta Chaudhuri, Thomas Decker, Cosmas Giallourakis, Jing Wang, Catherine Yan, Ali Zarrin, Shan Zha, and Yu Zhang for helpful discussions and comments; Lisa Acquaviva for mouse work; Natasha Barteneva for cell sorting; and Klaus Rajewsky (Immune Disease Institute, Harvard Medical School, Boston) for providing the Ig heavy chain knockin mice. T.P. is supported by a Boehringer Ingelheim Fonds PhD scholarship. This work was supported by National Institutes of Health Grants PO1CA092625-05 and 2PO1AI031541-15 (to F.W.A.). G.L. is a postdoctoral associate of the Howard Hughes Medical Institute. F.W.A. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 3661.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712291105/DC1.
References
- 1.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 2.Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat Rev Immunol. 2006;6(8):573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- 3.Chaudhuri J, et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- 4.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449(7161):478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 5.Catalan N, et al. The block in immunoglobulin class switch recombination caused by activation-induced cytidine deaminase deficiency occurs prior to the generation of DNA double strand breaks in switch mu region. J Immunol. 2003;171(5):2504–2509. doi: 10.4049/jimmunol.171.5.2504. [DOI] [PubMed] [Google Scholar]
- 6.Schrader CE, Linehan EK, Mochegova SN, Woodland RT, Stavnezer J. Inducible DNA breaks in Ig S regions are dependent on AID, UNG. J Exp Med. 2005;202:561–568. doi: 10.1084/jem.20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wuerffel RA, Du J, Thompson RJ, Kenter AL. Ig Sgamma3 DNA-specifc double strand breaks are induced in mitogen-activated B cells and are implicated in switch recombination. J Immunol. 1997;159:4139–4144. [PubMed] [Google Scholar]
- 8.Zarrin AA, et al. Antibody class switching mediated by yeast endonuclease-generated DNA breaks. Science. 2007;315:377–381. doi: 10.1126/science.1136386. [DOI] [PubMed] [Google Scholar]
- 9.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 10.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 11.Rada C, Di Noia JM, Neuberger MS. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell. 2004;16:163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 12.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 13.Fukita Y, Jacobs H, Rajewsky K. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity. 1998;9:105–114. doi: 10.1016/s1074-7613(00)80592-0. [DOI] [PubMed] [Google Scholar]
- 14.Jung S, Rajewsky K, Radbruch A. Shutdown of class switch recombination by deletion of a switch region control element. Science. 1993;259:984–987. doi: 10.1126/science.8438159. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Bottaro A, Li S, Stewart V, Alt FW. A selective defect in IgG2b switching as a result of targeted mutation of the I gamma 2b promoter and exon. EMBO J. 1993;12:3529–3537. doi: 10.1002/j.1460-2075.1993.tb06027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci USA. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaudhuri J, et al. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 18.Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 19.Sohail A, Klapacz J, Samaranayake M, Ullah A, Bhagwat AS. Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic Acids Res. 2003;31:2990–2994. doi: 10.1093/nar/gkg464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramiro AR, Stavropoulos P, Jankovic M, Nussenzweig MC. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat Immunol. 2003;4:452–456. doi: 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- 21.Shinkura R, et al. The influence of transcriptional orientation on endogenous switch region function. Nat Immunol. 2003;4:435–441. doi: 10.1038/ni918. [DOI] [PubMed] [Google Scholar]
- 22.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 23.Yu K, Roy D, Bayramyan M, Haworth IS, Lieber MR. Fine-structure analysis of activation-induced deaminase accessibility to class switch region R-loops. Mol Cell Biol. 2005;25:1730–1736. doi: 10.1128/MCB.25.5.1730-1736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–998. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- 25.Zarrin AA, et al. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat Immunol. 2004;5:1275–1281. doi: 10.1038/ni1137. [DOI] [PubMed] [Google Scholar]
