Abstract
An off-line mass spectrometry method that combines immuno-spin trapping and chromatographic procedures has been developed for selective detection of the nitrone spin trap 5,5-dimethyl-1-pyrroline-N-oxide (DMPO) covalently attached to proteins, an attachment which occurs only subsequent to DMPO trapping of free radicals. In this technique, the protein-DMPO nitrone adducts are digested to peptides with proteolytic agents, peptides from the enzymatic digest are separated by HPLC, and enzyme-linked immunosorbent assays (ELISA) using polyclonal anti-DMPO nitrone antiserum are used to detect the eluted HPLC fractions that contain DMPO nitrone adducts. The fractions showing positive ELISA signals are then concentrated and characterized by tandem mass spectrometry (MS/MS). This method, which constitutes the first liquid chromatography-ELISA-mass spectrometry (LC-ELISA-MS)-based strategy for selective identification of DMPO-trapped protein residues in complex peptide mixtures, facilitates location and preparative fractionation of DMPO nitrone adducts for further structural characterization. The strategy is demonstrated for human hemoglobin, horse heart myoglobin and sperm whale myoglobin, three globin proteins known to form DMPO-trappable protein radicals upon treatment with H2O2. The results demonstrate the power of the new experimental strategy to select DMPO-labeled peptides and identify sites of DMPO covalent attachments.
Keywords: immuno-spin trapping, DMPO, protein radical, amino-acid radical, mass spectrometry, hemoglobin, myoglobin
INTRODUCTION
Protein radicals have been detected as catalytic intermediates for an increasing number of enzymes and, as such, play an important role in the proper functioning of biochemical pathways [1]. Protein radicals, however, are also implicated in oxidative stress and are associated with a wide range of diseases and disorders including cancer, rheumatoid arthritis, atherosclerosis, diabetes mellitus, Alzheimer’s disease, Huntington’s disease, amyotrophic lateral sclerosis (ALS or Lou Gehrig’s disease) and general aging [2]. Protein radicals, particularly those not intrinsic to a catalytic mechanism, can cause cleavage of the protein backbone, cross-linking of the protein, and the formation of protein peroxy radicals and protein peroxides [3]. Yet, many aspects of how free radicals are formed, delocalized and propagated within protein structures remain only partly understood.
Protein radical species are generally detected using electron paramagnetic resonance spectroscopy (EPR). Direct EPR spectrometry can be used to detect and identify relatively stable protein-centered free radicals. However, the short lifetimes of these reactive species have generally required either that the samples containing the radical be rapidly frozen after formation [1] or that the radical be spin trapped to give an adduct more stable than the primary radical [4–5]. Nitrone spin traps, of which 5,5-dimethyl-1-pyrroline-N-oxide (DMPO) is the most commonly employed, often form long-lived adducts that can be seen by EPR for minutes. The lack of resolvable hyperfine coupling constants in the EPR spectra of many protein-derived radicals and/or protein radical adducts, however, makes it difficult to extract useful data regarding the site of radical formation. Consequently, additional experimental strategies such as site-directed mutagenesis and/or amino acid derivatization are often employed in conjunction with EPR to obtain information on the nature of the amino acid-derived radical(s) involved. As previously noted [4], these experimental strategies suffer from a number of potential artifacts and limitations. Moreover, EPR measurements require significant quantities of protein (100 μM or more) while relying on rather specialized and costly equipment.
Immuno-spin trapping, a technique developed in this laboratory [6], reduces the amount of protein needed for radical detection experiments from milligram to microgram quantities. In immuno-spin trapping, polyclonal antiserum that recognizes the nitrone spin trap DMPO is used to detect the presence of DMPO covalently attached to proteins. This approach does not involve EPR, but uses the immunology-based methods of enzyme-linked immunosorbent assay (ELISA) and/or Western blotting to measure antigen-antibody binding. Immuno-spin trapping has greatly increased the sensitivity of radical detection and has been used by a number of groups to demonstrate protein radical formation in different proteins exposed to a variety of oxidants [7–16]. Nonetheless, the technology does suffer from some of the drawbacks/limitations of spin trapping with EPR detection. In particular, immuno-spin trapping alone cannot be used to unequivocally localize the actual protein residues labeled by the spin trap.
To complement and overcome the limitations of EPR and immuno-spin trapping approaches, methods using mass spectrometry in combination with spin trapping have been developed. Here, the initial protein radicals are trapped with a spin trap molecule to form covalent adducts, and the protein adducts are digested to peptides with proteolytic agents (usually using the hydrolytic enzyme trypsin). The resulting digestion mixture is typically separated by reverse phase chromatography (RP-HPLC) and the eluted peptides analyzed by mass spectrometry (either off-line or on-line). The use of tandem mass spectrometry allows the unequivocal identification of the actual amino acid(s) labeled by the spin trap and has the advantage that only small (picomole or less) amounts of material are required. A series of publications [13–20] has illustrated the utility and applicability of this technique in investigating radicals produced by the interaction of various oxidants with myoglobin, hemoglobin, NADH dehydrogenase and cytochrome c peroxidase. In the majority of cases reported [13–17], identification of the amino acid trapped by the spin-trapping agent was obtained on-line with tandem mass spectrometry (MS/MS).
When proteins contain only a few sites to which the spin trap is attached, and the spin-trapping efficiency is high, standard LC-MS/MS approaches are generally successful in identifying those sites. However, even when EPR confirms that the spin trap is attached to a particular protein, LC-MS/MS analysis frequently fails to provide information about the sites of spin trap addition. One inherent reason is the low trapping efficiencies of the spin-traps for protein radicals. Consequently, the peptide adducts are generally grossly underrepresented in the general complex peptide mixtures. Thus, to improve the detection of low-abundance peptide adducts from proteolytic digests, selective enrichment methods must be employed prior to reverse phase LC-MS. Herein, we present an approach, which combines chromatographic procedures, immuno-spin trapping, and off-line tandem mass spectrometry for selective detection of protein residues labeled by the spin trap DMPO. This method adds a dimension of selectivity prior to the MS analyses, thereby facilitating the location of DMPO nitrone adducts for the unambiguous structural determination of modified sites in polypeptide chains. The strategy is demonstrated for human hemoglobin, horse heart myoglobin, and sperm whale myoglobin, three globin proteins that are known to form DMPO-trappable protein radicals upon treatment with H2O2.
MATERIALS AND METHODS
Materials
Horse heart metmyoglobin (HoMb) acquired from USB (Cleveland, OH) was purified before use by passage through a Sephadex G-25 gel filtration column (GE Healthcare Bio-Sciences, Piscataway, NJ) and elution with 50 mM potassium phosphate buffer, pH 6.8, containing 50 μM diethylenetriaminepentaacetic acid (DTPA). Recombinant sperm whale myoglobin (SwMb) protein was expressed, purified and oxidized to the met form as described previously [21, 22]. Human hemoglobin (HuHb) was a gift from Apex Biosciences, Inc. (Research Triangle Park, NC). The oxyferrous derivative was oxidized to the met form using a 20% excess of K3Fe(CN)6 and then passed over a Sephadex G-25 gel filtration column and eluted with 50 mM potassium phosphate buffer, pH 6.8, containing 50 μM DTPA. Enzyme concentrations (in heme) were estimated by measuring the absorbance of the heme Soret band using the following extinction coefficients: SwMb εMetMb, 409 = 157 M−1cm−1, HoMb εMetMb, 408 = 188 M−1cm−1 and HuHb εMetHb, 405 = 179 M−1cm−1 [23]. All aqueous solutions were prepared using water passed through a Picopure 2UV Plus system (Hydro Services and Supplies, Inc, RTP, NC) equipped with a 0.2 μm pore size filter. Diluted H2O2 solutions, obtained from a 30% solution (Fisher Scientific Co., Fairlawn, NJ), were used within 1 h of preparation. The H2O2 concentration was confirmed by absorbance measurements at 240 nm (εH202, 240 = 39.4 M−1cm−1) [24]. Catalase from beef liver (suspension, 64000 units mg), trypsin (from bovine pancreas, modified, sequencing grade) and chymotrypsin (from bovine pancreas, modified, sequencing grade) were obtained from Roche Molecular Biochemicals (Indianapolis, IN). Gelatin from cold water fish skin was obtained from Sigma (St Louis, MO). Prepacked Sephadex G-25 (PD-10) gel filtration cartridges were purchased from GE Healthcare Bio-Sciences (Piscataway, NJ). DMPO (high purity, ≥ 99%) from Alexis Biochemicals (San Diego, CA) was sublimed twice under vacuum at room temperature and stored under an argon atmosphere at −70 C before use. The DMPO concentration was measured at 228 nm, assuming a molar extinction coefficient of 7800 M−1cm−1 [25]. All other chemicals were of analytical grade and were purchased from Sigma (St Louis, MO) or Roche Molecular Biochemicals (Indianapolis, IN). Absorption spectra were recorded on a Cary 100 UV-visible spectrometer (Varian, Victoria, Australia). High pressure liquid chromatography (HPLC) was carried out on an Agilent Chemstation 1100 liquid chromatography system equipped with a control module, binary pump, manual injector, and diode-array UV-vis detector. HPLC fractions were collected using a fraction collector (Model 2110, Bio-Rad, Richmond, CA).
