Abstract
Electrochemistry measurements on DNA-modified electrodes are used to probe the effects of binding to DNA on the redox potential of SoxR, a transcription factor that contains a [2Fe-2S] cluster and is activated through oxidation. A DNA-bound potential of +200 mV versus NHE (normal hydrogen electrode) is found for SoxR isolated from Escherichia coli and Pseudomonas aeruginosa. This potential value corresponds to a dramatic shift of +490 mV versus values found in the absence of DNA. Using Redmond red as a covalently bound redox reporter affixed above the SoxR binding site, we also see, associated with SoxR binding, an attenuation in the Redmond red signal compared with that for Redmond red attached below the SoxR binding site. This observation is consistent with a SoxR-binding-induced structural distortion in the DNA base stack that inhibits DNA-mediated charge transport to the Redmond red probe. The dramatic shift in potential for DNA-bound SoxR compared with the free form is thus reconciled based on a high-energy conformational change in the SoxR–DNA complex. The substantial positive shift in potential for DNA-bound SoxR furthermore indicates that, in the reducing intracellular environment, DNA-bound SoxR is primarily in the reduced form; the activation of DNA-bound SoxR would then be limited to strong oxidants, making SoxR an effective sensor for oxidative stress. These results more generally underscore the importance of using DNA electrochemistry to determine DNA-bound potentials for redox-sensitive transcription factors because such binding can dramatically affect this key protein property.
Keywords: DNA electrochemistry, iron-sulfur proteins, oxidative stress
SoxR belongs to the MerR family of transcriptional regulators. The members of this family are defined by an N-terminal helix–turn–helix DNA binding motif, a coiled-coil dimerization region, and a C-terminal sensory domain (1–3). Although the DNA binding and dimerization regions are conserved among MerR-type regulators, their sensory domains are diverse (2). Typically, MerR type transcription factors occupy suboptimally spaced 19 ± 1-bp promoter elements in the inactivated state, often inducing a slight bend of the promoter DNA. Upon activation, these proteins are thought to undergo a conformational change that unwinds the promoter region, thereby allowing RNA polymerase to initiate transcription (2).
SoxR regulates an oxidative stress response to superoxide in the enterics Escherichia coli and Salmonella enterica (4, 5). This unique transcription factor is a 17-kDa polypeptide that binds DNA as a dimer and contains a [2Fe-2S] cluster in each monomer (4). Loss of this cluster does not affect protein folding, DNA binding, or promoter affinity (6–8), but oxidation of this cluster by either oxygen or superoxide-generating agents (e.g., methyl viologen) triggers expression of the transcription factor SoxS (8, 9). Subsequently, SoxS controls the expression of >100 genes in the SoxRS regulon that collectively act to repair or avoid oxidative damage (10).
The role of SoxR appears to vary dramatically across organisms. Whereas SoxR is conserved in both Gram-negative and Gram-positive bacteria, soxS is exclusively found in enterics, indicating that SoxR can be part of different regulatory networks (11, 12). Indeed, Pseudomonas putida and Pseudomonas aeruginosa do not rely on SoxR for an oxidative-stress response (13, 14). Instead, P. aeruginosa SoxR responds to phenazines, endogenous redox-active pigments, and activates transcription of two probable efflux pumps and a putative monooxygenase (15) that might aid in phenazine transport and modification. Considering that SoxR shows functional diversity between pseudomonads and enterics, it is surprising that the transcription factor is biochemically conserved: (i) Expression of P. putida SoxR in E. coli can complement a soxR deletion mutant (14), and (ii) the redox potentials of soluble SoxR from E. coli and P. aeruginosa in vitro are both approximately −290 mV (7, 8, 15).
That SoxR requires oxidation for its transcriptional activity seems biologically reasonable but also leads to a conundrum. Under normal physiological conditions, it is assumed that SoxR is kept in its reduced, inactive state by the intracellular NADPH/NADP+ redox potential of approximately −340 mV versus NHE (16, 17). Furthermore, it has been reported that NADPH-dependent SoxR reduction is enzyme-mediated, allowing for a rapid adjustment to changes in cellular conditions, although direct enzymatic interaction with SoxR has not yet been demonstrated (18, 19). The conundrum does not lie in the mechanism of SoxR reduction but rather in the specificity of its oxidation: At a low redox potential of −290 mV versus NHE (7, 8, 15), many cellular oxidants could react with SoxR, in particular glutathione (20), and therefore SoxR would be primarily in an oxidized form, even without imposing oxidative stress. Since this is not the case, how is SoxR maintained in its reduced and transcriptionally silent form?
The mechanism underlying the oxidation/activation of SoxR is also not well understood. For E. coli SoxR, it was first suggested that superoxide directly oxidizes the iron-sulfur cluster, but this has not been established (21, 22). Alternatively, the redox state of SoxR might be coupled to changes in the equilibrium of biologically relevant redox couples, such as NADPH or glutathione (16, 23). Recently, we have shown that the activation of SoxR in P. aeruginosa can occur in an oxygen-independent manner (14). Considering that both E. coli and P. aeruginosa SoxR can transfer electrons to the mediator safranin O, a phenazine derivative (7, 8, 15), it seems reasonable that endogenous phenazines may oxidize SoxR in pseudomonads. Alternatively, given that pseudomonad phenazines can also modulate the intracellular NADH/NAD+ ratio, the possibility that phenazines activate SoxR indirectly must also be considered (24).
One interesting possibility that has been suggested but never addressed experimentally is the effect of DNA binding on the redox potential of SoxR. The published redox potentials for SoxR were measured in the absence of DNA (4, 7, 8). This is particularly significant because SoxR activates transcription only in its DNA-bound state, so determining the redox potential of the DNA-bound form of SoxR becomes critical.
