Abstract
Purpose
We sought to identify the genetic defect in a four-generation Chinese family with autosomal dominant congenital coralliform cataracts and demonstrate the functional analysis of a candidate gene in the family.
Methods
Family history data were recorded. Clinical and ophthalmologic examinations were performed on affected and unaffected family members. All the members were genotyped with microsatellite markers at loci considered to be associated with cataracts. Two-point LOD scores were calculated using the Linkage software after genotyping. A mutation was detected by direct sequencing, using gene-specific primers. Wild-type and mutant proteins were analyzed with online software.
Results
Affected members of this family had coralliform cataracts. Linkage analysis was obtained at markers, D2S72 (LOD score [Z]=3.31, recombination fraction [θ]=0.0) and D2S1782 (Z=3.01, θ=0.0). Haplotype analysis indicated that the cataract gene was closely linked to these two markers. Sequencing the γD-crystallin gene (CRYGD) revealed a G>T transversion in exon 2, which caused a conservative substitution of Gly to Cys at codon 61 (P.G61C). This mutation co-segregated with the disease phenotype in all affected individuals and was not observed in any of the unaffected or 100 normal, unrelated individuals. Bioinformatic analyses showed that a highly conserved region was located around Gly61. Data generated with online software revealed that the mutation altered the protein’s stability, solvent-accessibility, and interactions with other proteins.
Conclusions
This is the first reported case of a congenital coralliform cataract phenotype associated with the mutation of Gly61Cys (P.G61C) in the CRYGD gene; it demonstrates a possible mechanism of action for the mutant gene.
Introduction
Hereditary congenital cataracts is a clinically and genetically heterogeneous lens disease responsible for a significant proportion of visual impairment and blindness in childhood [1,2] It can occur in an isolated fashion or as one component of a multi-system disorder. Non-syndromic congenital cataracts have an estimated incidence of 1–6 per 10,000 live births [3-6]; at least one-third of cases are familial.
From the first description of the cosegregation of inherited cataracts with the Duffy blood group locus [7], more than 30 loci have been mapped through linkage analysis and 17 genes have been characterized [8]. These include 10 genes encoding crystallins (CRYAA, CRYAB, CRYBA1/A3, CRYBA, CRYBB1, CRYBB2, CRYBB3, CRYGC, CRYGD, CRYGS), three genes encoding membrane transport proteins (MIP, GJA3, GJA8), one encoding a cytoskeletal protein (BSFP2), and three encoding transcription factors (HSF4, MAF, PITX3) [9]. The crystallin genes encode more than 90% of the water-soluble structural proteins present in the vertebrate crystallin lens and clearly represent compelling candidate genes for congenital cataracts.
Crystallins are subdivided into α-, β-, and γ-crystallins. γ- and β-crystallins are included in a superfamily of microbial stress proteins, which share a common two-domain structure, composed of four “Greek-key” motifs. They were thought to be specific to lens fiber cells, but it has been recently reported that some β- and γ-crystallin components were found in lens epithelial cells [10]. The unique spatial arrangement and solubility of the crystallins are essential to the optical transparency and high refractive index of the lens. Modification of the crystallins may disrupt their normal structure in the lens and cause cataracts [11].
Coralliform cataracts are an uncommon form of congenital cataract; it was first reported in 1895 [12] and subsequently described as an autosomal dominant trait in three British pedigrees circa 1910 [13]. Several studies [14-17] have shown that mutations in the CRYGD gene, located at 2q33–35, were responsible for coralliform, aceuliform, and fasciculiform phenotypes. To date, about 16 articles have reported CRYGD gene mutations that cause congenital cataracts [18-33] of which about five concern coralliform cataracts [21,23,27,33].
We report a four-generation Chinese family with congenital coralliform cataracts. Linkage analysis mapped the disease gene to 2q33–35, and a missense mutation (181G→C) in CRYGD was identified in this family, resulting in the substitution of Gly61Cys (P.G61C) in CRYGD. Analysis of the wild-type and mutant proteins suggested that increased stability, complexity, and decreased hydrophilicity of the mutant protein may be the cause of coralliform congenital cataracts.
