Abstract
We demonstrate a method for generating discretely structured protein nanotubes from the simple ring-shaped building block, homohexameric Hcp1 from Pseudomonas aeruginosa. Our design exploited the observation that the crystal lattice of Hcp1 contains rings stacked in a repeating head-to-tail pattern. High-resolution detail of the ring–ring interface allowed the selection of sites for specific cysteine mutations capable of engaging in disulfide bond formation across rings, thereby generating stable Hcp1 nanotubes. Protein nanotubes containing up to 25 subunits (≈100 nm in length) were self-assembled under simple conditions. Furthermore, we demonstrate that the tube ends and interior can be independently and specifically functionalized to generate nanocapsules.
Keywords: nanocapsule, protein engineering, dendrimer, disulfide, drug delivery
Proteins possess many properties that are valued in nanostructured materials: their function is template-encoded, they can self-assemble into complex supramolecular arrays, and they can mediate specific interactions with diverse substrates and other macromolecules. Protein nanotubes, in particular, hold the promise of ease of functionalization, intrinsic biocompatibility, and modular molecular recognition. These properties have been difficult to achieve with carbon or inorganically derived nanotubes.
A diverse group of protein nanotube systems has emerged in recent years. The bulk of research in this area makes use of naturally occurring protein scaffolds such as viral capsids (1–7), amyloid protein (8), actin (9), tubulin (10), pili (11), and flagella (12–15). Two studies—the engineering of tobacco mosaic virus to form an artificial light harvesting apparatus (7) and the use of a self-assembling amyloid protein fiber from Saccharomyces cerevisiae to template the synthesis of conducting nanowires (8)—demonstrate the diversity of applications for which these systems have been adapted. An alternative approach is to engineer tubular assemblies from proteins that have not evolved to form extended structures. Layer-by-layer assembly using nanoporous alumina templates and chemical crosslinking has successfully produced nanotubes composed of glucose oxidase (16) and cytochrome C (17).
Each protein nanotube system presents a characteristic set of strengths and weaknesses. Flagella, although very long, require in vivo assembly by a complex, multicomponent system, limiting the range of different components that can be included and precluding any preassembly chemical modification. Flagella and pili also lack an accessible, chemically derivatizable inner surface. Although the layer-by-layer chemical crosslinking strategy can likely be generalized to incorporate many proteins, this method does not offer any control over the nanostructure of the individual components within the tube.
Here, we report the synthesis of exceptionally structurally defined, nonhelical protein nanotubes composed of Hcp1 from Pseudomonas aeruginosa. Purified Hcp1 self-assembles into nanotubes in vitro, without the aid of assembly machinery. Hcp1 assembly is driven by steric and chemical complementarity and stabilized by engineered disulfide bonding, resulting in nanotubes that are remarkably stable yet can be readily disassembled. We show that the large inner cavity of Hcp1 nanotubes can be chemically modified and that the ends of Hcp1 nanotubes can be structurally differentiated. We exploited these features in the development of a reactive dendrimer “plug” that was capable of physically separating the interior of the Hcp1 nanotube from the exterior bulk environment, a critical first step toward generating Hcp1-based nanocapsules and nanoreactors.
Results
Design and Generation of Hcp1 Nanotubes.
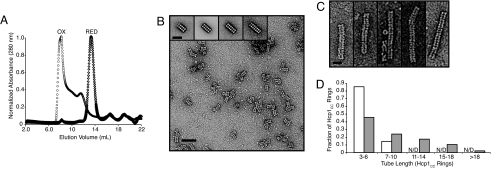
Hcp1 is a small (17.4-kDa) protein of unknown function that is a secreted component of the P. aeruginosa type VI secretion system (18–20). Recent x-ray crystallographic and electron micrographic analyses of Hcp1 [Protein Data Bank (PDB) ID code 1Y12] demonstrate that the protein adopts a homohexameric ring-shaped quaternary structure with an outer diameter of 9.0 nm, an inner diameter of 4.0 nm, and a height of 4.4 nm (Fig. 1A) (19). Subsequent detailed examination of the reported P6 crystal revealed that Hcp1 rings stack nonhelically, top to bottom, in extended parallel tubular structures to yield a honeycomb lattice (Fig. 1B). Although the biological significance of the Hcp1 crystal lattice is unknown, and solution studies and electron micrographs of the purified protein have provided no evidence of ring–ring association under physiological conditions, we hypothesized that this system could be exploited for the generation of defined protein nanotubes.
Fig. 1.