We have previously explored DNA-modified electrodes as flexible platforms for the study of DNA-mediated charge transport chemistry (25–27). Typically, self-assembled DNA monolayers on gold or graphite are interrogated electrochemically with the efficiency of charge transfer to an electroactive probe yielding information on the integrity of the intervening base pair stack. In fact, duplexes that are covalently modified with redox-active reporters at a fixed position provide particularly well defined systems for study of DNA charge transport at electrode surfaces, allowing for the electrochemical detection of even small perturbations in the intervening base pair stack (28–31).
In addition, DNA-modified electrodes have proven useful for probing redox centers within proteins bound to DNA (32–34). We have used DNA monolayers to probe the redox potential of MutY and Endonuclease III, base excision repair glycosylases that contain a [4Fe-4S] cluster. Initial studies of these enzymes had found no clear role for the clusters because, in the absence of DNA, they did not display redox activity within a physiologically relevant range of potentials (35–37). We found, however, that at DNA-modified Au surfaces, these repair enzymes display reversible, DNA-mediated electrochemistry with redox potentials of ≈90 mV (33). Moreover, experiments comparing directly the electrochemistry of Endonuclease III on bare and DNA-modified graphite demonstrated that binding to DNA shifts the redox potential of the protein by ≈200 mV into a physiologically relevant range, activating the cluster for oxidation (34). DNA binding thus changes the redox properties of the enzymes from being similar to ferredoxins to instead resembling high potential iron proteins. Based on these data, we have proposed a redox role for the [4Fe-4S] clusters in long-range DNA-mediated signaling as a first step in detecting damaged sites that are to be repaired in the genome (32–34, 38). Our ability to alter the redox states of these proteins in a DNA-mediated manner further suggests that DNA may be a medium through which oxidation/reduction reactions occur. This mechanism may also be important to consider in the context of SoxR.
Given the sensitivity of DNA-modified electrodes in probing redox centers of proteins bound to DNA and the precedent that DNA binding can alter redox potentials of the bound protein, here we explore the redox properties of the DNA-bound form of SoxR. Model studies have shown repeatedly the sensitivity of redox potentials of iron-sulfur clusters to environmental perturbations, which are expected to be significant for SoxR (39). Here, using self-assembled DNA monolayers on highly oriented pyrolytic graphite (HOPG), we address the effect of DNA binding on the redox potential of both E. coli and P. aeruginosa SoxR. The DNA-bound potential provides convincing evidence for the mechanism the cell uses to maintain SoxR in its reduced form in vivo.
Results
Experimental Strategy Used to Probe SoxR Electrochemically.
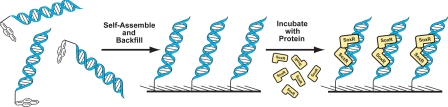
Fig. 1 illustrates the experimental strategy used to investigate the electrochemistry of DNA-bound SoxR from E. coli and P. aeruginosa PA14. DNA duplexes are prepared by hybridizing pyrene-modified single-stranded DNA with its complement (with or without covalently attached Redmond red). The duplexes are then self-assembled on HOPG in the absence of Mg2+ to form a loosely packed DNA monolayer, leaving room for SoxR to bind (32–34). The surface is backfilled with octane or decane to prevent direct charge transfer from the surface to the protein (40, 41). The electrode is subsequently incubated with protein, and electrochemical experiments are performed before and after protein addition. The DNA binding sites for SoxR are 18-bp symmetrical sequences that are conserved across species (15). For P. aeruginosa experiments, we chose the SoxR binding site found upstream of an operon that encodes the efflux pump MexGHI-OpmD in P. aeruginosa PA14.
Fig. 1.
Schematic illustration of the self-assembly/backfilling of a DNA monolayer followed by incubation with protein.
SoxR Binding Is Reported Through the Redmond Red Electrochemical Signal.
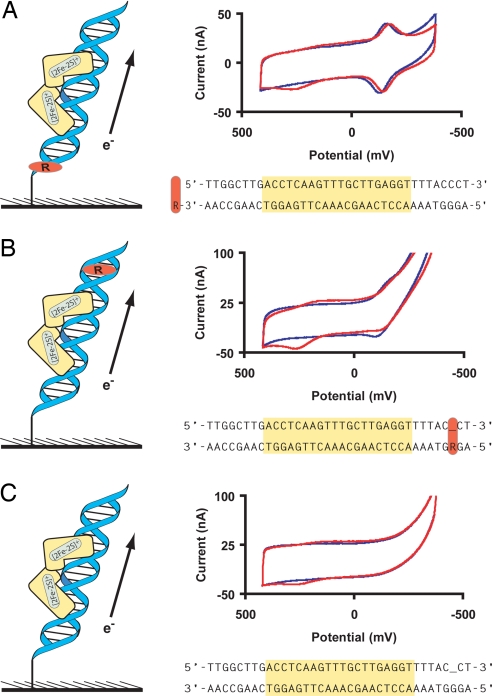
We can observe protein binding in electrochemistry experiments by monitoring DNA-mediated transport between the electrode and the redox active probe Redmond red that is attached at either end of the DNA duplex (Fig. 2). The midpoint potential of Redmond red is −160 mV versus NHE, and the linearity of the plot of peak current as a function of scan rate indicates that Redmond red behaves as a surface-bound species (42). Although small potential shifts (≈20 mV) in the Redmond red signal are occasionally observed upon addition of SoxR, Redmond red provides a convenient and reliable internal standard.
Fig. 2.