Methods
Clinical evaluation and DNA specimens
A four-generation family with non-syndromic congenital cataracts was recruited at the Beijing Tongren Eye Center, Capital Medical University, Beijing, China. Informed consent was obtained from each participant, consistent with the Declaration of Helsinki. Phenotype was documented by slit lamp photography. Genomic DNA was extracted from peripheral blood leukocytes using standard protocols.
Genotyping
Polymerase chain reactions (PCRs) were performed with microsatellite markers close to candidate loci associated with autosomal congenital cataracts. PCR products from each DNA sample were separated on a 6% polyacrylamide gel and analyzed. Pedigree and haplotype data were managed using Cyrillic (version 2.1) software. Exclusion analysis was performed by allele sharing in affected individuals.
Linkage analysis
A two-point linkage was calculated with the LINKAGE (version 5.1) package. Autosomal dominant cataracts were analyzed with full penetrance and a gene frequency of 0.001. The allele frequencies for each marker were assumed to be equal in both genders. The marker order and distances between the markers were taken from the NCBI and GDB databases.
DNA sequencing
Individual exons of the γ-crystallin gene cluster were amplified by PCR using primer pairs shown in Table 1 [34]. The PCR products were sequenced on an ABI3730 Automated Sequencer (PE Biosystems, Foster City, CA).
Table 1. Primer sequences used for sequencing CRYGA, CRYGB, CRYGC, and CRYGD.
| Gene (Exon) | Forward primers (5′→3′) | Reverse primers (5′→3′) |
|---|---|---|
|
CRYGA (1–2) |
TCCCTTTTGTGTTGTTTTTGCC |
TATGGCCATGGATCATTGATGC |
|
CRYGA (3) |
TCGTTGACACCCAAGGATGCATGC |
TACAAGAGCCACTTAGTGCAGGG |
|
CRYGB (1–2) |
TGCAAATCCCCTACTCACCAAAATGG |
TAAAAGATGGAAGGCAAAGACAGAGCC |
|
CRYGB (3) |
TAGGGACTGGAGCTTTAATTTCC |
TACTAGTGCCAGAAACACAAGC |
|
CRYGC (1–2) |
TGCAGGATGTTAAGAGATGC |
TTCTCTGATGTCCATCTAAGC |
|
CRYGC (3) |
TATTCCATGCCACAACCTACC |
TTGACAGAAGTCAGCAATTGC |
|
CRYGD (1–2) |
TCTTTTGTGCGGTTCTTGCCAACG |
TACCATCCAGTGAGTGTCCTGAGG |
| CRYGD (3) | TCTTTTTATTTCTGGGTCCGCC | TACAAGCAAATCAGTGCCAGG |
Denaturing HPLC
Denaturing HPLC was used to screen the mutation that was identified in the patients in the remaining patients, family members, and the 100 normal, unrelated control subjects in exon 2 of the CRYGD gene by using a commercial system (Wave DHPLC; Transgenomic, San Jose, CA).
Computer construction and analysis of protein models
The tertiary structure of the protein is highly conserved. Both mutant and wild-type versions of the protein structure were predicted and analyzed using the Swiss-model software (version 3.5), CLC protein workbench 3 (version 3.0.2), and the Phyre software (version 0.2).
Two-point LOD scores for chromosome 2q33–35 around the CRYGD locus. The highest observed LOD score was 3.31 (θ=0) for marker D2S72.
Results
Clinical data
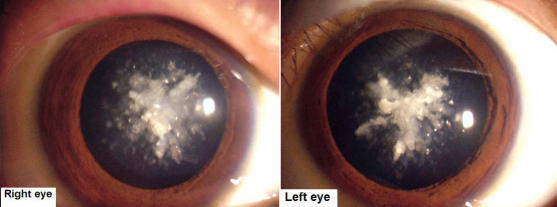
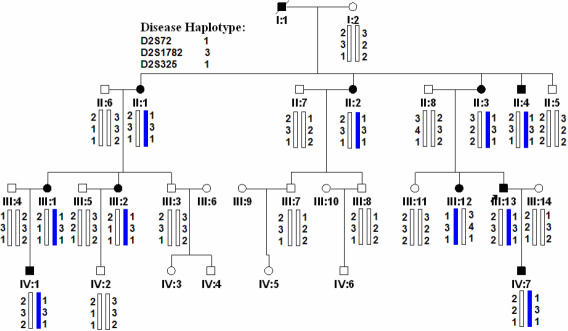
The proband was a 26-year-old male (III: 13) who had bilateral cataracts from birth. The form of the opacification was irregular, similar to sea coral, with crystal clumps radiating from the center to the capsule (Figure 1). All affected individuals showed a phenotype of coralliform cataracts. This four-generation family included 11 affected individuals with congenital coralliform cataracts and 20 unaffected individuals (Figure 2). The diagnosis was confirmed by ophthalmologists. There was no history of other ocular or systemic abnormalities in the family.