Structure-based design of Hcp1 nanotubes. (A) Overview of the Hcp1 hexameric ring structure. The six Hcp1 monomers are colored differently and shown in surface representation. Three monomers are excluded for the “inside” view. Key dimensions of the Hcp1 ring are given in nanometers. The “top” and “bottom” nomenclature is used within the text to highlight the polarity of the Hcp1 ring. Sites of cysteine substitutions used to generate the Hcp1 nanotubes (Arg-157 and Gly-90) are black, and the cysteine substitution site designed to introduce PAMAM-Mal (Q54) reactivity for “plugging” is white (Fig. 4). Table 1 provides a summary of Hcp1 amino acid substitutions and their utility. (B) Section of the Hcp1 honeycomb crystal lattice (PDB ID code 1Y12) viewed perpendicular to the crystallographic z-axis. Individual Hcp1 crystalline tubes are shown in black or gray to aid visualization. (C) Model of an Hcp1 nanotube containing 5 Hcp1CC ring subunits (gray) and the two capping subunits, G90C (green) and R157C (blue). The sites of each cysteine substitution (black), and the position of the C terminus on capping subunits (red) are indicated.
The strategy we chose for producing Hcp1 nanotubes uses engineered disulfide bonds to stabilize the crystallographic ring–ring interface. Molecular modeling of the crystallographic ring–ring interface led to the identification of Gly-90 and Arg-157 as sites on the top and bottom faces, respectively, for cysteine substitutions that could form ring-spanning disulfide bonds (Table 1 and Fig. 1). Criteria for the selection of these residues included their proximity and placement in secondary structural elements that were likely to tolerate substitution and allow conformations permissive of strict disulfide geometry.
Table 1.
Key attributes of Hcp1 mutants and nanotube subunits
| Hcp1 mutants | Utility |
|---|---|
| G90C, R157C (Hcp1CC) | Nanotube polymer subunit |
| G90C | Chain terminator; caps top end of Hcp1 nanotubes |
| R157C | Chain terminator; caps bottom end of Hcp1 nanotubes |
| G90C, Q54C | Top cap with plugging capacity |
| R157C, Q54C | Bottom cap with plugging capacity |
| G90C, C-terminal myc | Top cap; marks top end of Hcp1 nanotubes with myc tag |
| R157C, C-terminal FLAG | Bottom cap; marks bottom of Hcp1 nanotubes with FLAG tag |
The hcp1 Gly90Cys/Arg157Cys mutant (hcp1CC) and a wild-type control were generated using standard molecular biological techniques, expressed as His6-tagged proteins heterologously in Escherichia coli, and purified by Ni2+ chromatography to >95% homogeneity in a buffer containing 2 mM β-mercaptoethanol. After purification, the proteins were analyzed by size exclusion chromatography (SEC). As reported previously, the wild-type protein eluted in a single peak at a volume consistent with a homohexameric species (112 kDa). On the contrary, Hcp1CC eluted over a broad range that extended from the homohexameric elution point to the column void volume (>400 kDa) (Fig. 2A). To test whether the apparent oligomerization of Hcp1CC was mediated by the engineered disulfides, we repeated the SEC in the presence of excess reducing agent (5 mM DTT). Under these strongly reducing conditions, the higher mass species were not observed and the mutant protein eluted similarly to wild type (Fig. 2A). These data suggest that Hcp1CC is present in a hexameric form under reducing conditions and that these hexamers associate into high-order structures as a result of disulfide bond formation.
Fig. 2.
Characterization and optimization of Hcp1 nanotubes. (A) SEC of purified Hcp1CC under oxidizing (circles) and reducing (diamonds) conditions. The oxidized sample peak eluted at the column void volume (7 ml), whereas the reduced sample peak coeluted with wild-type Hcp1 at 14 ml (wild-type data not shown). (B) Negative stain TEM and single-particle analysis of Hcp1CC from the void volume fraction of the SEC in (A). (Scale bar, 45 nm.) (Inset) Averaged structures for Hcp1 nanotubes containing 4–7 ring subunits. (Scale bar, 20 nm.) (C) Electron micrographs of representative Hcp1 nanotubes prepared in vitro from completely reduced subunits. (Scale bar, 15 nm.) (D) Length distribution of Hcp1 nanotubes assembled in vivo (white) or in vitro (gray), expressed as the percentage of rings found in tubes of various length intervals. For both assembly methods, the population of tubes of each length (three or more rings) observed in representative transmission electron micrographs was counted (in vivo, n = 190; in vitro, n = 287). The sum of tubes of each length was multiplied by its respective length to yield the number of rings associated with each observed tube length. These subtotals are represented as percentages of the total number of rings observed in all tubes. Tubes with lengths >10 rings were not detected (N/D) in the in vivo assemblies.
To further characterize the oligomeric states of Hcp1CC suggested by SEC, we analyzed selected elution fractions by negative stain transmission electron microscopy (TEM). Fig. 2B shows a representative image of early-eluting Hcp1CC that illustrates the predominant topological arrangement observed: extended, roughly linear structures composed of regular repeating units [supporting information (SI) Fig. 5]. Reduction of early-eluting samples disrupted the extended structures and produced ring-shaped particles indistinguishable from wild-type Hcp1 (SI Fig. 6). These observations provided initial evidence that Hcp1CC disulfide bonds specifically promote ring–ring association into tubular structures.