Sample Preparation and Proteolytic Digestion
Typically, DMPO trapping reactions were conducted in 50 mM potassium phosphate buffer, pH 6.8. All buffers contained 50 μM diethylenetriaminepentaacetic acid (DTPA) to prevent trace-metal redox chemistry. All experiments were performed using 200 to 500 μM protein in the presence of 100 mM DMPO. The reactions were initiated by the addition of 1 eq of H2O2. After a 30-s incubation, the excess H2O2 was removed by adding catalase (26 units). The resulting solutions were cooled to 4 ºC and acidified to pH 2.5 by the addition of concentrated HCl. The solutions were then extracted with 1 volume of cold 2-butanone. The organic layers containing the extracted heme were discarded. The aqueous solutions were passed over a Sephadex G-25 gel filtration column previously equilibrated and eluted with 100 mM Tris-HCl, pH 8.5. Samples were diluted to a final concentration of ~50 μM with 100 mM Tris-HCl, pH 8.5, and were digested using a 20:1 substrate-to-protease ratio for 16 h at 37 ºC. Reactions were stopped by injecting the final mixture directly onto a reverse phase HPLC column. In the absence of the 2-butanone extraction, very few peptides were formed and proteolysis was incomplete (data not shown).
Reverse Phase HPLC Analysis of Proteolytic Hydrolysates
Digested samples (50 μL) were injected directly onto a Vydac 218TP54, 4.6 mm × 250 mm, C18 reverse phase HPLC column. Peptides were eluted at a flow rate of 0.8 mL/min with a linear gradient from 100% solvent A to 50 % solvent B over 80 min: solvent A, 0.1% trifluoroacetic acid in water; solvent B, 0.085% trifluoroacetic acid in acetonitrile. A rapid gradient to 90% solvent B and then back to 100% solvent A was used to regenerate the column. The eluent was monitored at 214 and 280 nm with an Agilent HP1100 diode array detector. Fractions were collected every 0.4 min in eppendorf tubes containing 200 μL of 100 mM NH4HCO3, pH 7.8, or other volatile buffers. 50 μL of each fraction was analyzed by ELISA, while the remaining amount was lyophilized and stored at −70 ºC for subsequent analyses.
ELISA
The development of the rabbit anti-DMPO nitrone adduct polyclonal antiserum has already been described [26]. Detection of protein-derived nitrone adducts was performed using standard ELISAs in 96-well plates (Greiner Labortechnik, Frickenhausen, Germany). A 50 μL aliquot of each HPLC fraction diluted in 150 μL of coating buffer (100 mM sodium bicarbonate, pH 9.6) was applied to each well and the plates were incubated for 60 min at 37 ºC. Plates wells were then washed once with 1X TBS washing buffer (0.05 % Tween-20, 0.2% cold-water fish skin gelatin, pH 7.4) and blocked with coating buffer (100 mM sodium bicarbonate, pH 9.6, containing 4% cold-water fish skin gelatin) for 60 min at 37 ºC. Thereafter, the rabbit anti-DMPO serum (1:5,000) in washing buffer was added and incubated at 37 ºC for 60 min. After three washes, the secondary antibody, anti-rabbit IgG-alkaline phosphatase (Pierce Chemical Co, Rockford, IL), 1:5000 in washing buffer, was added and incubated for 60 min at 37 ºC. After three additional washes, the antigen-antibody complexes were developed using a chemiluminescence system (CDP-Star, Roche Molecular Biochemicals, Indianapolis, IN) and Xfluor Software (Tecan US, Research Triangle Park, NC).
In experiments to determine the effect of solvent and/or modifiers on nitrone adduct stability, a sample containing 500 μM HoMb and 100 mM DMPO was incubated in 50 mM potassium phosphate buffer, pH 6.8, at 25 °C. The reaction was initiated by the addition of 3 eqs of H2O2, and the excess H2O2 was removed by adding 26 units catalase after 20 s. The resulting solution was passed over a Sephadex G-25 gel filtration column previously equilibrated and eluted with 50 mM potassium phosphate buffer, pH 6.8. Aliquots of this solution were diluted to a final concentration of ~1.6 μM with the appropriate concentration of chaotropes, organic solvents, or organic modifiers. After 14 hours at 37 °C, 50 μL of each of the above reaction mixtures was diluted in 150 μL of coating buffer (100 mM sodium bicarbonate, pH 9.6) and applied to each well of the ELISA plates.
Electrospray Mass Spectrometry
A Micromass Q-TOF Micro (Waters Micromass Corporation, Manchester, UK) hybrid tandem mass spectrometer was used for the acquisition of the electrospray ionization (ESI) mass spectra and tandem mass spectra. All experiments were performed in the positive ion mode. HPLC fractions showing positive ELISA signals were resuspended in a minimal volume (~50 μL) of 50:50 water:acetonitrile containing 0.1% formic acid and infused into the mass spectrometer at ~300 nL/min using a pressure injection vessel [27]. The needle voltage was ~3000 V and the collision energy was 10 eV for the MS analyses. Collision-induced dissociation experiments employed argon with collision energy between 30 and 50 eV. Data analysis was accomplished with a MassLynx data system and MaxEnt deconvolution software supplied by the manufacturer.
RESULTS
Immunochemical detection of globin-centered radicals
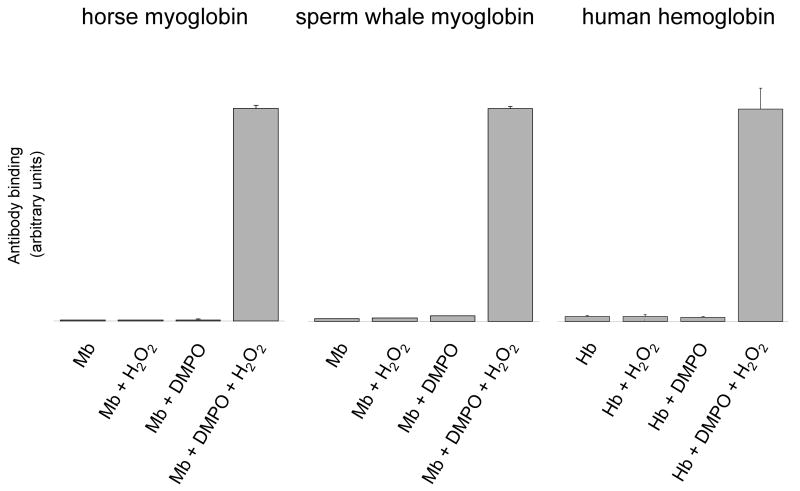
Immuno-spin trapping [26] was used to investigate protein-derived radical formations in horse heart metmyoglobin (HoMb), sperm whale metmyoglobin (SwMb) and human methemoglobin (HuHb) in the presence of H2O2. As previously described [6,7,14], the three globins react with H2O2 to form an oxyferryl (FeIV=O) heme and unstable protein radical species that can subsequently be trapped by DMPO. Anti-DMPO antibody binding to the globin-DMPO adduct was measured using ELISA (Fig. 1). When the three hemoproteins were treated with H2O2 in the presence of DMPO, substantial amounts of globin-derived nitrone adducts were detected. No nitrone adducts were observed in any of the control samples.
Figure 1.
Detection of myoglobin and hemoglobin radical-derived nitrone adducts by immuno-spin trapping. Reaction mixtures contained globin (500 μM), DMPO (100 mM) and H2O2 (500 μM), or subsets of these as indicated. The reactions were initiated by adding H2O2 to the mixture of globin and the spin trap. After 10 min incubation at 25 °C, reactions were diluted with deionized H2O and the globin radical-derived nitrone adducts were detected by ELISA. Values are mean ± S.D (n = 3).
Effects of solvents and/or modifiers on nitrone adduct stability
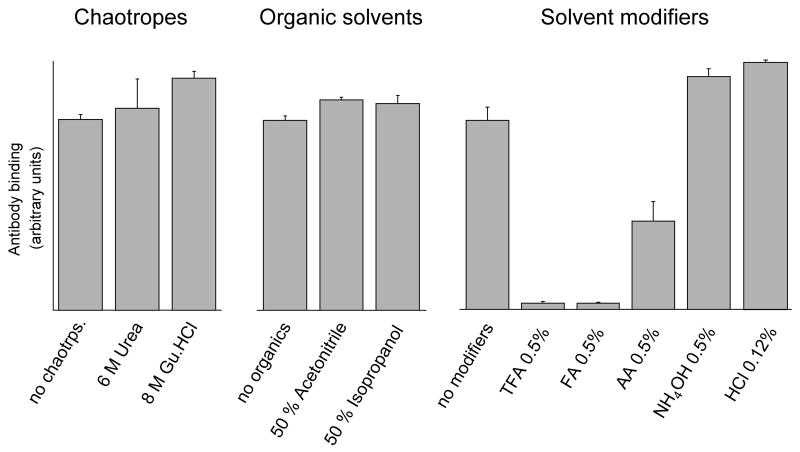
As the focus of this work is to add a dimension of selectivity (i.e., positive ELISA) prior to MS analyses, the stability of the protein nitrone adducts under the normal workup conditions used for protein denaturation, proteolysis, and peptide isolation were tested. It has been previously demonstrated [14] that the DMPO protein adduct is formed almost exclusively on a Tyr residue in horse heart myoglobin. ELISA analyses (Fig. 2) showed that horse heart Mb-derived nitrone adducts are not sensitive to overnight incubations in the most commonly used chaotropes and organic solvents such as urea, guanidine hydrochloride (Gu.HCl), acetonitrile and isopropanol. However, overnight incubations in formic acid (FA) or trifluoroacetic acid (TFA) decreased the ELISA signal intensities to control levels. In addition, a significant decrease in ELISA signal was observed after overnight incubations in acetic acid (AA). Thus these ELISA data imply that Tyr-DMPO addition products are unstable if collected and/or stored under acidic conditions.