Cyclic voltammetry at 50 mV/s of electrodes modified with DNA featuring Redmond red at the bottom (A), Redmond red above the binding site (B), and no Redmond red (C). Voltammograms before addition of SoxR are blue whereas those after addition of SoxR are red. The sequences used in the course of these experiments are illustrated with the binding sequence for SoxR highlighted, the location of Redmond red indicated by an “R,” and the location of abasic sites underlined.
It is expected that a redox-active probe located at the top of the DNA monolayer will report on perturbations of the base pair stack that intervene between the redox probe and the electrode, whereas the same probe located at the bottom of the monolayer near the electrode surface will not be affected by disruptions in base stacking above the probe. Previously, we have reported attenuation in charge accumulation by chronocoulometry for daunomycin covalently attached near the top of a DNA film due to perturbations in the intervening DNA structure by the base-flipping methylase M.HhaI and TATA binding protein (43). Here, as shown in Fig. 2, when Redmond red is incorporated above the SoxR binding site, a 16% decrease in the integrated cathodic charge of Redmond red is observed upon addition of SoxR. In contrast, when Redmond red is incorporated at the bottom of the DNA duplex below the SoxR binding site, there is little detectable change in the Redmond red signal in the presence of SoxR. Although the loss of signal observed upon addition of SoxR is far smaller than that found for TATA binding protein or M.HhaI, the decrease in electrochemical signal when the binding site is positioned between the probe and the electrode does provide evidence for SoxR binding.
Electrochemistry of P. aeruginosa SoxR.
As is evident in Fig. 2, besides the Redmond red probe, we also observe a second distinct and quasi-reversible electrochemical signal at +200 mV versus NHE upon addition of SoxR. The signal is observed only after SoxR addition and is not affected by Redmond red because it is also present in the absence of the probe. Note that no redox signature is observed at −290 mV versus NHE, the potential previously reported for SoxR in solution (Fig. 2). Incubation of the DNA-modified surface with PA2274, a control protein that lacks an iron-sulfur cluster, does not result in the appearance of any redox signature. Furthermore, experiments with SoxR stocks featuring low iron-sulfur content after purification do not lead to appreciable cyclic voltammetric signals (data not shown). Therefore, we can assign the new signal observed to the [2Fe-2S] cluster of SoxR.
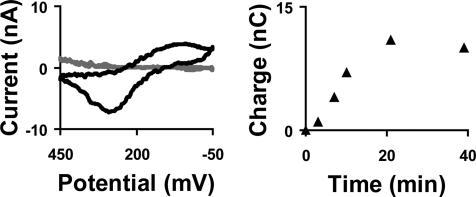
In a typical experiment, the observed SoxR signal increases over a period of ≈15 min and is stable for a minimum of 18 scans before slowly decaying (Fig. 3), although we have found a high variability in electrode stability upon addition of protein. High concentrations of protein (>10 μM) are required for these experiments, certainly concentrations higher than is required for site-specific binding, and both the high protein concentrations and long DNA sequences used make DNA/protein film formation difficult. It is important to note that the Redmond red signal is highly stable and exhibits no noticeable degradation during typical electrochemistry experiments.
Fig. 3.
Binding of SoxR to the DNA-modified film. (Left) Background subtracted cyclic voltammetry of P. aeruginosa SoxR at DNA-modified graphite electrodes at a 50 mV/s scan rate immediately after addition of SoxR (light) and 20 min after addition of SoxR (dark) revealing the signal observed. (Right) Integrated anodic charge for SoxR showing the growth of the signal as a function of time.
As can be seen in Figs. 2 and 3, the cathodic and anodic waves observed for SoxR are asymmetric: The oxidation wave is pronounced and substantially less broad compared with the reduction wave. In an ideal quasi-reversible system, the anodic to cathodic peak current ratio is unity (42, 44), but this is certainly not the case for SoxR. We find an anodic to cathodic peak current ratio of 3.0 for SoxR in Fig. 2, strongly indicative of a non-ideal and quasi-reversible electrochemical response. In contrast, the anodic to cathodic peak current ratio is 1.3 for the 3′-Redmond red on the same film, far closer to the ideal value for a fully reversible system. These data show that the electrochemistry of SoxR is complicated, hardly surprising given that SoxR binds DNA as a dimer.
Interestingly, the asymmetries in the reduction and oxidation waves of SoxR are qualitatively distinct from those previously observed for the DNA repair enzymes MutY and Endo III; the electrochemistry of those enzymes featured a reduction wave that was somewhat more pronounced than the oxidation wave (32). The better resolved anodic wave of SoxR integrates to very low surface coverages of 0.5 pmol/cm2. This apparent low coverage is comparable to that of 2 pmol/cm2 previously found for MutY at DNA monolayers on gold (33) and may reflect poor coupling of the iron-sulfur cluster with the base pair stack. However, the Redmond red probe at the bottom of the DNA monolayer integrates to surface coverages of 1 pmol/cm2 whereas the Redmond red probe at the top the film integrates to coverages that are 3-fold lower (over sample sizes of at least 10 electrodes). These values are far less than the ideal DNA surface coverage of 10 pmol/cm2 expected for a loosely packed DNA monolayer and indicate that the amount of DNA on the surface is the main determinant of the size of the SoxR signal.
Despite the broad cathodic wave, we can calculate an upper bound for the number of electrons transferred for the oxidation of SoxR. For an ideal surface-bound species, the slope derived from the plot of peak current as a function of scan rate divided by the integrated charge Q at any scan rate is equal to nF/4RT, where F is Faraday's constant, R is the gas constant, and T is the temperature (44); performing this operation for the Redmond red probe at the bottom of the monolayer (Qanodic/cathodic = ≈17 nC at any scan rate) yields a value of n = 2, as expected for 2 e− transfer to the resorufin moiety. The integrated charge for the anodic wave of SoxR on the same film varies from 3 to 9 nC, indicating that SoxR receives at most half the number of electrons transferred to the Redmond red. If we assume that all of the DNA is bound and that the Redmond red signal at the bottom of the monolayer corresponds perfectly to the number of DNA molecules on the surface (highly likely for the sparse films obtained), we can estimate that each DNA-bound SoxR dimer undergoes at most a one electron oxidation/reduction. In fact, all of these observations are consistent with titrations of free SoxR, which deviate from ideal reductions, but appear also to yield values of n = 1 (8, 45).