To gain greater structural insight into the observed Hcp1CC oligomers, we performed image averaging of manually picked particles composed of 4–7 repeating units (Fig. 2B, inset). In each group, averaged images showed the repeating unit had an hourglass shape with dimensions of 4.1 × 9.9 nm, with the long axis of the unit oriented perpendicular to the long axis of the particle. The dimensions and morphology of these Hcp1CC oligomers are consistent with those of wild-type Hcp1 tubes found in the P6 crystal lattice (Fig. 1C). Based on this congruence, our modeling of the Hcp1 crystal structure, and the sensitivity of these structures to reducing agents, we conclude that the engineered cysteine substitutions successfully stabilize the ring–ring interface found in the P6 crystal lattice.
Nanotube Length Optimization and Chain Termination.
Although in vivo assembly of Hcp1CC into higher-order species was encouraging, the oligomers were limited in length and included undesired structurally heterogeneous impurities (Fig. 2B). These shortcomings, combined with the constraints on subunit preparation and composition imposed by the in vivo assembly system, led us to seek an improved in vitro assembly method. Starting with fully reduced Hcp1CC hexamers, we used dialysis to exchange the protein into solutions of various ionic strength, pH, redox potential, and chaotropic reagents and screened for those that promoted Hcp1CC tubular oligomerization. These experiments showed that superior Hcp1 nanotubes, as judged by length and a lack of nonspecific aggregation, could be produced under conditions similar in composition yet milder than those used for crystallization of wild-type Hcp1: a solution containing 5% (wt/vol) PEG3350, 100 mM Hepes (pH 8.0), 50 mM trisodium citrate, and 2 mM β-mercaptoethanol. Optimal results were found when fully reduced Hcp1CC (at a concentration of 50–75 mg/ml) was exchanged into these conditions by dialysis over a multiday time course. This process produced a smooth white paste that was subsequently dissolved by dilution and sonication. This procedure markedly enriched the population of longer (>10-subunit) soluble protein nanotubes (Fig. 2C and SI Fig. 7). To quantify the difference in tube length distribution produced by the in vitro vs. the in vivo assembly method, we enumerated the population of nanotube lengths in representative TEM images of each sample (Fig. 2D). These data confirmed that in vitro assembly of Hcp1 nanotubes yields substantially longer tubes than in vivo assembly.
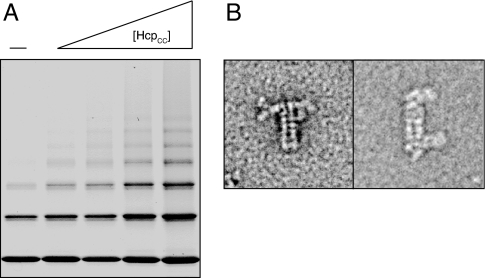
With an in vitro assembly method in hand, we next attempted to control Hcp1 oligomerization by chain termination. We hypothesized that two additional Hcp1 proteins, each containing either Gly90Cys or Arg157Cys, when included in the in vitro assembly system, would incorporate into the ends of Hcp1 nanotubes and terminate tube growth (Table 1). These single-cysteine mutants were cloned, expressed, and purified as above. Dialysis experiments to generate nanotubes were conducted with varying stoichimetries of single-cysteine and double-cysteine subunits. The extent of Hcp1 oligomerization was monitored by TEM (data not shown), and the ensemble of nanotube lengths was measured by nonreducing SDS/PAGE (Fig. 3A). As expected, both methods showed that increasing the proportion of Hcp1CC led to longer overall Hcp1 nanotubes. Thus, Hcp1 nanotube length can be controlled in vitro by the addition of specific chain-terminating subunits.
Fig. 3.
Chain termination and end differentiation of Hcp1 nanotubes. (A) Nonreducing SDS/PAGE analysis of capping subunits (–) or capping subunits containing various concentrations of Hcp1CC (total capping subunits:Hcp1CC, 3:1, 5:4, 2:3, 3:8) and the same total protein concentration. (B) TEM micrographs illustrating single-ended (Left) and double-ended (Right) antibody labeling of Hcp1 nanotubes capped with epitope-tagged subunits.
Nanotube End Differentiation.