Figure 2.
Effects of chaotropes, organic solvents and solvent modifiers on nitrone adduct stability. Reaction mixtures containing the Mb-derived nitrone adducts with and without the indicated concentration of chaotropes, organic solvents or organic modifiers were incubated for 14 hours at 37 ºC. Results obtained with HCl are for a 30 s incubation at 25 ºC. At the conclusion of the experiments, reactions were diluted with coating buffer for ELISA analysis as described under Materials and Methods. Gu.HCl, guanidine hydrochloride; FA, formic acid; TFA, trifluoroacetic acid; AA, acetic acid. Values are mean ± S.D (n = 3).
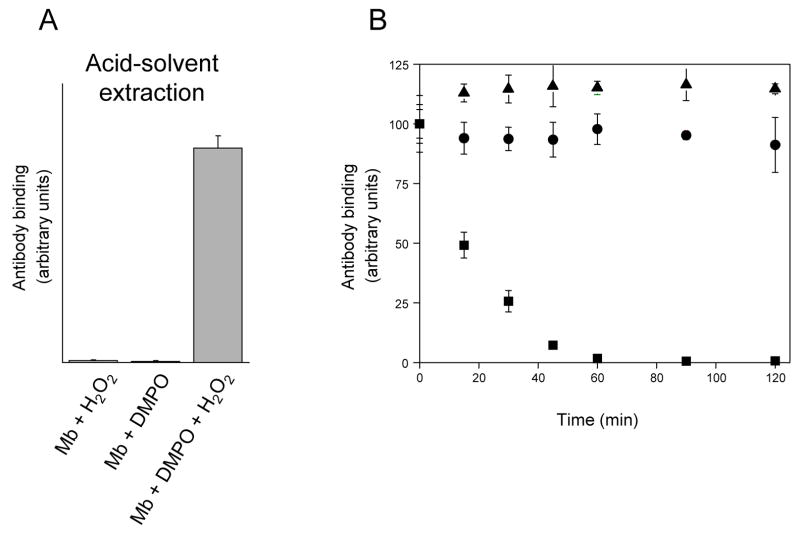
Preliminary experiments indicated that hemoglobin and myoglobin are relatively resistant to proteolytic attack and that extraction of their heme moieties makes them appreciably more sensitive to proteolysis (data not shown). On this basis, the heme was removed from the three globins by standard acid–solvent extraction prior to proteolytic digestion. This method involves a short-term (< 30 s) acidification to pH 2.5 using hydrochloric acid. Control experiments carried out with HoMb-derived nitrone adducts (Fig. 2) showed that this short-term acidification had little effect on the level of adduct detected. It can thus be reasonably assumed that hydrolysis of DMPO protein adducts during acid-solvent extraction procedure is minimal. Additional experiments were also performed to determine if the acid–solvent extraction protocol induced artifactual DMPO adduct formation. To investigate this possibility, reactions containing HoMb, H2O2, DMPO and subsets of these ingredients, were subjected to standard acid–solvent extraction prior to ELISA analysis. The results of these experiments are presented in Fig. 3A. Substantial amounts of globin-derived nitrone adducts were detected exclusively in the sample containing HoMb, H2O2 and DMPO. No nitrone adducts were observed in any of the control samples lacking DMPO or H2O2. The above experiments clearly indicate that standard acid–solvent extraction does not induce significant artifactual radical adduct formation.
Figure 3.
(A), detection of horse heart myoglobin radical-derived nitrone adducts by immuno-spin trapping: effect of acid-solvent extraction. Reaction mixtures contained horse heart myoglobin (500 μM), DMPO (100 mM) and H2O2 (500 μM), or subsets of these as indicated. The reactions were initiated by adding H2O2 to the mixture of myoglobin and the spin trap. After 10 min incubation at 25 °C, the heme was removed from the globin by acid-solvent extraction as described under Materials and Methods. At the conclusion of the solvent extraction process, reactions were diluted with coating buffer for subsequent ELISA analysis. Values are mean ± S.D (n = 3). (B), effect of trifluoroacetic acid on nitrone adduct stability: time course of adduct decay. DMPO-modified peptides were purified from the tryptic digest of hemoglobin, lyophilized and resuspended in H2O + 0.5% TFA. Excess NH4HCO3 was then added at various times after initiation of the reaction to stop further reaction. The zero time estimates refer to control experiments in which excess NH4HCO3 was added before TFA. At the conclusion of the experiments, reactions were diluted with deionized H2O for ELISA analysis. ▲, DMPO- cysteinyl adduct (peak 1 in Fig. 4); ■, DMPO-tyrosyl adduct (peak 4 in Fig. 4); ●, DMPO-histidinyl adduct (peak 6 in Fig. 4). Values are mean ± S.D (n = 3).
To extend the results obtained with protein-derived nitrone adducts, experiments were also carried out with peptides containing His-, Cys- or Tyr-DMPO addition products (Fig. 3B). These peptides were purified from the tryptic digest of HuHb treated with H2O2 in the presence of DMPO, lyophilized, and the sites of DMPO addition identified by tandem mass spectrometry, as shown later in the text. The lyophilized peptides were resuspended and incubated for various periods of time in H2O + 0.5 % TFA after which excess NH4HCO3 was added to neutralize the pH of the solution. The zero time points refer to control experiments in which NH4HCO3 was added before TFA. Quantitatively similar ELISA signals were observed when NH4HCO3 was added either before or at various times after initiation of the reaction with the samples containing His- or Cys-DMPO addition products (Fig. 3B). In contrast, a significant decrease in ELISA signal was observed with the Tyr-DMPO product (half-life of ~ 18 minutes). Thus, in contrast to the Tyr-DMPO addition product, both His- and Cys-DMPO adducts are stable if collected and/or stored under acidic conditions.
HPLC and ELISA analyses
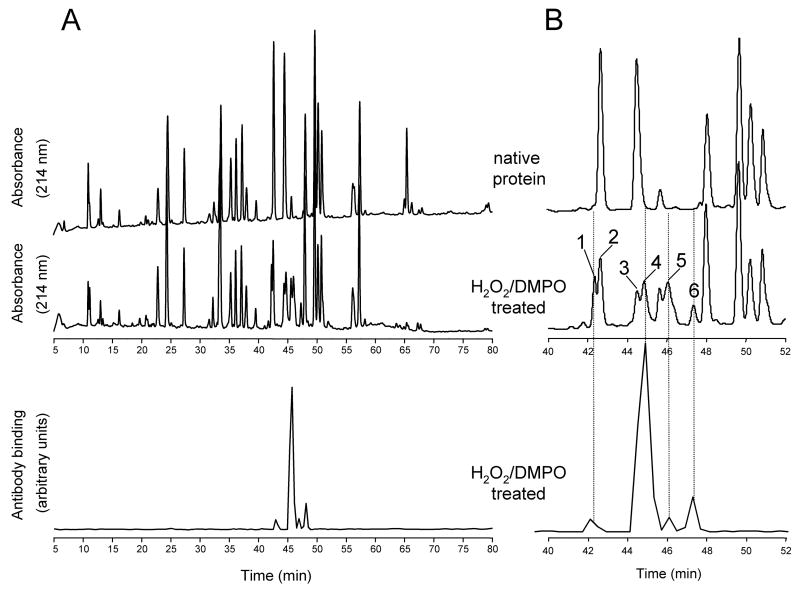
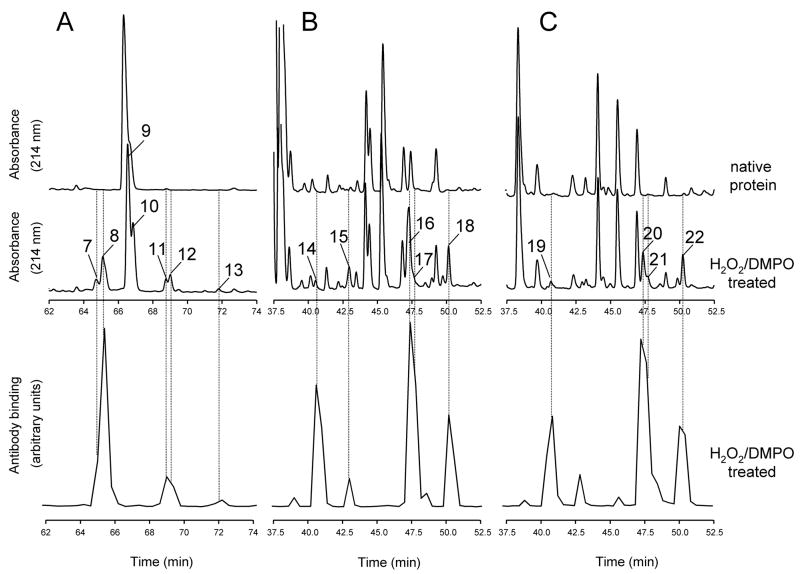
To determine whether the peptides modified with DMPO spin trap could be detected by ELISA following HPLC separation, equal amounts of native proteins and of proteins treated with H2O2 in the presence of DMPO were individually subjected to proteolytic digestion. After HPLC separation of the resulting peptides, the collected fractions were analyzed by ELISA using polyclonal anti-DMPO nitrone antiserum. In order to prevent acid hydrolysis of Tyr-DMPO adducts, fractions were collected in Eppendorf tubes containing 100 mM NH4HCO3, pH 7.8 (see Methods and Materials for details). The HPLC chromatograms for the tryptic digests of intact native HuHb and of HuMb treated with H2O2 in the presence of DMPO are shown in Fig. 4A (top and middle panels). The bottom panel shows the results of ELISA analyses of HPLC fractions from the digest mixture of HuMb treated with H2O2 in the presence of DMPO. Expanded views of the region of the chromatogram where selected HPLC peaks show a strong positive ELISA signal elute are shown in Fig. 4B. Similar analyses were performed for peptides derived from tryptic digests of HoMb (Fig. 5A) and from chymotryptic digests of both HoMb and SwMb (Fig. 5B and 5C, respectively). The chromatograms show only the time windows containing ELISA positive fractions. Comparison of the HPLC traces reveals several peaks from the digest mixtures of the proteins treated with H2O2 and DMPO that are not present in the controls (peak 1 and 4–6, Fig. 4B and peak 7, 8 and 11–22, Fig. 5). The changes in Fig. 4B are accompanied by significant decreases in intensity of several other peaks (peak 2 and 4) relative to control. Only the fractions showing positive ELISA signals or corresponding to chromatographic peaks showing marked decreases in intensity relative to controls were further concentrated and characterized by electrospray ionization mass spectrometry (ESI-MS).