Figure 1.
Slit lamp photographs of affected individual III:13. The photographs of the affected individual III:13 show that the opacities were coralliform cataract. The form of the opacification was irregular, similar to sea coral, with crystal clumps radiating from the center to the capsule.
Figure 2.
Pedigree and haplotype of the cataract family. A four-generation pedigree, segregating autosomal dominant coralliform cataract, is shown. Haplotyping shows segregation of two microsatellite markers on 2q. Squares and circles indicate males and females, respectively. Filled symbols and bars denote affected status.
Linkage and haplotype analysis
The CRYGD gene on chromosome 2 was linked to this family while other candidate genes were excluded by allele sharing and linkage analysis. Significant linkage was found with markers D2S72 and D2S1782 and the maximum LOD score was 3.31 (at θ=0). Haplotype analysis showed that the responsible locus was localized at chromosome 2q33–35, flanked by markers D2S72, D2S325, and D2S1782 (Figure 2 and Table 2).
Table 2. Two-point LOD scores for linkage between cataract locus and markers on chromosome.
| Marker | LOD scores by recombination fraction (θ) | ||||||
|---|---|---|---|---|---|---|---|
| 0 |
0.04 |
0.09 |
0.14 |
0.19 |
0.24 |
0.29 |
|
| D2S72 |
3.31 |
3.1 |
2.82 |
2.53 |
2.21 |
1.88 |
1.53 |
| D2S1782 |
3.01 |
2.67 |
2.24 |
1.82 |
1.41 |
1.04 |
0.71 |
| D2S325 | 1.81 | 1.65 | 1.44 | 1.23 | 1.01 | 0.78 | 0.5 |
Mutation analysis for CRYGA, CRYGB, CRYGC, and CRYGD
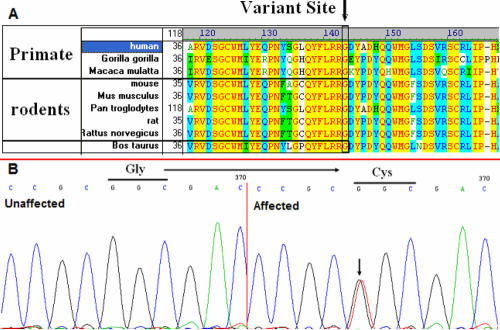
Direct cycle sequencing of the amplified fragments of CRYGD in two affected individuals identified a single base alteration, C.G181T (Figure 3B), in exon 2 of the CRYGD gene (GI: 181106), resulting in the substitution of Gly to Cys at codon 61 (P.G61C). The remainder of the coding sequence showed no other change.
Figure 3.
Multiple-sequence alignment and DNA sequence chromatograms of the CRYGD gene. A: Multiple-sequence alignment of CRYGD from primates, rodents, and cattle, to humans (Homo sapiens). The Gly61 residue is located within a highly conserved region. B: DNA sequence chromatograms of the P.G61C mutation in CRYGD. The G→T transversion at position 181 resulted in the P.G61C mutation.
Multiple-sequence alignment and mutation analysis
From the NCBI and UCSC websites we obtained the CRYGD family protein-sequences and using the Vector NTI software, we obtained multiple-sequence alignments of CRYGD family proteins in various species, including primates, rodents and cattle (Figure 3A). We found that codon 61, where the mutation (P.G61C) occurred, was located in a highly conserved region of the protein.