A particularly desirable feature of a nanotube system is the ability to differentially distinguish the tube ends. The results of the chain termination experiments described above suggested that single-cysteine chain terminator subunits endow the Hcp1 protein nanotube with this property (Table 1). To test this, we cloned, expressed, and purified two epitope-tagged terminator subunits that could be incorporated into Hcp1 nanotubes and detected by immuno-TEM: Gly90Cys-myc and Arg157Cys-FLAG. These epitope tag sequences were introduced into the C terminus of the Hcp1 expression construct after the native sequence and immediately before the His6 tag. In the crystal structure of Hcp1, the C terminus of the protein was disordered and extended into solution from the exterior face of the hexameric ring (Fig. 1C, red). We reasoned that epitope tags located at these positions would allow unobstructed access to antibodies. When Hcp1 nanotubes were prepared with the epitope-tagged terminator subunits and exposed to antibodies specific to either the FLAG or myc epitopes, tubes were labeled exclusively at their ends (Fig. 3B and SI Fig. 8) This observation confirmed that the epitope-tagged single-cysteine mutants indeed terminate Hcp1 nanotube growth. Hcp1 nanotubes labeled with antibody at both ends were not observed when the anti-FLAG antibody was applied alone; however, doubly end-labeled tubes were observed when both antibodies were applied together (Fig. 3C and SI Fig. 9). These data show that each single-cysteine terminator subunit only incorporates into one end of the Hcp1 nanotube. This further confirms that the disulfide-stabilized ring–ring association observed in Hcp1 nanotubes reproduces the polar head-to-tail stacking observed in the wild-type crystal lattice. Thus, terminating subunits can be used to distinguish tube ends in the Hcp1 nanotube system.
Design of an Hcp1-Reactive Dendrimer “Plug.”
Biological nanoscale capsules are potential vectors for drug delivery, vessels for nanoscale reaction containment, and scaffolds for a variety of other biotechnology applications (21, 22). As a first step toward the generation of Hcp1 nanocapsules, we sought a method for physically separating the interior of Hcp1 nanotubes from their external environment. To accomplish this, we chose to introduce a reactive site into the chain-terminating subunits that could covalently trap a macromolecular “plug” capable of occluding the interior space. Examining the crystal structure of Hcp1, we identified Gln-54 as a location for the introduction of chemical reactivity (Table 1 and Fig. 1A). This site is located on the interior vertical face of the Hcp1 hexameric ring on a small loop that we anticipated would tolerate mutation. Additionally, the site is inaccessible to the engineered ring-linking cysteine residues (Fig. 1A). For the plug component of the Hcp1 nanocapsules, we selected the widely used, commercially available polyamidoamine (PAMAM) dendrimer. We chose thiol-maleimide chemistry as a means to generate a specific link between the dendrimer and the protein. To this end, Gln-54 was mutated to cysteine and the PAMAM dendrimer was perfunctionalized with maleimide groups.
To functionalize PAMAM with maleimide groups, we reacted a generation 2 PAMAM (G2 PAMAM) containing 16 amino termini with stoichiometric quantities (relative to the amino groups) of the heterobifunctional linker maleimidopropionic acid N-hydroxysuccinimide ester (MPANHS), producing G2 PAMAM-Mal16. The extent of this reaction was confirmed by MALDI-MS (SI Figs. 10 and 11). After purification by SEC, G2 PAMAM-Mal16 was reacted with Hcp1 Gln54Cys and wild-type Hcp1 (see Materials and Methods). Analysis of the products of these reactions by SDS/PAGE showed a ladder of six bands of higher mass than monomeric Hcp1 present only in the reaction containing Hcp1 Gln54Cys and G2 PAMAM-Mal16 (Fig. 4A). We interpret this result as evidence of specific reactivity of hexameric rings of Gln54Cys with G2 PAMAM-Mal16 (Fig. 4A, schematic). Importantly, wild-type Hcp1, which contains a buried cysteine residue (Cys-73), does not react with G2 PAMAM-Mal16 under these conditions.
Fig. 4.
PAMAM-Mal dendrimers react specifically and efficiently with Hcp1 Q54C. (A) SDS/PAGE analysis of Hcp1 Q54C and wild-type Hcp1 (rings in schematic) model reactions with G2 PAMAM-Mal16 (filled sphere in schematic). (B) Optimization of the PAMAM-Mal dendrimer system for reaction with Hcp1 Q54C. Values in parentheses indicate the number of Hcp1 Q54C monomers reacted with dendrimer. (C) PAMAM-Mal reacts inside the central hole of Hcp1 Q54C. TEM micrographs of Hcp1 (control) and Hcp1 Q54C G3-PAMAM-Mal32. (D) Comparison of thiol Sepharose reactivity of Hcp1 wild-type and Hcp1 proteins containing surface (R157C) and inner (Q54C) cysteine substitutions. After incubation, equal quantities of thiol Sepharose bead-associated (B) and unreacted flow-although (F) were analyzed by SDS/PAGE. (E) G3 PAMAM-Mal16 reaction with Hcp1 nanotubes containing the Q54C capping subunits (total capping subunits:Hcp1CC, 3:2).