Figure 4.
Comparison of peptide maps of human hemoglobin. (A), reversed-phase HPLC of the tryptic peptides derived from intact native Hb and from Hb reacted with H2O2 in the presence of DMPO; (B), expanded view of panel A. Absorbance was monitored at 214 nm. Fractions were collected every 0.4 min in Eppendorf tubes containing excess NH4HCO3, and an aliquot of each fraction was analyzed by ELISA. The fractions showing a strong positive ELISA signal or corresponding to chromatographic peaks showing marked decreases in intensity relative to controls were further characterized by tandem mass spectrometry. Peak assignment and mass analysis are summarized in Table 1.
Figure 5.
Comparison of peptide maps from horse heart and sperm whale myoglobin. Reversed-phase HPLC of peptides derived from intact Mb and from Mb reacted with H2O2 in the presence of DMPO. Absorbance was monitored at 214 nm. Fractions were collected every 0.4 min in Eppendorf tubes containing excess NH4HCO3, and an aliquot of each fraction was analyzed by ELISA. Chromatograms show only the time window in which some peptides exhibiting a strong positive ELISA signal eluted. (A), tryptic maps for HoMb; (B), chymotryptic maps for HoMb; (C), chymotryptic maps for SwMb. Peak assignment and mass analysis are summarized in Table 1.
MS analyses of DMPO-modified peptides
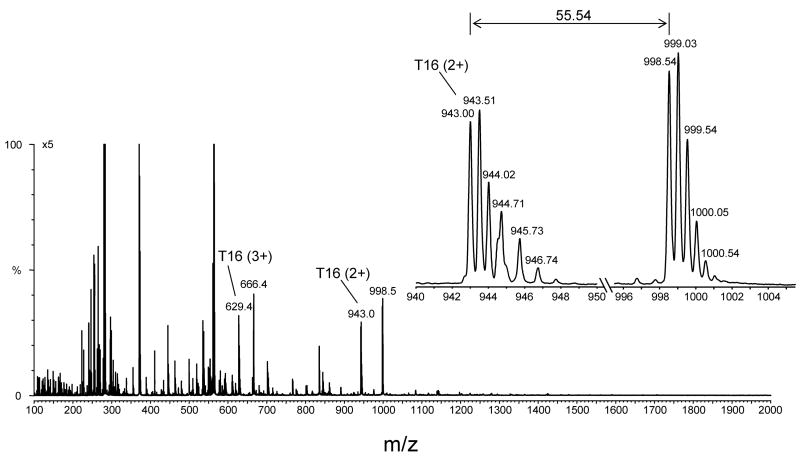
ELISA-positive HPLC fractions were initially characterized using flow injection analysis coupled to electrospray ionization mass spectrometry (ESI-MS). From these analyses, pairs of ions separated by 111.07 Da (111.07, theoretical mass shift) were observed in some of the ELISA-positive fractions, but not all. This mass corresponds to the net addition of one molecule of DMPO to the unmodified peptide and depends on the charge state, i.e., Δm = 111.07 (+1), 55.54 (+2), 37.02 (+3) or 27.77 (+4). These ion pairs were used to identify potential ions of interest. This is exemplified in Fig. 6, which displays the full scan mass spectrum of the ELISA-positive fraction from the tryptic digest of HoMb (Fig. 5B) corresponding to peak 7 and eluting at 64.9 min. The ions of m/z 629.0 (+3) and 943.0 (+2) correspond in mass to tryptic peptide T16 (residue 103–118), which has a theoretical mass of 1884.0 Da. The ions of m/z 666.0 (+3) and 998.5 (+2) correspond in mass to the addition of one molecule of DMPO to tryptic peptide T16. The insert in Fig. 6 depicts one pair of doubly charged ions, which shows a mass-to-charge (m/z) difference of 55.5. These pairs of ions were observed in all ELISA-positive fractions containing Tyr-DMPO addition products. By contrast, such pairs were not detected with His-DMPO and/or Cys-DMPO addition products (data not shown).
Figure 6.
Full scan mass spectrum of one HPLC fraction from the peptide map of horse heart myoglobin showing a strong ELISA signal (peak 7 in Fig. 5A eluting at approximately 64.8 min). The fraction was lyophilized, resuspended in 50:50 acetonitrile/H2O + 0.1 % formic acid, and directly infused into the electrospray source of a Micromass Q-TOF hybrid tandem mass spectrometer. An expanded view of a pair of ions having the same charge state (+2 ion) and a mass-to-charge (m/z) difference of 55.5 is shown. This mass difference corresponds to the loss of one molecule of DMPO.
MS/MS fragmentation of DMPO-modified peptides
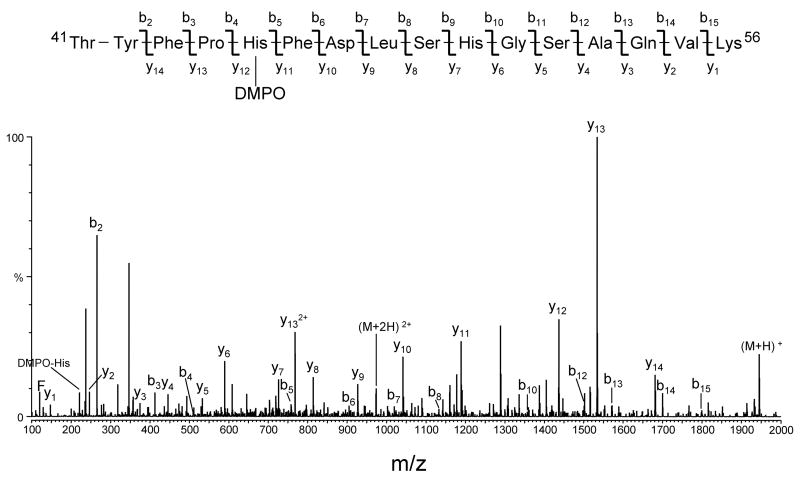
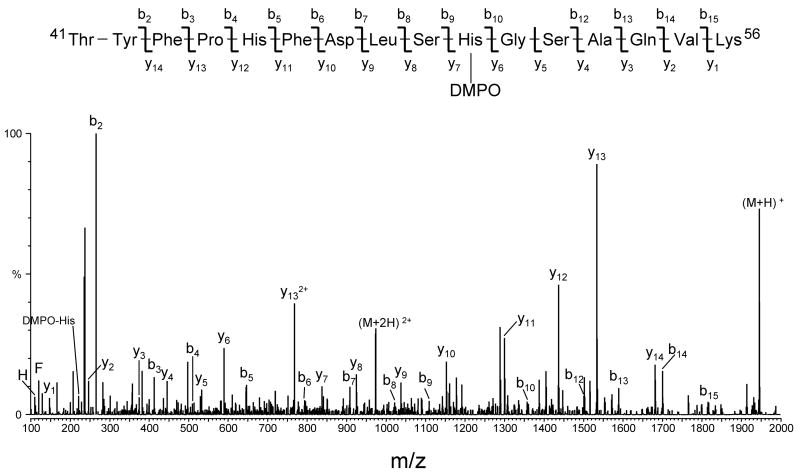
The sites of DMPO addition on the peptides were identified by tandem mass spectrometry (MS/MS) using collision-induced dissociation. Examples of MS/MS data which allowed the determination of the site of DMPO modification in peptides from human hemoglobin are presented in Figs. 7 and 8. The two MS/MS spectra obtained by analyzing ELISA-positive fractions from the tryptic digest of HuHb (Fig. 4B) correspond, respectively, to peak 5 eluting at 46.0 min and peak 6 eluting at 47.4 min. The two spectra were acquired from the molecular ion of m/z 973.0 (+2 ion), which corresponds in mass to tryptic peptide T16 (amino acid residues 41–56) of the human hemoglobin α chain, plus DMPO. The spectra show an almost complete series of both carboxy-terminal y ions (y2 through y14) and amino-terminal b ions (b2 through b14), thus accounting for all the amino acids in the Thr41-Lys56 sequence. The observation of abundant y11 and y12 ions in Fig. 7 and of y6 and y7 ions in Fig. 8 provides the necessary data to assign the location of the DMPO adduct: the mass difference between y11 and y12 ions (in Fig. 7) and between y6 and y7 ions (in Fig. 8) corresponds to the mass of a DMPO-modified histidine residue. In addition, ions corresponding in mass to the immonium ion of histidine plus a DMPO were observed (labeled as DMPO-His). These data allow for the identification of His-45 and His-50 as two sites of modification by DMPO in the spin-trap experiment.