Denaturing HPLC
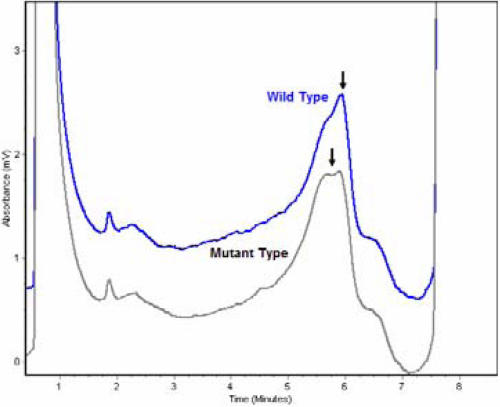
Denaturing HPLC analysis confirmed this mutation (Figure 4), which co-segregated with all affected individuals in the family. Further, this mutation was not observed in any of the unaffected family members or the 100 normal controls.
Figure 4.
Denaturing high-performance liquid chromatography results of wild-type and mutated CRYGD. Denaturing HPLC results show variant traces for CRYGD compared to the wild-type (WT) trace. The profile in black is the mutant protein; the profile in blue is the wild-type protein.
Discussion
We identified a new mutation, P.G61C, in the CRYGD gene in a four-generation Chinese family with autosomal dominant congenital coralliform cataracts. The disease gene was linked to 2q33–35 with a maximum LOD score of 3.31, spanning the γD-crystallin gene cluster, which includes CRYGA, CRYGB, CRYGC, and CRYGD. Mutation analysis of the candidate gene detected a new mutation, P.G61C, in CRYGD that co-segregated with the disease phenotype in all affected individuals but was not present in the unaffected family members or 100 normal control subjects. The result of multiple-sequence alignments showed that Gly61 was a highly conserved residue.
The lens crystallins constitute 80%–90% of the soluble proteins in the lens cells, and in most species, α-, β-, and γ-crystallins constitute the three main families. The human γ-crystallin gene cluster comprises six genes: CRYGA, CRYGB, CRYGC, CRYGD, CRYGE, and CRYGF, as well as a gene fragment of CRYGG [35]. In mammals, each of these genes consists of three exons; only CRYGC and CRYGD encode abundant lens γ-crystallins in humans [36,37]. CRYGD is one of the only two γ-crystallins to be expressed at high concentrations in the fiber cells of the embryonic human lens, which subsequently forms lens nucleus fibers [38-42]. For this reason and with the phenotype observed, we focused our attention on CRYGD. After screening for mutations in CRYGA, CRYGB, CRYGC, and CRYGD by direct cycle sequencing, we identified a G→T transversion in exon 2 of GRYGD, which was present only in affected members of the family. The transversion C.G181T located in exon 2 was predicted to cause a conservative substitution of Gly to Cys at codon 61 (P.G61C).
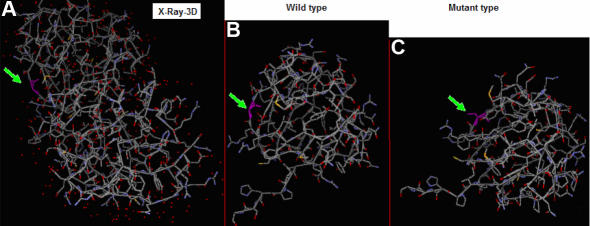
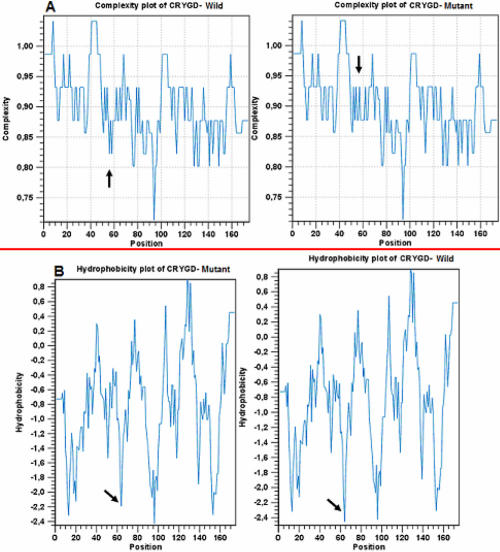
We used the online database PDB to study the three-dimensional (3D) structure of CRYGD (Figure 5A). This showed that Gly61 is an exposed surface residue on a strand. The online Phyre software (version 0.2) was used to compare the 3D-structures of the wild-type (Figure 5B) and mutant proteins (Figure 5C); the 3D-structure did not change much. CLC protein workbench 3 (version 3.0.2) predicted that the substitution in CRYGD would increase the complexity (Figure 6A) and hydrophobicity (Figure 6B) of the protein.