The SDS/PAGE band pattern of Hcp1 Gln54Cys-G2 PAMAM-Mal16 suggested that the dendrimer did not react completely with all six of the engineered cysteine residues available on the inner surface of each Hcp1 Gln54Cys hexameric ring. We reasoned that the radius of the G2-Mal16 dendrimer may be insufficient to span the ≈4-nm Hcp1 interior diameter. To obtain more a complete dendrimer–ring reaction, we next performed experiments with a larger dendrimer starting material, G3 PAMAM. The shape of this dendrimer is approximately spherical, with a diameter of 3 nm and 32 terminal amino groups (23). We prepared G3 PAMAM derivatized with 8, 16, or 32 maleimide groups and tested the reactivity of these dendrimers with Hcp1 Gln54Cys. SDS/PAGE analysis of these reactions showed a clear correlation between the number of maleimide groups on the dendrimer and the extent of reaction with the cysteine residues (Fig. 4B). The improved reactivity of the G3 dendrimers is also attributable to their greater diameter. A direct comparison of equally maleimide-derivatized G2 and G3 dendrimers showed a marked enrichment for the more reacted product species when the larger G3 dendrimer was used (Fig. 4B).
To ensure that the reactive dendrimer did not grossly affect the ring structure of Hcp1 Gln54Cys, and to confirm the localization of the dendrimer to the inner cavity of the ring, we compared TEM images of Hcp1 Gln54Cys-G3 PAMAM-Mal32 and unreacted Hcp1 (Fig. 4C). Although the overall ring structure and rough dimensions of unreacted Hcp1 were maintained in Hcp1 Gln54Cys-G3 PAMAM-Mal32, the dendrimer-reacted rings displayed a smaller and off-center hole. This decrease in area of the hole in Hcp1 Gln54Cys-G3 PAMAM-Mal32 is consistent with the additional mass of the dendrimer localizing to the region. The off-center appearance of G3 PAMAM-Mal32 in the Hcp1 Gln54Cys hole is likely attributable to a proximity effect: tethering of the dendrimer to the initial reaction site favors secondary reactions near that site. Once these reactions take place, which could involve any branch of the dendrimer, the center-of-mass of the dendrimer becomes constrained and the remaining free branches must extend to react with distal Hcp1 monomers. Evidence for this effect is found in Fig. 4B, which shows that despite ample maleimide reactivity (32 sites) and a sufficiently wide radius (> 4 nm including linkers), asymmetric reaction of the dendrimer with Hcp1 Gln54Cys results in ≈50% of the dendrimers being unable to reach all six subunits of the hexamer.
Nanotube Plugging for Capsule Formation.
The model reactions of G3 PAMAM-Mal with Hcp1 Gln54Cys suggested that this dendrimer could effectively plug the ends of appropriately terminated Hcp1 nanotubes, thus producing nanocapsules. To extend the model ring-plugging reaction into the Hcp1 nanotube system, we introduced the Gln54Cys mutation into the Hcp1 Gly90Cys and Hcp1 Arg157Cys chain terminator subunits (Table 1). A concern in using the thiol-reactive dendrimer as the tube plug is the potential for side reactions of the dendrimer with free rings or unterminated tube fragments. These reaction contaminants contain highly accessible free thiols (Gly90Cys and Arg157Cys), whereas the desired product only contains free thiols at the relatively inaccessible, inward facing position Cys-54. Based on these structural constraints, we reasoned that the undesired products could be selectively removed by capture onto activated thiol Sepharose resin. This hypothesis was tested on model subunits either containing no surface cysteine residues (wild-type) or containing Cys-54 or Cys-157. The results of this experiment confirmed our expectation that Cys-54 is sufficiently inaccessible to avoid capture by the resin, whereas the outer surface-exposed Cys-157 is efficiently retained (Fig. 4D). Employing this technique facilitated the preparation of relatively pure samples of Cys-54-containing chain-terminated Hcp1 nanotubes.
The final step in the construction of Hcp1 nanocapsules was to demonstrate that the dendrimer could successfully react with intact Cys-54-containing chain-terminated Hcp1 nanotubes. G3 PAMAM-Mal16 was reacted with thiol Sepharose-purified, Cys-54-terminated Hcp1 nanotubes in reaction conditions similar to the model reactions described above. The desired product of this reaction contains two key linkages: thioether crosslinks between the PAMAM dendrimer and the Cys-54-containing chain-terminating protein subunits and disulfide bonds between the protein subunits that constitute the tubes. Of these two linkages, only the disulfide bonds between Hcp1 subunits are susceptible to thiol-based reducing agents such as DTT. Evidence for the successful reaction of the dendrimer with the Hcp1 nanotubes can be seen in the distinctive population of high-mass species present in lane 3 of Fig. 4E. Unlike the high-mass bands seen in the sample without dendrimer added (lanes 1 and 2), these bands are not completely eliminated under reducing conditions (lane 4). The partial susceptibility to reducing agents of the high-mass bands in lane 3 implies that these bands represent the cumulative effect of DTT-insensitive dendrimer–protein thioether bonds on the terminator subunits and DTT-sensitive protein–protein disulfide bonds between the dendrimer-plugged terminator subunits and the Hcp1CC rings.