Figure 7.
Deconvoluted MS/MS spectrum of human hemoglobin DMPO-modified peptide 41–56 (peak 5 in Fig. 4B eluting at approximately 46.0 min). The MS/MS spectrum was acquired from parent at m/z 973.0 (+2 ion), which corresponds in mass to tryptic peptide αT16 plus DMPO. For clarity, not all identified fragment ions are labeled on the spectrum.
Figure 8.
Deconvoluted MS/MS spectrum of human hemoglobin DMPO-modified peptide 41–56 (peak 6 in Fig. 4B eluting at approximately 47.4 min). The MS/MS spectrum was acquired from parent at m/z 973.0 (+2 ion), which corresponds in mass to tryptic peptide αT16 plus DMPO. For clarity reasons, not all identified fragment ions are labeled on the spectrum.
Table 1 summarizes the sites of DMPO modification identified in this study. As shown in the table, eighteen DMPO-labeled peptides were detected in the digest mixtures of the proteins treated with H2O2 in the presence of DMPO. MS/MS analyses of these peptides conclusively identified ten sites of covalent attachment of DMPO on the three globin proteins, six of which were previously uncharacterized. Specifically, DMPO spin adduct formation was observed on three tyrosine, one cysteine and six histidine residues. Interestingly, none of these His addition products had been previously detected by direct on-line LC MS/MS measurements. This latter result demonstrates the power of our new experimental strategy to select DMPO-labeled peptides and to identify sites of DMPO modification.
Table 1.
Summary of identified peptides originating from proteolytic digestion of human hemoglobin, horse heart myoglobin and sperm whale myoglobin.
| Peaka | Elution Time (min) | Proteinb | Proteasec | Amino acid sequence d | Res.Mod.e | Observed f (m/z) | Calculated f (m/z) |
|---|---|---|---|---|---|---|---|
| 1 | 42.34 | HuHb | Try | Gly83-Lys95 (α) | Cys93 | 766.86 | 766.87 |
| 2 | 42.65 | HuHb | Try | Gly83-Lys95 (α) | n.m. | 711.34 | 711.34 |
| 3 | 44.50 | HuHb | Try | Thr41-Lys56 (α) | n.m. | 917.43 | 917.45 |
| 4 | 44.84 | HuHb | Try | Thr41-Lys56 (α) | Tyr42 | 972.96/917.43 g | 972.98/917.45 g |
| 5 | 46.06 | HuHb | Try | Thr41-Lys56 (α) | His45 | 972.99 | 972.98 |
| 6 | 47.37 | HuHb | Try | Thr41-Lys56 (α) | His50 | 973.01 | 972.98 |
| 7 | 64.73 | HoMb | Try | Tyr103-Lys118 | Tyr103 | 998.55/943.09 g | 998.55/943.02 g |
| 8 | 65.11 | HoMb | Try | Tyr103-His116 | Tyr103 | 890.95/835.56 g | 890.99/835.45 g |
| 9 | 66.56 | HoMb | Try | Tyr103-Lys118 | n.m. | 942.80 | 943.02 |
| 10 | 66.86 | HoMb | Try | Tyr103-His116 | n.m. | 835.26 | 835.45 |
| 11 | 68.76 | HoMb | Try | Tyr103-Lys118 | His116 | 998.52 | 998.55 |
| 12 | 69.01 | HoMb | Try | Tyr103-His116 | His113 | 890.98 | 890.99 |
| 13 | 71.80 | HoMb | Try | Tyr103-His116 | His116 | 890.98 | 890.99 |
| 14 | 40.52 | HoMb | Chy | Ala90-Leu104 | Tyr103 | 923.51/867.98 g | 923.54/868.00 g |
| 15 | 42.96 | HoMb | Chy | Lys98-Leu104 | Tyr103 | 493.28/437.73 g | 493.33/437.79 g |
| 16 | 47.27 | HoMb | Chy | Ala90-Phe106 | Tyr103 | 1061.55/1006.03 g | 1061.59/1006.06 g |
| 17 | 47.61 | HoMb | Chy | Lys96-Phe106 | Tyr103 | 763.94/708.40 g | 763.96/708.42 g |
| 18 | 50.16 | HoMb | Chy | Lys98-Phe106 | Tyr103 | 631.34/575.80 g | 631.38/575.85 g |
| 19 | 40.65 | SwMb | Chy | Ala90-Leu104 | Tyr103 | 923.61/868.08 g | 923.54/868.00 g |
| 20 | 47.29 | SwMb | Chy | Ala90-Phe106 | Tyr103 | 1061.69/1006.16 g | 1061.59/1006.06 g |
| 21 | 47.63 | SwMb | Chy | Lys96-Phe106 | Tyr103 | 764.02/708.49 g | 763.96/708.42 g |
| 22 | 50.20 | SwMb | Chy | Lys98-Phe106 | Tyr103 | 631.42/575.87 g | 631.38/575.85 g |
| n.p. | 67.28 | SwMb | Try | Tyr103-His116 | Tyr103 | 897.98/842.47 g | 897.99/842.46 g |
| n.p. | 67.28 | SwMb | Try | Tyr103-Arg118 | Tyr103 | 1019.57/964.03 g | 1019.56/964.03 g |
| n.p. | 71.23 | SwMb | Try | Tyr103-Arg118 | His116 | 1019.55 | 1019.56 |
| n.p. | 71.23 | SwMb | Try | Tyr103- His116 | His113 | 898.00 | 897.99 |
Peak numbers are assigned in the order of retention time as shown in Fig. 4 and 5; n.p., peptide maps not presented
Protein: HuHb, Human hemoglobin; HoMb, Horse myoglobin; SwMb; Sperm whale myoglobin.
Protease: Try, Trypsin; Chy, Chymotrypsin.
(α), α chain; (β), β chain.
Res. Mod., residue modified by DMPO; n.m., not modified.
Observed and calculated m/z values: value are for +2 ions
Pairs of ions showing a mass-to-charge difference of approximately 55.5
Retention time of DMPO-modified peptides
HPLC peaks of DMPO-labeled tryptic peptides were generally well separated from the unmodified complements, suggesting that the covalently linked DMPO had a marked impact on the retention time of the peptide. The data in Table 1 indicate that, compared with unmodified peptides, retention times were shifted between 0.34 and 4.94 min. All peptides containing an adduct at a His eluted at a longer retention time, suggesting that the covalently linked DMPO increased the hydrophobicity. However, when the point of attachment was a Tyr, the adducted peptide eluted either before or after the non-adducted peptide, depending on the peptide investigated. No conclusion can be drawn for DMPO-Cys because only one peptide having this point of attachment was identified. Similarly, no conclusion can be drawn from chymotryptic maps because of the complexity and considerable peak overlaps in the elution profiles.
UV absorbance of DMPO-modified peptides
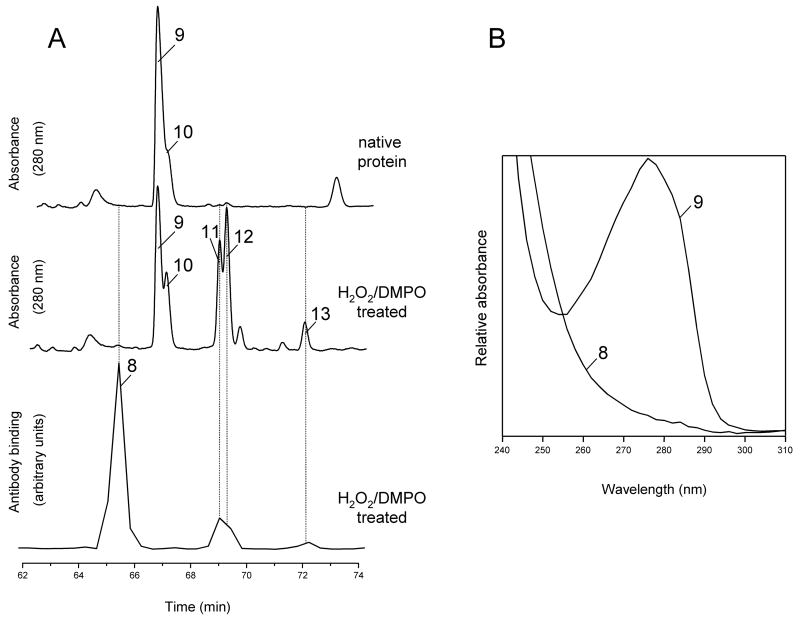
DMPO-labeled peptides may also be identified by comparing the peptide maps at 280 nm. This is exemplified in Fig. 9A (top and middle panels), which displays expanded views of selected HPLC peaks from the tryptic digests of intact native HoMb and of HoMb treated with H2O2 and DMPO. The bottom panel shows the result of ELISA analyses of HPLC fractions from the digest mixture of HoMb treated with H2O2 in the presence of DMPO. Peak numbers refer to compounds in table 1. The two chromatographic peaks, which absorb at 280 nm and elute at 66.6 and 66.9 min, are due to the peptides Tyr103-Lys118 and Tyr103-His116 (marked with 9 and 10, respectively). The absorbance spectra of these peaks, recorded by the diode array detector of the HPLC system, are typical of tyrosine-containing peptides (spectrum of peak 9 shown in Fig. 9B). Upon DMPO/H2O2 treatment of HoMb, the two peaks were attenuated and three new peaks appeared (peaks 11, 12 and 13) with absorbance at 280 nm and showing weak positive ELISA signals. These three peaks are all DMPO-modified peptides containing DMPO attachment sites at either His-113 or His-116 (see Table 1 for peak assignments). The absorbance spectra of these peaks showed the same λmax = 280 nm and shape as their unmodified complements and are typical of tyrosine-containing peptide (spectra not shown). The peak at 65.1 min (peak 8), which shows a strong positive ELISA signal, is assigned to a Tyr-103 containing peptides, in which Tyr-103 has been modified by DMPO. This peptide showed a dramatic reduction in absorbance at 280 nm and only minor changes in absorbance at 214 nm compared with its unmodified cognate (spectrum of peak 8 shown in Fig. 9B). DMPO has a strong absorption maximum at 228 nm (εDMPO, 228 = 7800 M−1cm−1) [25]. However, none of the DMPO-labeled peptides identified in this study showed λmax at 228 nm or significant changes in absorbance at this wavelength (data not shown).