Figure 5.
Comparison of wild-type and mutant CRYGD 3D-structures. A: The CRYGD 3D-structure is from PDB. Gly61 is indicated in pink. B: Wild-type CRYGD 3D-structure is shown using the Phyre software; Gly61 is indicated in pink. C: Mutant CRYGD 3D-structure is displayed using the Phyre software; Cys61 is indicated in pink. Comparing mutant and wild-type CRYGD, the 3D-structure did not significantly differ.
Figure 6.
Comparison of complexity and hydrophobicity between wild-type and mutant CRYGD. CLC protein workbench 3 (version 3.0.2) predicted the effect of the substitution on CRYGD complexity (A) and hydrophobicity (B) of the protein. Complexity and hydrophobicity of the mutant protein increased (black arrow).
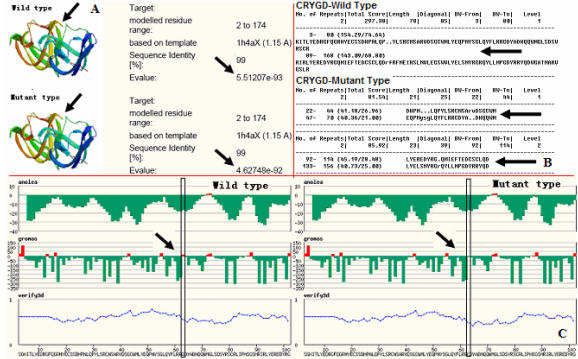
The online bioinformatics Swiss-model software (version 3.5) predicted both wild-type and mutant CRYGD structures; the P.G61C mutation exerted little effect on the tertiary structure of the protein but decreased the E-value and grooms value (Figure 7A,C). That is, the mutation is expected to stabilize the protein and affect the protein surface solvent accessibility and interactions with other proteins.
Figure 7.
Using the online Radar software to predict the effect of the substitution in CRYGD. Effect on alignment of repeats in protein sequences is shown in B. The Swiss-model software (version 3.5) predicted that the P.G61C mutation exerted little effect on the tertiary structure of the protein but decreased the E-value and grooms value (A,C).
Furthermore, we used Radar to predict the effect that the substitution would have in the wild-type protein with an increase from one to two repeats (Figure 7B). Many large proteins have evolved by internal duplication, and many internal sequence repeats correspond to functional and structural units.
The alteration had little effect on the backbone or 3D-structure of the protein; complexity and hydrophobicity of the mutant protein increased while the E-value and grooms value decreased. It is known that γ-crystallin is one of three major lens crystallin components (α-, β-, and γ-crystallins) [43]. They form heterogeneous oligomers in the lens and have molecular weights ranging from 40 to 200 kDa [44]. The predicted new characteristics of the mutant protein, specifically decreased water solubility and increased stability of the oligomers, may be the cause of the disease.
Acknowledgments
The authors thank the family for their participation in this project and Dr. Siquan Zhu and Dr. Shuzhen Wang (Beijing Tongren Hospital, Capital University of Medical Sciences, Beijing, China) for phenotype identification. This work is partly supported by grants from the National Natural Science Foundation of China (No. 30471864). This study was supported by the National Basic Research Program of China (No. 2007CB5119005), the National Infrastructure Program of Chinese Genetic Resources (No. 2006DKA21301), and the National Natural Science Foundation of China (No. 30471864).