Comparison of the Voltammetry of E. coli and P. aeruginosa SoxR.
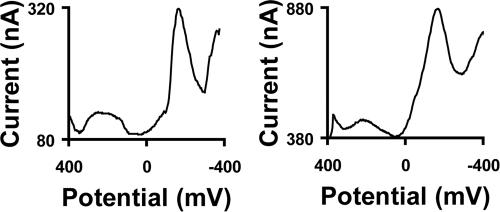
To expand our work to multiple organisms, we directly compared the electrochemistry of E. coli and P. aeruginosa SoxR. Only weak cyclic voltammetry for E. coli SoxR is obtained, irrespective of the source, likely because of poor solubility. Therefore, the comparison was made using square wave voltammetry, which is a more sensitive technique and allows for better discrimination of small signals. As can be seen in Fig. 4, the potentials, referenced to the Redmond red internal standard, are nearly identical for P. aeruginosa SoxR and E. coli SoxR. This observation is consistent with the in vitro redox titrations of SoxR in the absence of DNA that found their potentials to differ from one another by ≈10 mV (7, 8, 15). Here, we see that the DNA-bound potentials are indistinguishable. The identical redox potentials of the free and DNA-bound E. coli and P. aeruginosa SoxR should allow both to activate transcription upon oxidation. In fact, previous work has shown that P. putida SoxR is functional and can complement an E. coli SoxR deletion mutant, lending credence to the in vivo importance of these observations (13).
Fig. 4.
Square wave voltammetry of P. aeruginosa SoxR (Left) and E. coli (Right) at DNA-modified graphite electrodes at a frequency of 15 Hz showing both the Redmond red and SoxR signals. The 5′ Redmond red-modified sequence was 5′-AGR GTA AAA CCT CAA GCA AAC TTG AGG TCA AGC CAA-3′ plus pyrene-modified complement, where “R” indicates the position of the probe.
Discussion
The activity of SoxR, a transcription factor containing an Fe-S cofactor, is regulated via a redox switch: SoxR triggers transcription in its oxidized state (4, 5). However, the redox potentials of free E. coli and P. aeruginosa SoxR have previously been determined to be approximately −290 mV in solution (7, 8, 15). Although the redox potential of SoxR can explain how it is maintained in its reduced state by coupling it to the cellular NADPH/NADP+ pool (−40 mV), it was unclear how the relatively low potential of −290 mV would allow for specificity in vivo. To understand the activation of SoxR at a mechanistic level, it is crucial to determine its redox potential within an appropriate context.
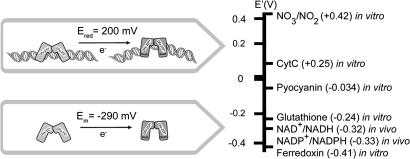
Here, using DNA-modified HOPG electrodes, we have demonstrated that DNA association positively shifts the redox potential of SoxR to 200 mV versus NHE. The +490-mV shift between the free and DNA-bound states of SoxR is functionally crucial because it keeps SoxR reduced across a range of intracellular potential. For example, Fig. 5 shows standard and free midpoint potentials for a variety of cellular redox pairs, and where DNA-bound and free SoxR are positioned along this series. Although numerous redox couples, ranging from glutathione to FADH, are oxidants for soluble SoxR, they are reductants to DNA-bound SoxR. In fact, the positive shift in potential associated with DNA binding means that DNA-bound SoxR is primarily in the reduced, transcriptionally silent form in vivo. Oxidative stress serves to promote oxidation of DNA-bound SoxR, activating the numerous genes required to protect the organism. This provides a rationale for how DNA-bound SoxR can serve as an effective sensor of oxidative stress in E. coli.
Fig. 5.
Redox potentials of free and DNA-bound SoxR along with those of cellular oxidants/reductants at pH 7.
In P. aeruginosa, the paradigm for SoxR activation may be different. Here, activation may be promoted by pyocyanin. When we consider the standard potential of the phenazine pyocyanin (Em = −34 mV at pH 7 and Em = −110 mV at pH 8), we predict it would also act as a reductant for DNA-bound SoxR (46). However, pyocyanin is an extracellular electron shuttle that reacts readily with oxygen, as indicated by the bright blue color of P. aeruginosa cultures. Uptake of oxidized pyocyanin increases the intracellular ratio of the oxidized versus the reduced form and thus favors the oxidation of SoxR. Considering a one-electron transfer under physiological conditions (pH 7 and 37°C), to shift the redox equilibrium of DNA-bound SoxR toward its oxidized state would require a ratio of oxidized to reduced pyocyanin of at least 6,500:1. It remains to be determined whether this is of physiological relevance in P. aeruginosa.
The substantial shift in SoxR potential on DNA binding of ≈500 mV is striking but understandable. The significance of the molecular environment for tuning the redox potentials of Fe-S clusters is well documented (47–49): Each hydrogen-bonding interaction with the cluster can cause a potential shift of ≈80 mV. Moreover, for [4Fe-4S] clusters in proteins, all with the same ligating residues, cluster potentials vary from approximately −600 mV for ferredoxins to approximately +400 mV for high potential iron proteins (50). We have previously observed a negative shift of at least ≈200 mV for Endo III in the presence of DNA (34). Because the structures of Endo III with and without DNA were known and showed no significant distortion in the protein (51–53), thermodynamically this shift was interpreted as a favorable shift in the binding affinity of the protein in the oxidized form relative to the reduced form (34), perhaps not so surprising on binding to the DNA polyanion.