Figure 9.
Tryptic maps and absorbance spectra from intact horse heart Mb and from Mb reacted with H2O2 in the presence of DMPO. (A), HPLC tryptic maps monitored at 280 nm. Fractions were collected every 0.4 min in Eppendorf tubes containing excess NH4HCO3, and an aliquot of each fraction was analyzed by ELISA. Chromatograms show only the time window in which some peptides exhibiting a strong positive ELISA signal eluted. Peak assignment and mass analysis are summarized in Table 1. (B), absorbance spectra recorded by the diode-array detector in the eluent of the HPLC system. The spectra are scaled arbitrarily.
DISCUSSION
Methods using mass spectrometry in combination with spin-trapping methodology have been used by this laboratory [13–15,28,29] and by others [16–20] to detect and characterize peptide- and protein-based radicals. A variant to this approach, presented here, combines chromatographic procedures and an immunology-based method to facilitate detection of peptide residues labeled by the nitrone spin-trap DMPO.
Long-term stability of DMPO nitrone adducts
When DMPO traps a protein radical, a nitroxide radical adduct is formed with a covalent bond from DMPO to the protein [26]. The radical adduct then decays over time to the EPR-silent nitrone adduct and, potentially, other product(s). The nitrone adduct is the most stable end product and preserves the chemical bond formed between DMPO and the original free radical [26]. Previous investigations in this laboratory [13–15] have established that the protein-DMPO nitrone adducts are stable enough to permit peptide mass mapping and the identification of the spin-trapped species using standard on-line LC-MS/MS analysis. However, there is a lack of information regarding the long-term stability of those adducts. The focus of this work is to add a dimension of selectivity (i.e., positive ELISA) prior to MS analyses. Thus, the first step in this investigation was to determine the stability of the protein nitrone adducts under the normal workup conditions used for protein denaturation, proteolysis and off-line MS analysis. The results of this experiment (Fig. 2) indicated protein-derived nitrone adducts are not sensitive to overnight incubations in the most commonly used chaotropes and organic solvents. However, overnight incubations of Tyr-DMPO nitrone adducts in formic acid (FA) or trifluoroacetic acid (TFA) reduced the protein-derived nitrone adducts signal intensities to control levels. Experiments were also carried out with peptides containing His-, Cys- or Tyr-DMPO nitrone adducts (Fig. 3B). These experiments indicate that both His- and Cys-DMPO nitrone adducts are stable if collected and/or stored under acidic conditions. In contrast, Tyr-DMPO nitrone adducts are unstable under acidic conditions (half-life of ~ 18 minutes in 0.5 % TFA), but are stable when stored under neutral or basic conditions.
Selective identification of DMPO nitrone adducts
With these observations in mind, peptide mappings of proteolytic digests were carried out by RP-HPLC using TFA in the mobile phase. In order to prevent acid hydrolysis of Tyr-DMPO adducts, HPLC fractions were routinely collected and/or stored in Eppendorf tubes containing ammonium bicarbonate or other volatile ammonium buffers. The fractions that contain DMPO-modified peptide adducts were detected by ELISA using polyclonal anti-DMPO nitrone serum. Only the fractions showing positive ELISA signals or corresponding to chromatographic peaks showing marked decreases in intensity relative to controls were further concentrated by lyophilization and characterized by electrospray ionization mass spectrometry (ESI-MS).
Although this approach to detecting adducted peptides through nanospray analysis of collected fractions is time-consuming, it is compensated by several advantages. Decoupling the HPLC separation from the MS/MS analysis allows independent optimization of separation and detection conditions without the time constraints of an online method. Typically, ESI-MS experiments are performed in positive ion mode using an acidic solvent to create multiple charged species. Decoupling of the separation and detection conditions enables peptide separation at basic as well as acidic pH. The ion-pairing agent TFA, which has a suppressive effect on ion generation [30], can be utilized during the chromatographic separation to ensure the highest efficiency of peptide fractionation. Also, in contrast to on-line coupling where the sample is consumed during the MS/MS analysis, each HPLC fraction can be examined for many minutes and stored for further structural characterization if needed. In addition, an analytical HPLC column having a much greater loading capacity than the microcapillary LC column can be used and, consequently, more material can be analyzed. Finally, the HPLC fractions that contain DMPO adducts can easily be detected by ELISA, which adds a dimension of selectivity prior to the MS analyses. As a consequence of all these factors, there is a significant increase in the number of peptide adducts identified and in the quality of the data.
Our approach can provide exact identification of the specific amino acid trapped by DMPO but it can be difficult to determine which atom forms a covalent bond with the spin trap. Another limitation is that it is difficult to estimate relative amounts of different DMPO-peptide adducts by ELISA. This is because the intrinsic affinity of the antibody for each of the three types of DMPO adducts identified in this study (e.g. Tyr-DMPO, His-DMPO and Cys-DMPO) is not known. The required experiments would be very laborious (e.g. subdigestions sufficient to isolate each DMPO-amino acid adducts individually for further ELISA characterization) and have not been undertaken. Thus, strong positive ELISA signal may reflect a high concentration of the adduct or a high affinity of the antibody for this adduct, or both. Also, the DMPO-adducted peptides may display different affinities for the antibody depending on how they are bound to the ELISA plate and this may also affect the ELISA signal intensity.
DMPO-labeled peptides can sometimes be identified by pairs of ions separated by 111.07 Da (111.07, theoretical mass shift), as exemplified in Fig. 6. This mass corresponds to the net addition of one molecule of DMPO to the unmodified peptide and depends on the charge state, i.e., Δm = 111.07 (+1), 55.54 (+2), 37.02 (+3) or 27.77 (+4). These pairs of ions can greatly facilitate the selection of DMPO-containing ions of interest. DMPO-labeled peptides may also be identified by comparing the peptide maps at 280 nm, as shown in Fig. 9. Tyr-DMPO addition products show a dramatic reduction in absorbance at 280 nm and only minor change in absorbance at 214 nm compared with their unmodified cognate. A decrease of conjugation between the ring and the Tyr hydroxyl group associated with the binding of DMPO could conceivably cause such an effect. This explanation is consistent with earlier EPR spin-trapping experiments [31], demonstrating that the Tyr-DMPO radical adduct in the horseradish peroxidase/Tyr/H2O2 system is formed by trapping of the Tyr phenolic oxygen.
Examples of the MS/MS data obtainable from the liquid chromatography-ELISA-mass spectrometry (LC-ELISA-MS)-based strategy are presented in Fig. 7 and 8. In both cases, the MS/MS fragmentation spectra showed a nearly complete series of both carboxy-terminal y ions and amino-terminal b ions, thus providing the necessary data to unequivocally assign the sites of DMPO attachment.
LC-ELISA-MS analysis of protein model systems
Human hemoglobin (HuHb), horse heart myoglobin (HoMb) and sperm whale myoglobin (SwMb) were used as protein model systems. The three globin proteins are known to form (oxo)ferryl (FeIV=O) hemes and DMPO-trappable protein radical species upon treatment of the met form (i.e., the FeIII heme state) with H2O2 [32–34]. Stopped-flow UV-vis rapid-scan experiments suggest that the reaction of metMb with H2O2 may first produce a transient Compound I-like intermediate in which the ferryl species is associated with a porphyrin rather than a protein radical [35]. Electron transfer from the protein to the porphyrin radical cation would then produce the observed protein radicals. Table 1 summarizes the sites of modification by DMPO identified in this study. As shown in the table, ten sites of covalent attachment of DMPO on the three globin proteins were identified. Specifically, DMPO spin adduct formation was observed on three tyrosine, one cysteine and six histidine residues.