References
- 1.Scott MH, Hejtmancik JF, Wozencraft LA, Reuter LM, Parks MM, Kaiser-Kupfer MI. Autosomal dominant congenital cataract. Interocular phenotypic variability. Ophthalmology. 1994;101:866–71. doi: 10.1016/s0161-6420(94)31246-2. [DOI] [PubMed] [Google Scholar]
- 2.Hejtmancik JF. The genetics of cataract: our vision becomes clearer. Am J Hum Genet. 1998;62:520–5. doi: 10.1086/301774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambert SR, Drack AV. Infantile cataracts. Surv Ophthalmol. 1996;40:427–58. doi: 10.1016/s0039-6257(96)82011-x. [DOI] [PubMed] [Google Scholar]
- 4.Francis PJ, Berry V, Bhattacharya SS, Moore AT. The genetics of childhood cataract. J Med Genet. 2000;37:481–8. doi: 10.1136/jmg.37.7.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renwick JH, Lawler SD. Probable linkage between a congenital cataract locus and the Duffy blood group locus. Ann Hum Genet. 1963;27:67–84. doi: 10.1111/j.1469-1809.1963.tb00782.x. [DOI] [PubMed] [Google Scholar]
- 6.Reddy MA, Francis PJ, Berry V, Bhattacharya SS, Moore AT. Molecular genetic basis of inherited cataract and associated phenotypes. Surv Ophthalmol. 2004;49:300–15. doi: 10.1016/j.survophthal.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Mackay DS, Andley UP, Shiels A. Cell death triggered by a novel mutation in the alphaA-crystallin gene underlies autosomal dominant cataract linked to chromosome 21q. Eur J Hum Genet. 2003;11:784–93. doi: 10.1038/sj.ejhg.5201046. [DOI] [PubMed] [Google Scholar]
- 8.Litt M, Kramer P, LaMorticella DM, Murphey W, Lovrien EW, Weleber RG. Autosomal dominant congenital cataract associated with a missense mutation in the human alpha crystallin gene CRYAA. Hum Mol Genet. 1998;7:471–4. doi: 10.1093/hmg/7.3.471. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Ma X, Gu F, Liu NP, Hao XL, Wang KJ, Wang NL, Zhu SQ. A missense mutation S228P in the CRYBB1 gene causes autosomal dominant congenital cataract. Chin Med J (Engl) 2007;120:820–4. [PubMed] [Google Scholar]
- 10.Wang X, Garcia CM, Shui YB, Beebe DC. Expression and regulation of alpha-, beta-, and gamma-crystallins in mammalian lens epithelial cells. Invest Ophthalmol Vis Sci. 2004;45:3608–19. doi: 10.1167/iovs.04-0423. [DOI] [PubMed] [Google Scholar]
- 11.Van Rens GL, Driessen HP, Nalini V, Slingsby C, de Jong WW, Bloemendal H. Isolation and characterization of cDNAs encoding beta A2- and beta A4-crystallins: heterologous interactions in the predicted beta A4-beta B2 heterodimer. Gene. 1991;102:179–88. doi: 10.1016/0378-1119(91)90076-n. [DOI] [PubMed] [Google Scholar]
- 12.Gunn RM. Peculiar coralliform cataract with crystals in the lens. Trans Ophthalmol Soc. 1895;XV:119. [Google Scholar]
- 13.Harman NB. Ten pedigrees of congenital and infantile cataract; lamellar, coralliform, discoid, and posterior polar with microphthalmia. Trans Ophthalmol Soc U K. 1910;30:251–74. [Google Scholar]
- 14.Heon E, Priston M, Schorderet DF, Billingsley GD, Girard PO, Lubsen N, Munier FL. The gamma-crystallins and human cataracts: a puzzle made clearer. Am J Hum Genet. 1999;65:1261–7. doi: 10.1086/302619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mackay DS, Andley UP, Shiels A. A missense mutation in the gammaD crystallin gene (CRYGD) associated with autosomal dominant “coral-like” cataract linked to chromosome 2q. Mol Vis. 2004;10:155–62. [PubMed] [Google Scholar]