By contrast, although the binding affinities of oxidized and reduced SoxR are comparable (6, 7), SoxR and other MerR-type transcriptional regulators have been shown to induce conformational changes of the promoter region (1, 54–59). In particular, copper phenanthroline footprinting studies have provided strong evidence that SoxR significantly distorts its promoter sequence (60, 61). Although this experiment only reports on the reduced form of SoxR, the observed loss of signal for Redmond red found here strongly supports a DNA-distortion mechanism. We propose that the more positive reduction potential for DNA-bound SoxR yields a higher energy complex, which may drive a conformational change in the protein/DNA complex. If this were the case, it would constitute an effective means of allosteric regulation in vivo.
It is important to note that the crystal structure of SoxR in any form has not been reported (62). Therefore, the structural difference between the free (low energy) and DNA-bound (high energy) complexes is not clear, but a positive shift of the magnitude we observe has been associated with bulk folding of other metalloproteins from P. aeruginosa (63). These energetic differences have been attributed to burying the cofactor in a more hydrophobic environment. Consequently, we predict a large structural difference between the free and DNA-bound SoxR to provide a rationale for the dramatic shift in potential associated with binding.
Within a broader context, these data illustrate that it is critical to take the effect of DNA binding into account when considering the redox characteristics of DNA binding proteins. It is also likely that it is the DNA-bound potential of these proteins that is most relevant within the crowded environment of the cell, and this potential may be altered even further upon recruitment of RNA polymerase. Therefore, in many cases, as with SoxR, it is perhaps the redox characteristics within a multiprotein/DNA complex that must be considered. In fact, because several transcription factors feature iron-sulfur clusters as sensor elements, a change in the redox potential of these cofactors upon binding DNA may generally be an important trait to consider.
Experimental Procedures
Materials.
All phosphoramidites and reagents for DNA synthesis were purchased from Glen Research. 1,6-Diaminohexane was obtained from Acros Organics. All organic solvents and other reagents were purchased from Aldrich in the highest available purity.
Oligonucleotide Synthesis.
Oligonucleotides were prepared using standard phosphoramidite chemistry on an ABI 394 DNA synthesizer. DNA was purified by HPLC on a reverse-phase C18 column with acetonitrile and ammonium acetate as eluents. The desired products were characterized by HPLC, UV-visible spectroscopy, and MALDI-TOF mass spectrometry. For experiments on HOPG, DNA was modified with pyrene at the 5′ terminus by following the procedure reported in ref. 27. In brief, oligonucleotides were prepared by solid phase synthesis using standard reagents with an unprotected hydroxyl group at the 5′ terminus. The 5′-OH was treated with a 120 mg/ml solution of carbonyldiimidazole in dioxane for 2 h followed by an 80 mg/ml solution of 1,6-diaminohexane for 30 min. Subsequently, the free amine was treated with 1-pyrenebutyric acid, N-hydroxysuccinimide ester, resulting in the desired pyrene moiety linked to the 5′ terminus. The oligonucleotides were deprotected with concentrated NH4OH at 60°C for 8 h.
DNA modified with Redmond red at the 3′ terminus or 3 bases in from the 5′ terminus was prepared according to the ultra-mild protocols outlined on the Glen Research web site (www.glenres.com). Pac-protected bases and ultra mild reagents were used. The oligonucleotides were deprotected in 0.05 M potassium carbonate in methanol at room temperature for 12–14 h to prevent degradation of the Redmond red moiety under harsh conditions.
Expression and Purification.
E. coli SoxR was prepared as described in ref. 64. N-terminally histidine-tagged SoxR from P. aeruginosa PA14 were expressed from plasmid pET16b in E. coli strain BL21 (DE3). Cells were grown in 1 liter of LB medium with 100 μg/ml ampicillin at 37°C. At an OD600nm of 0.3, protein expression was induced by the addition of 1 mM IPTG and the cultures were incubated for an additional 10 h at 16°C. All subsequent steps were performed at 4°C. Cells were pelleted, resuspended in buffer A [50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, 10% glycerol] containing 10 mM imidazole and PIC (protease inhibitor mixture without EDTA; Roche), and lysed using a French Press. The cell extract was centrifuged at 14,000 × g for 20 min. The supernatant was incubated with Talon-beads (Clontech) for 30 min and then transferred to a column. The beads were washed with buffer A containing 50 mM imidazole and PIC. Histidine-tagged SoxR was eluted from the column with buffer A containing 250 mM imidazole and PIC. Peak fractions and purity were determined by SDS/PAGE with Coomassie blue staining. Purified protein was dialyzed against SoxR storage buffer [50 mM Pi (pH 8.0), 500 mM NaCl, 20% glycerol].
To generate expression plasmid pET16b-soxR, soxR (PA14_35170) was PCR-amplified from genomic DNA of P. aeruginosa PA14 using primers A (CGC catatg AAG AAT TCC TGC GCA TC) and B (GGC gga tcc CTA GCC GTC GTG CTC G). Primer A contains an NdeI restriction site (small letters) and soxR's start codon (italicized). Primer B contains a BamHI site (small letters) and soxR's stop codon. The PCR fragment was ligated into NdeI/BamHI-digested pET16b.
Formation of DNA Monolayers and Electrochemical Measurements.