DMPO-tyrosyl adducts
In this work, DMPO spin adduct formation was observed on Tyr-42 in the α-chain of HuHb, on Tyr-103 of HoMb and on Tyr-103 of SwMb. The α and β hemoglobin subunits of HuHb each have three tyrosine residues (Tyr-24, Tyr-42, and Tyr-140 in the α- and Tyr-35, Tyr-130, and Tyr-145 in the β-chain, respectively). From the crystal structure of HuHb, three of the tyrosyl residues exhibit at least some degree of solvent accessibility (i.e., Tyr-24 and Tyr-42 in the α-, and Tyr-35 in the β-chain). In this structure, Tyr-42 in the α-chain is the tyrosine residue nearest to the heme group, and the shortest distance separating these two groups is 4.03 Å. Tyr-42 is thus optimally placed for oxidation by the porphyrin radical cation species and production of DMPO-trappable tyrosyl radical. The most exposed tyrosine residue of the HuHb tetramer is Tyr-35 in the β-chain, which is not detectably modified by DMPO. HoMb and SwMb have very similar tertiary structures. SwMb differs from HoMb and many other myoglobins in that it has three tyrosine residues (Tyr-103, Tyr-146 and Tyr-151), whereas the equine homologue has only two (Tyr-103 and Tyr-146). In SwMb and HoMb, Tyr-103 is the tyrosine residue nearest to the heme group (shortest distance to heme is 3.32 Å, compatible with van der Waals contact) and is exposed to solvent, while Tyr-146 is an internal residue. Tyr-151, which is unique to SwMb, is the most exposed tyrosine residue but also the most distant from the heme (shortest distance to heme is 9.06 Å). Interestingly, the π–orbitals of Tyr-103 are nearly coplanar with the unsaturated heme system, and this relative orientation has been shown to kinetically favor electron transfer [36]. Thus, our observation that DMPO forms a detectable nitrone adduct with Tyr-103 could have been anticipated and is consistent with previous mass spectrometric (HoMb and SwMb) [14] and site-directed mutagenicity studies (SwMb) [31].
DMPO-cysteinyl adducts
The α and β hemoglobin subunits of HuHb possess three cysteine residues (Cys-104 in the α- and Cys-93 and 112 in the β-chain, respectively). In contrast, both HoMb and SwMb are completely devoid of cysteine residues. Cys-93 is the only residue on the β-chain of HuHb that forms a detectable amount of DMPO trappable protein radicals. In addition, Cys-93 possesses the only sulfhydryl group in the hemoglobin tetramer that reacts with N-ethylmaleimide, a reagent specific for sulfhydryl groups [37], indicating a freely accessible environment. Not unexpectedly, crystal structures of HuHb confirm that Cys-93 is the only cysteine residue in hemoglobin tetramers having a sulfhydryl sulfur exposed to solvent. The distance of closest approach of the Cys-93 sulfhydryl sulfur to the β-chain heme is 12.52 Å. Cys-93 is separated from the β-chain heme by only a single residue, the proximal histidine (His-92), which could constitute a conduit for electron transfer. Tyr-145 could also serve as a conduit for electron transfer because it is located in the immediate vicinity of Cys-93 (shortest distance to the sulfhydryl sulfur is 3.8 Å) and is closest to the heme (shortest distance to heme is 7.53 Å). Alternatively, thiyl radicals could be generated through reduction of tyrosine-derived radicals on one HuHb molecule by the sulfhydryl at Cys-93 on a second HuHb molecule. Similar intermolecular electron transfer reactions between Tyr and Cys residues have recently been reported for the human myoglobin/H2O2 [38] and human hemoglobin/peroxynitrite systems [39]. Regardless of the exact electron-transfer pathway, our observation that Cys-93 is the only residue on the β-chain of HuHb that forms DMPO trappable protein radicals confirms previous on-line LC MS/MS measurements performed in this laboratory [13] and is of significant interest. It reinforces the contention that cysteine residues in polypeptide chains can act as ‘sinks’ by providing reducing equivalents that limit heme-induced oxidative damage.
DMPO-histidinyl adducts
HuHb α and β subunits have ten and nine histidine residues, respectively. However, only two residues in the α-chain (i.e., His-45 and His-50) were detectably modified by DMPO. His-45 is nearest to the heme group (shortest distance to heme pyrrole rings is 5.66 Å) and moderately exposed to solvent. His-50 is the most exposed His residue of the α-chain, but is most distant from the heme (shortest distance to heme is 19.63 Å). SwMb has twelve histidine residues, five of which are inside the protein and inaccessible to the solvent. In comparison to SwMb, HoMb lacks His-12, which is replaced by Gln. The other histidine residues, together with most amino-acid residues in their immediate environments, are strictly conserved in the two myoglobin chains. Under the experimental conditions used in this investigation, in both SwMb and HoMb, His-113 and His-116 were the only histidine residues detectably modified by DMPO. These two histidines are close to each other (shortest distance between imidazole rings is 6.64 Å) and are located far from the heme (shortest distances to heme are 14.92 Å and 16.79 Å for His-113 and His-116, respectively). The two residues are either moderately (His-113) or highly (His-116) exposed to the solvent. They are quite readily alkylated in SwMb, indicating a freely accessible environment [40].
Whether the histidinyl radicals detected in this study arise through internal and/or external electron transfer mediated by the heme group or are generated via Fenton-type metal-catalyzed oxidation remains to be established. Histidine is known to form transition metal coordination sites in proteins, which makes this residue a likely target for Fenton-type metal-catalyzed oxidation. However, there are good reasons to believe that Fenton-type reactions do not play an important role, at least under the experimental conditions used in this study. First, no nitrone adducts were detected by ELISA when the apoprotein form of HoMb was treated with H2O2 in the presence of DMPO (peptide map not shown). Substantial amounts of globin-derived nitrone adducts were only detected when the holoprotein was exposed to H2O2. Thus, formation of histidine radicals in native HoMb reported here appears to be mediated by the heme group. Second, diethylenetriaminepentaacetic acid (DTPA), a strong chelator for metals, was included in all solutions prior to the addition of H2O2 to minimize the possibility of free transition metal-mediated decomposition. We therefore tentatively conclude that the observed histidinyl radicals arise through internal and/or external electron transfer mediated by the heme group.
Acknowledgments
This work has been supported by the Intramural Research Program of the National Institutes of Health and the National Institute of Environmental Health Sciences. The authors thank Dr. Jason Williams and Dr. Marilyn Ehrenshaft for their careful review of the manuscript and Ms. Jean Corbett, Mrs. Mary J. Mason and Dr. Ann Motten for their valuable assistance in the preparation of this manuscript. We thank Dr. Robert Petrovich and all the members of the protein expression core facility for their kind access to PCR machines and shakers and for their comments and suggestions. The synthetic gene of sperm whale myoglobin in pUC18 was a generous gift from Prof. Paul Ortiz de Montellano of the University of California, San Francisco.