- 16.Shentu X, Yao K, Xu W, Zheng S, Hu S, Gong X. Special fasciculiform cataract caused by a mutation in the gammaD-crystallin gene. Mol Vis. 2004;10:233–9. http://www.molvis.org/molvis/v10/a29/ [PubMed] [Google Scholar]
- 17.Gu JZ, Qi YH, Lin H, Li X, Wang J, Meng WM, Su H. Autosomal dominant congenital golden crystal nuclear cataract caused by a missense mutation in gammaD crystallin gene (CRYGD) in a Chinese family. Zhonghua Yan Ke Za Zhi. 2006;42:913–7. [PubMed] [Google Scholar]
- 18.Kmoch S, Brynda J, Asfaw B, Bezouska K, Novák P, Rezácová P, Ondrová L, Filipec M, Sedlácek J, Elleder M. Link between a novel human gammaD-crystallin allele and a unique cataract phenotype explained by protein crystallography. Hum Mol Genet. 2000;9:1779–86. doi: 10.1093/hmg/9.12.1779. [DOI] [PubMed] [Google Scholar]
- 19.Nandrot E, Slingsby C, Basak A, Cherif-Chefchaouni M, Benazzouz B, Hajaji Y, Boutayeb S, Gribouval O, Arbogast L, Berraho A, Abitbol M, Hilal L. Gamma-D crystallin gene (CRYGD) mutation causes autosomal dominant congenital cerulean cataracts. J Med Genet. 2003;40:262–7. doi: 10.1136/jmg.40.4.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burdon KP, Wirth MG, Mackey DA, Russell-Eggitt IM, Craig JE, Elder JE, Dickinson JL, Sale MM. Investigation of crystallin genes in familial cataract, and report of two disease associated mutations. Br J Ophthalmol. 2004;88:79–83. doi: 10.1136/bjo.88.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu WZ, Zheng S, Xu SJ, Huang W, Yao K, Zhang SZ. Localization and screening of autosomal dominant coralliform cataract associated gene. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2004;21:19–22. [PubMed] [Google Scholar]
- 22.Shentu X, Yao K, Xu W, Zheng S, Hu S, Gong X. Special fasciculiform cataract caused by a mutation in the gammaD-crystallin gene. Mol Vis. 2004;10:233–9. [PubMed] [Google Scholar]
- 23.Xu WZ, Zheng S, Xu SJ, Huang W, Yao K, Zhang SZ. Autosomal dominant coralliform cataract related to a missense mutation of the gammaD-crystallin gene. Chin Med J (Engl) 2004;117:727–32. [PubMed] [Google Scholar]
- 24.Shentu XC, Yao K, Sun ZH, Xu W. Study on ultrastructure changes and the genetic locus for a special phenotype cataract. Zhonghua Yan Ke Za Zhi. 2004;40:306–10. [PubMed] [Google Scholar]
- 25.Zenteno JC, Morales ME, Moran-Barroso V, Sanchez-Navarro A. CRYGD gene analysis in a family with autosomal dominant congenital cataract: evidence for molecular homogeneity and intrafamilial clinical heterogeneity in aculeiform cataract. Mol Vis. 2005;11:438–42. [PubMed] [Google Scholar]
- 26.Gu J, Qi Y, Wang L, Wang J, Shi L, Lin H, Li X, Su H, Huang S. A new congenital nuclear cataract caused by a missense mutation in the gammaD-crystallin gene (CRYGD) in a Chinese family. Mol Vis. 2005;11:971–6. [PubMed] [Google Scholar]
- 27.Gu F, Li R, Ma XX, Shi LS, Huang SZ, Ma X. A missense mutation in the gammaD-crystallin gene CRYGD associated with autosomal dominant congenital cataract in a Chinese family. Mol Vis. 2006;12:26–31. [PubMed] [Google Scholar]
- 28.Messina-Baas OM, Gonzalez-Huerta LM, Cuevas-Covarrubias SA. Two affected siblings with nuclear cataract associated with a novel missense mutation in the CRYGD gene. Mol Vis. 2006;12:995–1000. [PubMed] [Google Scholar]
- 29.Gu JZ, Qi YH, Lin H, Li X, Wang J, Meng WM, Su H. Autosomal dominant congenital golden crystal nuclear cataract caused by a missense mutation in gammaD crystallin gene (CRYGD) in a Chinese family. Zhonghua Yan Ke Za Zhi. 2006;42:913–7. [PubMed] [Google Scholar]