DNA films were self-assembled on SPI-1 grade HOPG electrodes (SPI) with an estimated surface area of 0.08 cm2 defined by an o-ring. Duplex DNA was formed in (pH 8) 50 mM Pi/500 mM NaCl/20% glycerol buffer (SoxR storage buffer) by combining equimolar amounts of the pyrene-modified strand with its complement. Loosely packed DNA monolayers were allowed to form over a period of 24–48 h. The electrodes were then thoroughly rinsed with SoxR storage buffer before being backfilled for 2–4 h with 10% by volume octane or decane solutions in SoxR storage buffer. The electrodes were then thoroughly rinsed with SoxR storage buffer again and moved into a nitrogen atmosphere for electrochemistry experiments.
Electrochemical data were collected with a Bioanalytical Systems CV-50W potentiostat using the inverted drop cell configuration. All measurements reported for the working electrode were taken versus a platinum (Pt) auxiliary and a silver/silver chloride (Ag/AgCl) reference. The Ag/AgCl reference was frequently standardized versus SCE, and all reported potentials have an experimental uncertainty of <40 mV. Electrochemical experiments were performed at ambient temperature and under an anaerobic atmosphere in SoxR storage buffer. In a typical experiment, background electrochemical scans were performed before SoxR was added to the storage buffer, resulting in an ≈15–35 μM monomer concentration within the cell. Further scans were then performed in the presence of SoxR, typically over a period of 30–45 min.
ACKNOWLEDGMENTS.
This work was supported by National Institutes of Health Grant GM61077 (to J.K.B.), the Howard Hughes Medical Institute (D.K.N.), and a European Molecular Biology Organization Long-Term Fellowship (to L.E.P.D.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Brown NL, Stoyanov JV, Kidd SP, Hobman JL. The MerR family of transcriptional regulators. FEMS Microbiol Rev. 2003;27:145–163. doi: 10.1016/S0168-6445(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 2.Hobman JL, Wilkie J, Brown NL. A design for life: Prokaryotic metal-binding MerR family regulators. Biometals. 2005;18:429–436. doi: 10.1007/s10534-005-3717-7. [DOI] [PubMed] [Google Scholar]
- 3.Giedroc DP, Arunkumar AI. Metal sensor proteins: Nature's metalloregulated allosteric switches. J Chem Soc Dalton Trans. 2007:3107–3120. doi: 10.1039/b706769k. [DOI] [PubMed] [Google Scholar]
- 4.Pomposiello PJ, Demple B. Redox-operated genetic switches: The SoxR and OxyR transcription factors. Trends Biotechnol. 2001;19:109–114. doi: 10.1016/s0167-7799(00)01542-0. [DOI] [PubMed] [Google Scholar]
- 5.Demple B, Ding H, Jorgensen M. Escherichia coli SoxR protein: Sensor/transducer of oxidative stress and nitric oxide. Methods Enzymol. 2002;348:355–364. doi: 10.1016/s0076-6879(02)48654-5. [DOI] [PubMed] [Google Scholar]
- 6.Hidalgo E, Demple B. An iron-sulfur center essential for transcriptional activation by the redox sensing SoxR protein. EMBO J. 1994;13:138–146. doi: 10.1002/j.1460-2075.1994.tb06243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaudu P, Weiss B. SoxR, a [2Fe-2S] transcription factor, is active only in its oxidized form. Proc Natl Acad Sci USA. 1996;93:10094–10098. doi: 10.1073/pnas.93.19.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding H, Hidalgo E, Demple B. The redox state of the [2Fe-2S] clusters in SoxR protein regulates its activity as a transcription factor. J Biol Chem. 1996;271:33173–33175. doi: 10.1074/jbc.271.52.33173. [DOI] [PubMed] [Google Scholar]
- 9.Ding H, Demple B. In vivo kinetics of a redox-regulated transcriptional switch. Proc Natl Acad Sci USA. 1997;94:8445–8449. doi: 10.1073/pnas.94.16.8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pomposiello J, Bennik MHJ, Demple B. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J Bacteriol. 2001;183:3890–3902. doi: 10.1128/JB.183.13.3890-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ha U, Jin S. Expression of the soxR gene of Pseudomonas aeruginosa is inducible during infection of burn wounds in mice and is required to cause efficient bacteremia. Infect Immun. 1999;67:5324–5331. doi: 10.1128/iai.67.10.5324-5331.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stover CK, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 13.Park W, Pena-Llopis S, Lee Y, Demple B. Regulation of superoxide stress in Pseudomonas putida KT2440 is different from the SoxR paradigm in Escherichia coli. Biochem Biophys Res Commun. 2006;341:51–56. doi: 10.1016/j.bbrc.2005.12.142. [DOI] [PubMed] [Google Scholar]
- 14.Dietrich LEP, Price-Whelan A, Petersen A, Whiteley M, Newman DK. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol Microbiol. 2006;61:1308–1321. doi: 10.1111/j.1365-2958.2006.05306.x. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi K, Tagawa S. Activation of SoxR-dependent transcription in Pseudomonas aeruginosa. J Biochem. 2004;136:607–615. doi: 10.1093/jb/mvh168. [DOI] [PubMed] [Google Scholar]
- 16.Zheng M, Storz G. Redox sensing by prokaryotic transcription factors. Biochem Pharmacol. 2000;59:1–6. doi: 10.1016/s0006-2952(99)00289-0. [DOI] [PubMed] [Google Scholar]