Discussion
By engineering the quaternary structure of Hcp1, we have added a new, higher level of organization to the system: supramolecular assembly into extended tubes. The hierarchical self-assembly of these Hcp1 nanotubes allows precise control of the positioning of moieties in space, a desirable property that is difficult to achieve at the nanoscale (24). We exploited the self-assembly features of the system to combine distinct Hcp1 subunits in vitro, including chain-terminating subunits. Furthermore, we show that by varying the concentration of these chain-terminating subunits relative to the chain-extending double mutant, we can control the extent of polymerization and thereby nanotube length. Perhaps most intriguingly, we show that the polarity of Hcp1 nanotubes enables differentiation of the tube ends by the incorporation of unique capping subunits. Finally, by using a third engineered cysteine residue and a thiol-reactive dendrimer plug molecule, we provided the first steps in the development of a nanoencapsulation system.
Hcp1 nanotubes have several features that distinguish them from other protein nanotube systems. Many of the unique features of Hcp1 nanotubes stem from the building block Hcp1. Hcp1 is a superb scaffold for engineering. It is small, easily expressed in large quantities, tolerant of amino acid substitutions, and structurally characterized (in its tubular array) to atomic resolution. Furthermore, the N and C termini of Hcp1 are ideally positioned in the nanotube for placing fusions inside and outside the structure, respectively. Hcp1 nanotubes also have a large interior volume relative to their mass and are not viral in origin. Although Hcp1 is a component of a secretion system that has been linked to virulence in many bacteria, Hcp1 itself is likely not toxic to humans, because it appears to be a structural subunit rather than a toxin of the secretion system (19, 25–27).
The ring–ring disulfide linkage endows Hcp1 nanotubes with two distinctive properties: covalent strength and redox control of assembly and disassembly. The stability of the Hcp1 nanotube disulfide linkage is attested to by our observation that the nanotubes did not show signs of degradation over weeks of storage at room temperature. The redox control of assembly and disassembly is important when considering the potential utility of Hcp1 nanocapsules, because this feature would provide a release mechanism for the nanocapsule contents once in contact with the reducing environment of the cytoplasm.
Hcp1 nanotubes can be formed in vitro without the need for biological assembly machinery. This feature allows for a wide spectrum of specific functionalities to be introduced at two stages: in the primary sequence before production and by chemical modification before assembly. Mixtures of subunits with distinct sequences and chemical modifications can create a powerful combinatorial array of Hcp1 nanotubes. In vitro assembly is particularly valuable for the construction of nanocapsules, because it facilitates chemical modification both before and after capsule assembly. For instance, cargo could be covalently linked to the interior of free Hcp1 rings before incorporation, and external modifications could be performed after nanotube assembly. The intersection of so many desirable qualities in a single, simple protein building block makes Hcp1 a uniquely powerful platform for future bionanoengineering.
Materials and Methods
Plasmid Construction.
P. aeruginosa hcp1 (PA0085) was cloned into the pET29b vector (Novagen) by using the XhoI and NdeI cut sites to produce a C-terminally His6-tagged Hcp1 protein. The G90C, R157C and Q54C mutations and combinations thereof were introduced via PCR amplification of mutant plasmid followed by DpnI degradation of the wild-type plasmid. FLAG and Myc tags preceded by a Ser, Gly, Ala linker sequence were introduced before the pET29b His6 tag by ligation of annealed oligos into the XhoI cut site. All constructs were confirmed by sequencing.
Protein Expression and Purification.
E. coli BL21 DE3 were grown with shaking in 2YT medium at 37°C until the OD600 reached 0.5, at which point Hcp1 overexpression was induced with the addition of IPTG to 500 μM. Expression was allowed to continue at 37°C for 4–5 h. Bacteria were harvested by centrifugation and resuspended in 25 ml of buffer A (500 mM NaCl, 50 mM Tris 7.5, 10% glycerol) plus 2 mM β-mercaptoethanol and 10 μg/ml lysozyme per liter of culture volume. The bacteria were lysed by sonication, and the crude lysate was clarified by centrifugation and loaded directly onto a 1- or 5-ml HisTrap FF column (GE Health Sciences). Hcp1 was eluted with a 30–300 mM imidazole gradient in the same buffer. The final step of purification was SEC on a Sephadex 200 16/60 column (GE Health Sciences) in buffer A. Analytical chromatography was performed on a Superdex 200 10/300 column (GE Health Sciences). Samples were run in buffer A with or without the addition of 5 mM DTT.
Electron Microscopy and Image Processing.