- 26.Milstein C, Neuberger MS, Staden R. Both DNA strands of antibody genes are hypermutation targets. Proc Natl Acad Sci USA. 1998;95:8791–8794. doi: 10.1073/pnas.95.15.8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue K, Rada C, Neuberger MS. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2−/− ung−/− mice. J Exp Med. 2006;203:2085–2094. doi: 10.1084/jem.20061067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Besmer E, Market E, Papavasiliou FN. The transcription elongation complex directs activation-induced cytidine deaminase-mediated DNA deamination. Mol Cell Biol. 2006;26:4378–4385. doi: 10.1128/MCB.02375-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen HM, Storb U. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc Natl Acad Sci USA. 2004;101:12997–13002. doi: 10.1073/pnas.0404974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ronai D, et al. Detection of chromatin-associated single-stranded DNA in regions targeted for somatic hypermutation. J Exp Med. 2007;204:181–190. doi: 10.1084/jem.20062032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sonoda E, et al. B cell development under the condition of allelic inclusion. Immunity. 1997;6:225–233. doi: 10.1016/s1074-7613(00)80325-8. [DOI] [PubMed] [Google Scholar]
- 32.Bolland DJ, et al. Antisense intergenic transcription in V (D)J recombination. Nat Immunol. 2004;5:630–637. doi: 10.1038/ni1068. [DOI] [PubMed] [Google Scholar]
- 33.Lennon GG, Perry RP. C mu-containing transcripts initiate heterogeneously within the IgH enhancer region and contain a novel 5′-nontranslatable exon. Nature. 1985;318:475–478. doi: 10.1038/318475a0. [DOI] [PubMed] [Google Scholar]
- 34.Reth MG, Alt FW. Novel immunoglobulin heavy chains are produced from DJH gene segment rearrangements in lymphoid cells. Nature. 1984;312:418–423. doi: 10.1038/312418a0. [DOI] [PubMed] [Google Scholar]
- 35.Alt FW, Rosenberg N, Enea V, Siden E, Baltimore D. Multiple immunoglobulin heavy-chain gene transcripts in Abelson murine leukemia virus-transformed lymphoid cell lines. Mol Cell Biol. 1982;2:386–400. doi: 10.1128/mcb.2.4.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perlot T, Alt FW, Bassing CH, Suh H, Pinaud E. Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc Natl Acad Sci USA. 2005;102:14362–14367. doi: 10.1073/pnas.0507090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Julius MA, et al. Translocated c-myc genes produce chimeric transcripts containing antisense sequences of the immunoglobulin heavy chain locus in mouse plasmacytomas. Oncogene. 1988;2:469–476. [PubMed] [Google Scholar]
- 38.Apel TW, Mautner J, Polack A, Bornkamm GW, Eick D. Two antisense promoters in the immunoglobulin mu-switch region drive expression of c-myc in the Burkitt's lymphoma cell line BL67. Oncogene. 1992;7:1267–1271. [PubMed] [Google Scholar]
- 39.Morrison AM, et al. Deregulated PAX-5 transcription from a translocated IgH promoter in marginal zone lymphoma. Blood. 1998;92:3865–3878. [PubMed] [Google Scholar]
- 40.Capaccioli S, et al. A bcl-2/IgH antisense transcript deregulates bcl-2 gene expression in human follicular lymphoma t(14;18) cell lines. Oncogene. 1996;13:105–115. [PubMed] [Google Scholar]
- 41.Bolland DJ, et al. Antisense intergenic transcription precedes Igh D-to-J recombination and is controlled by the intronic enhancer Emu. Mol Cell Biol. 2007;27:5523–5533. doi: 10.1128/MCB.02407-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakraborty T, et al. Repeat organization and epigenetic regulation of the DH-Cmu domain of the immunoglobulin heavy-chain gene locus. Mol Cell. 2007;27:842–850. doi: 10.1016/j.molcel.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 43.Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 44.Lapidot M, Pilpel Y. Genome-wide natural antisense transcription: Coupling its regulation to its different regulatory mechanisms. EMBO Rep. 2006;7:1216–1222. doi: 10.1038/sj.embor.7400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang PK, Kuroda MI. Noncoding RNAs and intranuclear positioning in monoallelic gene expression. Cell. 2007;128:777–786. doi: 10.1016/j.cell.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 46.Werner A. Natural antisense transcripts. RNA Biol. 2005;2:53–62. doi: 10.4161/rna.2.2.1852. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.