- 17.Liochev SI, Fridovich I. Fumarase C, the stable fumarase of Escherichia coli, is controlled by the soxRS regulon. Proc Natl Acad Sci USA. 1992;89:5892–5896. doi: 10.1073/pnas.89.13.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi K, Tagawa S. Isolation of reductase for SoxR that governs an oxidative response regulon from Escherichia coli. FEBS Lett. 1999;451:227–230. doi: 10.1016/s0014-5793(99)00565-7. [DOI] [PubMed] [Google Scholar]
- 19.Koo M-S, et al. A reducing system of the superoxide sensor SoxR in E. coli. EMBO J. 2003;22:2614–2622. doi: 10.1093/emboj/cdg252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang C, Lodish HF, Sinskey AJ. Measurement of glutathione redox state in cytosol and secretory pathway of cultured cells. Methods Enzymol. 1995;251:212–221. doi: 10.1016/0076-6879(95)51123-7. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi K, Tagawa S. A direct demonstration of reaction of superoxide anion with SoxR as studied by pulse radiolysis. J Inorg Biochem. 1997;67:258. [Google Scholar]
- 22.Liochev SI, Benov L, Touati D, Fridovich I. Induction of the SoxRS regulon of Escherichia coli by superoxide. J Biol Chem. 1999;274:9479–9481. doi: 10.1074/jbc.274.14.9479. [DOI] [PubMed] [Google Scholar]
- 23.Ding H, Demple B. Thiol-mediated disassembly and reassembly of [2Fe-2S] clusters in the redox-regulated transcription factor SoxR. Biochemistry. 1998;37:17280–17286. doi: 10.1021/bi980532g. [DOI] [PubMed] [Google Scholar]
- 24.Price-Whelan A, Dietrich LEP, Newman DK. Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189:6372–6381. doi: 10.1128/JB.00505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelley SO, Boon EM, Barton JK, Jackson NM, Hill MG. Single-base mismatch detection based on charge transduction through DNA. Nucleic Acids Res. 1999;27:4830–4837. doi: 10.1093/nar/27.24.4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boon EM, Ceres DM, Drummond TG, Hill MG, Barton JK. Mutation detection by electrocatalysis at DNA-modified electrodes. Nat Biotechnol. 2000;18:1096–1100. doi: 10.1038/80301. [DOI] [PubMed] [Google Scholar]
- 27.Gorodetsky AA, Barton JK. Electrochemistry using self-assembled DNA monolayers on highly oriented pyrolytic graphite. Langmuir. 2006;22:7917–7922. doi: 10.1021/la0611054. [DOI] [PubMed] [Google Scholar]
- 28.Kelley SO, Jackson NM, Hill MG, Barton JK. Long-range electron transfer through DNA films. Angew Chem Int Ed. 1999;38:941–945. doi: 10.1002/(SICI)1521-3773(19990401)38:7<941::AID-ANIE941>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 29.Inouye M, Ikeda R, Takase M, Tsuri T, Chiba J. Single-nucleotide polymorphism detection with “wire-like” DNA probes that display quasi “on–off” digital action. Proc Natl Acad Sci USA. 2005;102:11606–11610. doi: 10.1073/pnas.0502078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okamoto A, Kamei T, Saito I. DNA hole transport on an electrode: Application to effective photoelectrochemical SNP typing. J Am Chem Soc. 2006;128:658–662. doi: 10.1021/ja057040t. [DOI] [PubMed] [Google Scholar]
- 31.Gorodetsky AA, Green O, Yavin E, Barton JK. Coupling into the base pair stack is necessary for DNA-mediated electrochemistry. Bioconjugate Chem. 2007;18:1434–1441. doi: 10.1021/bc0700483. [DOI] [PubMed] [Google Scholar]
- 32.Boon EM, Livingston AL, Chmiel NH, David SS, Barton JK. DNA-mediated charge transport for DNA repair. Proc Natl Acad Sci USA. 2003;100:12543–12547. doi: 10.1073/pnas.2035257100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boal AK, et al. DNA-bound redox activity of DNA repair glycosylases containing [4Fe-4S] clusters. Biochemistry. 2005;44:8397–8407. doi: 10.1021/bi047494n. [DOI] [PubMed] [Google Scholar]
- 34.Gorodetsky AA, Boal AK, Barton JK. Direct electrochemistry of endonuclease III in the presence and absence of DNA. J Am Chem Soc. 2006;128:12082–12083. doi: 10.1021/ja064784d. [DOI] [PubMed] [Google Scholar]
- 35.Cunningham RP, et al. Endonuclease III is an iron-sulfur protein. Biochemistry. 1989;28:4450–4455. doi: 10.1021/bi00436a049. [DOI] [PubMed] [Google Scholar]
- 36.Asahara H, Wistort PM, Bank JF, Bakerian RH, Cunningham RP. Purification and characterization of Escherichia coli endonuclease III from the cloned nth gene. Biochemistry. 1989;28:4444–4449. doi: 10.1021/bi00436a048. [DOI] [PubMed] [Google Scholar]
- 37.Fu W, O'Handley S, Cunningham RP, Johnson MK. The role of the iron-sulfur cluster in Escherichia coli endonuclease III. A resonance Raman study. J Biol Chem. 1992;267:16135–16137. [PubMed] [Google Scholar]
- 38.Boal AK, Yavin E, Barton JK. DNA repair glycosylases with a [4Fe-4S] cluster: A redox cofactor for DNA-mediated charge transport? J Inorg Biochem. 2007;101:1913–1921. doi: 10.1016/j.jinorgbio.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stephens PJ, Jollie DR, Warshel A. Protein control of redox potentials of iron-sulfur proteins. Chem Rev. 1996;96:2491–2513. doi: 10.1021/cr950045w. [DOI] [PubMed] [Google Scholar]
- 40.Kim Y, Long EC, Barton JK, Lieber CM. Imaging of oligonucleotide-metal complexes by scanning tunneling microscopy. Langmuir. 1992;8:496–500. [Google Scholar]