ABBREVIATIONS
- DMPO
5,5-dimethyl-1-pyrroline-N-oxide
- ELISA
Enzyme-Linked ImmunoSorbent Assay
- ESI
electrospray ionization
- Mb
myoglobin
- metMb
metmyoglobin
- HoMb
Horse heart myoglobin
- SwMb
Sperm whale myoglobin
- HuHb
Human hemoglobin
- heme
iron protoporphyrin IX regardless of the oxidation and ligation states
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stubbe J, van der Donk WA. Protein radicals in enzyme catalysis. Chem Rev. 1998;98:705–762. doi: 10.1021/cr9400875. [DOI] [PubMed] [Google Scholar]
- 2.Dalle-Donne I, Scaloni A, Giustarini D, Cavarra E, Tell G, Lungarella G, Colombo R, Rossi R, Milzani A. Proteins as biomarkers of oxidative/nitrosative stress in diseases: the contribution of redox proteomics. Mass Spec Rev. 2005;24:55–99. doi: 10.1002/mas.20006. [DOI] [PubMed] [Google Scholar]
- 3.Davies MJ, Fu S, Wang H, Dean RT. Stable markers of oxidant damage to proteins and their application in the study of human disease. Free Radic Biol Med. 1999;27:1151–1163. doi: 10.1016/s0891-5849(99)00206-3. [DOI] [PubMed] [Google Scholar]
- 4.Davies MJ, Hawkins CL. EPR spin trapping of protein radicals. Free Radic BiolMed. 2004;36:1072–1086. doi: 10.1016/j.freeradbiomed.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 5.Augusto O, Vaz SM. EPR spin-trapping of protein radicals to investigate biological oxidative mechanisms. Amino Acids. 2007;32:535–542. doi: 10.1007/s00726-006-0429-4. [DOI] [PubMed] [Google Scholar]
- 6.Detweiler CD, Deterding LJ, Tomer KB, Chignell CF, Germolec D, Mason RP. Immunological identification of the heart myoglobin radical formed by hydrogen peroxide. Free Radic Biol Med. 2002;33:364–369. doi: 10.1016/s0891-5849(02)00895-x. [DOI] [PubMed] [Google Scholar]
- 7.Ramirez DC, Chen YR, Mason RP. Immunochemical detection of hemoglobin-derived radicals formed by reaction with hydrogen peroxide: involvement of a protein-tyrosyl radical. Free Radic Biol Med. 2003;34:830–839. doi: 10.1016/s0891-5849(02)01437-5. [DOI] [PubMed] [Google Scholar]
- 8.Ramirez DC, Gomez Mejiba SE, Mason RP. Mechanism of hydrogen peroxide-induced Cu,Zn-superoxide dismutase-centered radical formation as explored by immuno-spin trapping: the role of copper- and carbonate radical anion-mediated oxidations. Free Radic Biol Med. 2005;38:201–214. doi: 10.1016/j.freeradbiomed.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Ehrenshaft M, Mason RP. Protein radical formation on thyroid peroxidase during turnover as detected by immuno-spin trapping. Free Radic Biol Med. 2006;41:422–430. doi: 10.1016/j.freeradbiomed.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 10.Demicheli V, Quijano C, Alvarez B, Radi R. Inactivation and nitration of human superoxide dismutase (SOD) by fluxes of nitric oxide and superoxide. Free Radic Biol Med. 2007;42:1359–1368. doi: 10.1016/j.freeradbiomed.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 11.Bonini MG, Siraki AG, Atanassov BS, Mason RP. Immunolocalization of hypochlorite-induced, catalase-bound free radical formation in mouse hepatocytes. Free Radic Biol Med. 2007;42:530–540. doi: 10.1016/j.freeradbiomed.2006.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vlasova II, Tyurin VA, Kapralov AA, Kurnikov IV, Osipov AN, Potapovich MV, Stoyanovsky DA, Kagan VE. Nitric oxide inhibits peroxidasactivity of cytochrome c cardiolipin complex and blocks cardiolipin oxidation. J Biol Chem. 2006;281:14554–14562. doi: 10.1074/jbc.M509507200. [DOI] [PubMed] [Google Scholar]
- 13.Deterding LJ, Ramirez DC, Dubin JR, Mason RP, Tomer KB. Identification of free radicals on hemoglobin from its self-peroxidation using mass spectrometry and immuno-spin trapping: observation of a histidinyl radical. J Biol Chem. 2004;279:11600–11607. doi: 10.1074/jbc.M310704200. [DOI] [PubMed] [Google Scholar]
- 14.Detweiler CD, Lardinois OM, Deterding LJ, Ortiz de Montellano PR, Tomer KB, Mason RP. Identification of the myoglobin tyrosyl radical by immuno-spin trapping and its dimerization. Free Radic Biol Med. 2005;38:969–976. doi: 10.1016/j.freeradbiomed.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 15.Deterding LJ, Bhattacharjee S, Ramirez DC, Mason RP, Tomer KB. Top-down and bottom-up mass spectrometric characterization of human myoglobin-centered free radicals induced by oxidative damage. Anal Chem. 2007 doi: 10.1021/ac070935z. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen YR, Chen CL, Zhang L, Green-Church KB, Zweier JL. Superoxide generation from mitochondria NADH dehydrogenase induces self-inactivation with specific protein radical formation. J Biol Chem. 2005;280:37339–37348. doi: 10.1074/jbc.M503936200. [DOI] [PubMed] [Google Scholar]
- 17.Tsaprailis G, English AM. Different pathways of radical translocation in yeast cytochrome c peroxidase and its W191F mutant on reaction with H2O2 suggest an antioxidant role. J Biol Inorg Chem. 2003;8:248–255. doi: 10.1007/s00775-002-0407-6. [DOI] [PubMed] [Google Scholar]
- 18.Harris MN, Burchiel SW, Winyard PG, Engen JR, Mobarak CD, Timmins GS. Determining the site of spin trapping of the equine myoglobin radical by combined use of EPR, electrophoretic purification, and mass spectrometry. Chem Res Toxicol. 2002;15:1589–1594. doi: 10.1021/tx025594t. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, He S, Mauk AG. Radical formation at Tyr39 and Tyr153 following reaction of yeast cytochrome c peroxidase with hydrogen peroxide. Biochemistry. 2002;41:13507–13513. doi: 10.1021/bi026122g. [DOI] [PubMed] [Google Scholar]
- 20.Fenwick CW, English AM. Trapping and LC-MS identification of protein radicals formed in the horse heart metmyoglobin- H2O2 reaction. J Am Chem Soc. 1996;118:12236–12237. [Google Scholar]
- 21.Wilks A, Ortiz de Montellano PR. Intramolecular translocation of the protein radical formed in the reaction of recombinant sperm whale myoglobin with H2O2. J Biol Chem. 1992;267:8827–8833. [PubMed] [Google Scholar]
- 22.Lardinois OM, Ortiz de Montellano PR. H2O2-mediated cross-linking between lactoperoxidase and myoglobin. Elucidation of protein-protein radical transfer reactions. J Biol Chem. 2001;276:23186–23191. doi: 10.1074/jbc.M102084200. [DOI] [PubMed] [Google Scholar]
- 23.Antonini E, Brunori M, editors. Hemoglobin and myoglobin in their reactions with ligands. Amsterdam: Elsevier; 1971. [Google Scholar]
- 24.Nelson DP, Kiesow LA. Enthalpy of decomposition of hydrogen peroxide by catalase at 25 degrees C (with molar extinction coefficients of H2O2 solutions in the UV) Anal Biochem. 1972;49:474–478. doi: 10.1016/0003-2697(72)90451-4. [DOI] [PubMed] [Google Scholar]
- 25.Buettner GR. On the reaction of superoxide with DMPO/.OOH. Free Radic Res Commun. 1990;10:11–15. doi: 10.3109/10715769009145928. [DOI] [PubMed] [Google Scholar]
- 26.Mason RP. Using anti-5,5-dimethyl-1-pyrroline N-oxide (anti-DMPO) to detect protein radicals in time and space with immuno-spin trapping. Free Radical Biol Med. 2004;36:1214–1223. doi: 10.1016/j.freeradbiomed.2004.02.077. [DOI] [PubMed] [Google Scholar]
- 27.Deterding LJ, Moseley MA, Tomer KB, Jorgenson JW. Coaxial continuous flow fast atom bombardment in conjunction with tandem mass spectrometry for the analysis of biomolecules. Anal Chem. 1989;61:2504–2511. doi: 10.1021/ac00197a011. [DOI] [PubMed] [Google Scholar]
- 28.Deterding LJ, Barr DP, Mason RP, Tomer KB. Characterization of cytochrome c free radical reactions with peptides by mass spectrometry. J Biol Chem. 1998;273:12863–12869. doi: 10.1074/jbc.273.21.12863. [DOI] [PubMed] [Google Scholar]
- 29.Barr DP, Gunther MR, Deterding LJ, Tomer KB, Mason RP. ESR spin-trapping of a protein-derived tyrosyl radical from the reaction of cytochrome c with hydrogen peroxide. J Biol Chem. 1996;271:15498–15503. doi: 10.1074/jbc.271.26.15498. [DOI] [PubMed] [Google Scholar]
- 30.Annesley TM. Ion suppression in mass spectrometry. Clin Chem. 2003;49:1041–1044. doi: 10.1373/49.7.1041. [DOI] [PubMed] [Google Scholar]
- 31.Gunther MR, Tschirret-Guth RA, Witkowska HE, Fann YC, Barr DP, Ortiz de Montellano PR, Mason RP. Site-specific spin trapping of tyrosine radicals in the oxidation of metmyoglobin by hydrogen peroxide. Biochem J. 1998;330:1293–1299. doi: 10.1042/bj3301293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibson JF, Ingram DJE, Nicholls P. Free radical producted in the reaction of metmyoglobin with hydrogen peroxide. Nature. 1958;181:1398–1399. doi: 10.1038/1811398a0. [DOI] [PubMed] [Google Scholar]
- 33.King NK, Winfield ME. The mechanism of metmyoglobin oxidation. J Biol Chem. 1963;238:1520–1528. [PubMed] [Google Scholar]
- 34.Shiga T, Imaizumi K. Electron spin resonance study on peroxidase- and oxidase-reactions of horse radish peroxidase and methemoglobin. Arch Biochem Biophys. 1975;167:469–479. doi: 10.1016/0003-9861(75)90489-0. [DOI] [PubMed] [Google Scholar]
- 35.Egawa T, Shimada H, Ishimura Y. Formation of compound I in the reaction of native myoglobins with hydrogen peroxide. J Biol Chem. 2000;275:34858–34866. doi: 10.1074/jbc.M004026200. [DOI] [PubMed] [Google Scholar]
- 36.Makinen MW, Schichman SA, Hill SC, Gray HB. Heme-heme orientation and electron transfer kinetic behavior of multisite oxidation-reduction enzymes. Science. 1983;222:929–931. doi: 10.1126/science.6415814. [DOI] [PubMed] [Google Scholar]
- 37.Guidotti G, Konigsberg W. The characterization of modified human hemoglobinI. Reaction with iodoacetamide and N-ethylmaleimide. J Biol Chem. 1964;239:1474–1484. [PubMed] [Google Scholar]
- 38.Witting PK, Mauk AG. Reaction of human myoglobin and H2O2. Electron transfer between tyrosine 103 phenoxyl radical and cysteine 110 yields a protein-thiyl radical. J Biol Chem. 2001;276:16540–16547. doi: 10.1074/jbc.M011707200. [DOI] [PubMed] [Google Scholar]
- 39.Bhattacharjee S, Deterding LJ, Jiang J, Bonini MG, Tomer KB, Ramirez DC, Mason RP. Electron transfer between a tyrosyl radical and a cysteine residue in hemoproteins: spin-trapping analysis. J Am Chem Soc. 2007 doi: 10.1021/ja073349w. In press. [DOI] [PubMed] [Google Scholar]
- 40.Hugli TE, Gurd FRN. Carboxymethylation of sperm whale myoglobin in the dissolved state. J Biol Chem. 1970;245:1939–1946. [PubMed] [Google Scholar]