Samples were prepared for TEM analysis by dilution in buffer B [200 mM NaCl, 20 mM Tris (pH 7.5), 2% glycerol]. Because the particle density of a given sample depends on the extent of oligomerization, appropriate dilutions were determined empirically for each sample. For free hexameric Hcp1, a final concentration of 50 μg/ml (0.45 μM) produced a suitable distribution of particles on the grid. All samples were negatively stained by uranyl formate following the established protocol of Ohi et al. and imaged at room temperature in a Tecnai T20 microscope (FEI) operated at 120 kV at a magnification of ×50,000 (28). Images were recorded on a 4K × 4K UltraScan CCD camera (Gatan Inc.) with a defocus of −1.5 μm. All images were binned by a factor of 2 to a final pixel size of 4.5 A per pixel at specimen level. A total of 375 tubes of different length were manually selected by using program Ximdisp, the display program associated with the MRC program suite, from a total of 104 CCD images (29). Image processing was performed using SPIDER (30). All particles were windowed into 120 × 120 pixel images and treated with standard multireference alignment and classification protocols specifying 10 classes. For antibody labeling, monoclonal M2 anti-FLAG antibody (Sigma) and polyclonal rabbit anti-myc antibody (Abcam) were mixed to a final concentration of 3.3 μg/ml with myc- and/or FLAG- tagged Hcp1 nanotubes (75 μg/ml) in a 1:1 mixture of buffer A and buffer B and incubated at 4°C overnight, followed by staining and imaging as above.
Nanotube Generation.
Purified Hcp1 at a concentration of 50–75 mg/ml was treated with 4 mM DTT to fully reduce all subunits. Protein (either pure Hcp1CC or mixtures with capping subunits) was dialyzed into a buffer containing 5% PEG 3350, 50 mM trisodium citrate, 100 mM Hepes (pH 7.5), and 2 mM β-mercaptoethanol until a smooth paste formed (2–4 days). This paste was diluted 50- to 100-fold in buffer A, followed by microtip sonication for 10–20 s or until the solution appeared clear.
Dendrimer Reactions.
Generation 2 and Generation 3 ethylenediamine core PAMAM dendrimers (Sigma) were reacted with 3-(maleimido)propionic acid N-succinimidyl ester (MPANHS). Typical reactions used 2.5 μmol of dendrimer diluted in 300 μl of DMSO plus 600 μl of H2O. Stoichiometric amounts of MPANHS were dissolved in 0.6–1.0 ml of DMSO and added drop-wise with intermittent vortexing to the PAMAM/DMSO/H2O solution. The reaction was allowed to proceed for 5 min with vortexing before dilution to a 2.5-ml total volume with H2O and purification on PD-10 column (GE Health Sciences). The starting materials and purified reaction products were characterized by MALDI-TOF MS using a 2-(4-hydroxy-phenylazo)-benzoic acid matrix (SI Figs. 10 and 11). Hcp1 protein rings and tubes (in buffer A) were mixed with an equal volume of reactive dendrimers (in water) at an ≈1:10 molar ratio of rings:dendrimers. The reaction was allowed to proceed for 20 min at room temperature.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Paul Ashby for assistance with atomic force microscopy experiments, Christopher Petzold for assistance with mass spectrometry, Melike Firat for technical assistance, Kamil Godula and Ramesh Jasti for chemistry advice, John Mekalanos for valuable discussions, Scott Classen and Juergen Bosch for assistance with molecular modeling, and members of the Biological Nanostructures facility at the Molecular Foundry for their support. This work was supported in part by a grant from the Sandler Family Supporting Foundation (Sandler Program in Basic Science) (Y.C.). The work was performed in part at the Molecular Foundry, Lawrence Berkeley National Laboratory, with support from the Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
Conflict of interest statement: A preliminary patent detailing these findings has been filed.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712247105/DC1.
References
- 1.Lee SW, Mao C, Flynn CE, Belcher AM. Ordering of quantum dots using genetically engineered viruses. Science. 2002;296:892–895. doi: 10.1126/science.1068054. [DOI] [PubMed] [Google Scholar]
- 2.Mao C, et al. Virus-based toolkit for the directed synthesis of magnetic and semiconducting nanowires. Science. 2004;303:213–217. doi: 10.1126/science.1092740. [DOI] [PubMed] [Google Scholar]
- 3.Huang Y, et al. Programmable assembly of nanoarchitectures using genetically engineered viruses. Nano Lett. 2005;5:1429–1434. doi: 10.1021/nl050795d. [DOI] [PubMed] [Google Scholar]
- 4.Nam KT, et al. Virus-enabled synthesis and assembly of nanowires for lithium ion battery electrodes. Science. 2006;312:885–888. doi: 10.1126/science.1122716. [DOI] [PubMed] [Google Scholar]