- 41.Giancarlo LC, Flynn GW. Scanning tunneling and atomic force microscopy probes of self-assembled, physisorbed monolayers: Peeking at the peaks. Annu Rev Phys Chem. 1998;49:297–336. doi: 10.1146/annurev.physchem.49.1.297. [DOI] [PubMed] [Google Scholar]
- 42.Bard AJ, Faulkner LR. Electrochemical Methods. 2nd Ed. New York: Wiley; 2001. [Google Scholar]
- 43.Boon EM, Salas JE, Barton JK. An electrical probe of protein–DNA interactions on DNA-modified surfaces. Nat Biotechnol. 2002;20:282–286. doi: 10.1038/nbt0302-282. [DOI] [PubMed] [Google Scholar]
- 44.Udit AK, Hill MG, Bittner VG, Arnold FM, Gray HB. Reduction of dioxygen catalyzed by pyrene-wired heme domain cytochrome P450 BM3 electrodes. J Am Chem Soc. 2004;126:10218–10219. doi: 10.1021/ja0466560. [DOI] [PubMed] [Google Scholar]
- 45.Hidalgo E, Ding H, Demple B. Redox signal transduction via iron-sulfur clusters in the SoxR transcription activator. Trends Biochem Sci. 1997;22:207–210. doi: 10.1016/s0968-0004(97)01068-2. [DOI] [PubMed] [Google Scholar]
- 46.Price-Whelan A, Dietrich LEP, Newman DK. Rethinking “secondary” metabolism: Physiological roles for phenazine antibiotics. Nat Chem Biol. 2006;2:71–78. doi: 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- 47.Rusling JF. Enzyme bioelectrochemistry in cast biomembrane-like films. Acc Chem Res. 1998;31:363–369. [Google Scholar]
- 48.Carter CW, Jr, et al. A comparison of Fe4S4 clusters in high-potential iron protein and in ferredoxin. Proc Natl Acad Sci USA. 1972;69:3526–3529. doi: 10.1073/pnas.69.12.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heering HA, Bulsink YBM, Hagen WR, Meyer TE. Influence of charge and polarity on the redox potentials of high-potential iron-sulfur proteins: Evidence for the existence of two groups. Biochemistry. 1995;34:14675–14686. doi: 10.1021/bi00045a008. [DOI] [PubMed] [Google Scholar]
- 50.Beinert H. Iron-sulfur proteins: Ancient structures, still full of surprises. J Biol Inorg Chem. 2000;5:2–15. doi: 10.1007/s007750050002. [DOI] [PubMed] [Google Scholar]
- 51.Kuo CF, et al. Atomic structure of the DNA repair [4Fe-4S] enzyme endonuclease III. Science. 1992;258:434–440. doi: 10.1126/science.1411536. [DOI] [PubMed] [Google Scholar]
- 52.Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer JA. Novel DNA-binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fromme JC, Verdine GL. Structure of a trapped endonuclease III–DNA covalent intermediate. EMBO J. 2003;22:3461–3471. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ansari AZ, Bradner JE, O'Halloran TV. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature. 1995;374:371–375. doi: 10.1038/374370a0. [DOI] [PubMed] [Google Scholar]
- 55.Outten CE, Outten FW, O'Halloran TV. DNA distortion mechanism for transcriptional activation by ZntR, a Zn(II)-responsive MerR homologue in Escherichia coli. J Biol Chem. 1999;274:37517–37524. doi: 10.1074/jbc.274.53.37517. [DOI] [PubMed] [Google Scholar]
- 56.Zheleznova-Heldwein EE, Brennan RG. Crystal structure of the transcription activator BmrR bound to DNA, a drug. Nature. 2001;409:378–382. doi: 10.1038/35053138. [DOI] [PubMed] [Google Scholar]
- 57.Godsey MH, Baranova NN, Neyfakh AA, Brennan RG. Crystal structure of MtaN, a global multidrug transporter gene activator. J Biol Chem. 2001;276:47178–47184. doi: 10.1074/jbc.M105819200. [DOI] [PubMed] [Google Scholar]
- 58.Changela A, et al. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science. 2003;301:1383–1387. doi: 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- 59.Newberry KJ, Brennan RG. The structural mechanism for transcription activation by MerR family member multidrug transporter activation, N terminus. J Biol Chem. 2004;279:20356–20362. doi: 10.1074/jbc.M400960200. [DOI] [PubMed] [Google Scholar]
- 60.Hidalgo E, Bollinger JM, Jr, Bradley TM, Walsh CT, Demple B. Binuclear [2Fe-2S] clusters in the Escherichia coli SoxR protein and role of the metal centers in transcription. J Biol Chem. 1995;270:28908–28914. doi: 10.1074/jbc.270.36.20908. [DOI] [PubMed] [Google Scholar]
- 61.Hidalgo E, Demple B. Spacing of promoter elements regulates the basal expression of the soxS gene and converts SoxR from a transcriptional activator into a repressor. EMBO J. 1997;16:1056–1065. doi: 10.1093/emboj/16.5.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watanabe S, Kita A, Kobayashi K, Takahashi Y, Miki K. Crystallization and preliminary x-ray crystallographic studies of the oxidative-stress sensor SoxR and its complex with DNA. Act Crystallogr F. 2006;62:1275–1277. doi: 10.1107/S1744309106048482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wittung-Stafshede P, et al. Reduction potentials of blue and purple copper proteins in their unfolded states: A closer look at rack-induced coordination. J Biol Inorg Chem. 1998;3:367–370. [Google Scholar]
- 64.Chander M, Demple B. Functional analysis of SoxR residues affecting transduction of oxidative stress signals into gene expression. J Biol Chem. 2004;279:41603–41610. doi: 10.1074/jbc.M405512200. [DOI] [PubMed] [Google Scholar]