- 5.Mukherjee S, Pfeifer CM, Johnson JM, Liu J, Zlotnick A. Redirecting the coat protein of a spherical virus to assemble into tubular nanostructures. J Am Chem Soc. 2006;128:2538–2539. doi: 10.1021/ja056656f. [DOI] [PubMed] [Google Scholar]
- 6.Tsukamoto R, Muraoka M, Seki M, Tabata H, Yamashita I. Synthesis of CoPt and FePt3 nanowires using the central channel of tobacco mosaic virus as a biotemplate. Chem Mater. 2007;19:2389–2391. [Google Scholar]
- 7.Miller RA, Presley AD, Francis MB. Self-assembling light-harvesting systems from synthetically modified tobacco mosaic virus coat proteins. J Am Chem Soc. 2007;129:3104–3109. doi: 10.1021/ja063887t. [DOI] [PubMed] [Google Scholar]
- 8.Scheibel T, et al. Conducting nanowires built by controlled self-assembly of amyloid fibers and selective metal deposition. Proc Natl Acad Sci USA. 2003;100:4527–4532. doi: 10.1073/pnas.0431081100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong GC, et al. Hierarchical self-assembly of F-actin and cationic lipid complexes: Stacked three-layer tubule networks. Science. 2000;288:2035–2039. doi: 10.1126/science.288.5473.2035. [DOI] [PubMed] [Google Scholar]
- 10.Raviv U, et al. Cationic liposome-microtubule complexes: Pathways to the formation of two-state lipid-protein nanotubes with open or closed ends. Proc Natl Acad Sci USA. 2005;102:11167–11172. doi: 10.1073/pnas.0502183102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Audette GF, vanSchaik EJ, Hazes B, Irvin RT. DNA-binding protein nanotubes: Learning from nature's nanotech examples. Nano Lett. 2004;4:1897–1902. [Google Scholar]
- 12.Kumara MT, Srividya N, Muralidharan S, Tripp BC. Bioengineered flagella protein nanotubes with cysteine loops: Self-assembly and manipulation in an optical trap. Nano Lett. 2006;6:2121–2129. doi: 10.1021/nl060598u. [DOI] [PubMed] [Google Scholar]
- 13.Kumara MT, Tripp BC, Muralidharan S. Self-assembly of metal nanoparticles and nanotubes on bioengineered flagella scaffolds. Chem Mater. 2007;19:2056–2064. [Google Scholar]
- 14.Kumara MT, Tripp BC, Muralidharan S. Exciton energy transfer in self-assembled quantum dots on bioengineered bacterial flagella nanotubes. J Phys Chem C. 2007;111:5276–5280. [Google Scholar]
- 15.Woods RD, et al. Bifunctional nanotube scaffolds for diverse ligands are purified simply from Escherichia coli strains coexpressing two functionalized flagellar genes. Nano Lett. 2007;7:1809–1816. doi: 10.1021/nl0702968. [DOI] [PubMed] [Google Scholar]
- 16.Hou S, Wang J, Martin CR. Template-synthesized protein nanotubes. Nano Lett. 2005;5:231–234. doi: 10.1021/nl048305p. [DOI] [PubMed] [Google Scholar]
- 17.Tian Y, He Q, Cui Y, Li J. Fabrication of protein nanotubes based on layer-by-layer assembly. Biomacromolecules. 2006;7:2539–2542. doi: 10.1021/bm060412l. [DOI] [PubMed] [Google Scholar]
- 18.Economou A, et al. Secretion by numbers: Protein traffic in prokaryotes. Molecular microbiology. 2006;62:308–319. doi: 10.1111/j.1365-2958.2006.05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mougous JD, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yahr TL. A critical new pathway for toxin secretion? N Engl J Med. 2006;355:1171–1172. doi: 10.1056/NEJMcibr063931. [DOI] [PubMed] [Google Scholar]
- 21.Uchida M, et al. Biological containers: Protein cages as multifunctional nanoplatforms. Adv Mater. 2007;19:1025–1042. [Google Scholar]
- 22.Pinto Reis C, Neufeld RJ, Ribeiro AJ, Veiga F. Nanoencapsulation II. Biomedical applications and current status of peptide and protein nanoparticulate delivery systems. Nanomedicine. 2006;2:53–65. doi: 10.1016/j.nano.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 23.Esfand R, Tomalia DA. Poly(amidoamine) (PAMAM) dendrimers: From biomimicry to drug delivery and biomedical applications. Drug Discov Today. 2001;6:427–436. doi: 10.1016/s1359-6446(01)01757-3. [DOI] [PubMed] [Google Scholar]
- 24.Elemans JAW, Rowan AE, Nolte RJM. Mastering molecular matter: Supramolecular architectures by hierarchical self-assembly. J Mater Chem. 2003;13:2661–2670. [Google Scholar]
- 25.Kulasekara HD, Miller SI. Threonine phosphorylation times bacterial secretion. Nat Cell Biol. 2007;9:734–736. doi: 10.1038/ncb0707-734. [DOI] [PubMed] [Google Scholar]
- 26.Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol. 2007;9:797–803. doi: 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- 27.Schell MA, et al. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol Microbiol. 2007;64:1466–1485. doi: 10.1111/j.1365-2958.2007.05734.x. [DOI] [PubMed] [Google Scholar]
- 28.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification - Powerful tools in modern electron microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crowther RA, Henderson R, Smith JM. MRC image processing programs. J Struct Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 30.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.