- 30.Plotnikova OV, Kondrashov FA, Vlasov PK, Grigorenko AP, Ginter EK, Rogaev EI. Conversion and compensatory evolution of the gamma-crystallin genes and identification of a cataractogenic mutation that reverses the sequence of the human CRYGD gene to an ancestral state. Am J Hum Genet. 2007;81:32–43. doi: 10.1086/518616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen L, Yao W, Eiberg H, Kjaer KW, Baggesen K, Hejtmancik JF, Rosenberg T. Genetic heterogeneity in microcornea-cataract: five novel mutations in CRYAA, CRYGD, and GJA8. Invest Ophthalmol Vis Sci. 2007;48:3937–44. doi: 10.1167/iovs.07-0013. [DOI] [PubMed] [Google Scholar]
- 32.Xu WZ, Zheng S, Dong Q, Cai SR, Yao K, Zhang SZ. Ultrastructure and crystallin mutant molecular modeling of hereditary coralliform cataract. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2005;34:243–7. doi: 10.3785/j.issn.1008-9292.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 33.Evans P, Wyatt K, Wistow GJ, Bateman OA, Wallace BA, Slingsby C. The P23T cataract mutation causes loss of solubility of folded gammaD-crystallin. J Mol Biol. 2004;343:435–44. doi: 10.1016/j.jmb.2004.08.050. [DOI] [PubMed] [Google Scholar]
- 34.Mackay DS, Andley UP, Shiels A. A missense mutation in the γD crystallin gene (CRYGD) associated with autosomal dominant “coral-like” cataract linked to chromosome 2q. Mol Vis. 2004;10:155–62. [PubMed] [Google Scholar]
- 35.Xu WZ, Zheng S, Xu SJ, Huang W, Yao K, Zhang SZ. Autosomal dominant coralliform cataract related to a missense mutation of the γD-crystallin gene. Chin Med J (Engl) 2004;117:727–32. [PubMed] [Google Scholar]
- 36.Russell P, Meakin SO, Hohman TC, Tsui LC, Breitman ML. Relationship between proteins encoded by three human gamma-crystallin genes and distinct polypeptides in the eye lens. Mol Cell Biol. 1987;7:3320–3. doi: 10.1128/mcb.7.9.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brakenhoff RH, Aarts HJ, Reek FH, Lubsen NH, Schoenmakers JG. Human gamma-crystallin genes. A gene family on its way to extinction. J Mol Biol. 1990;216:519–32. doi: 10.1016/0022-2836(90)90380-5. [DOI] [PubMed] [Google Scholar]
- 38.David LL, Shearer TR, Shih M. Sequence analysis of lens beta-crystallins suggests involvement of calpain in cataract formation. J Biol Chem. 1993;268:1937–40. [PubMed] [Google Scholar]
- 39.Riazuddin SA, Yasmeen A, Yao W, Sergeev YV, Zhang Q, Zulfiqar F, Riaz A, Riazuddin S, Hejtmancik JF. Mutations in betaB3-crystallin associated with autosomal recessive cataract in two Pakistani families. Invest Ophthalmol Vis Sci. 2005;46:2100–6. doi: 10.1167/iovs.04-1481. [DOI] [PubMed] [Google Scholar]
- 40.Willoughby CE, Shafiq A, Ferrini W, Chan LL, Billingsley G, Priston M, Mok C, Chandna A, Kaye S, Heon E. CRYBB1 mutation associated with congenital cataract and microcornea. Mol Vis. 2005;11:587–93. [PubMed] [Google Scholar]
- 41.Mackay DS, Boskovska OB, Knopf HL, Lampi KJ, Shiels A. A nonsense mutation in CRYBB1 associated with autosomal dominant cataract linked to human chromosome 22q. Am J Hum Genet. 2002;71:1216–21. doi: 10.1086/344212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bloemendal H, de Jong W, Jaenicke R, Lubsen NH, Slingsby C, Tardieu A. Ageing and vision: structure, stability and function of lens crystallins. Prog Biophys Mol Biol. 2004;86:407–85. doi: 10.1016/j.pbiomolbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 43.Harding J. Cataract: biochemistry, epidemiology, and pharmacology.1st ed. London: Chapman and Hall; 1991. [Google Scholar]
- 44.Lampi KJ, Ma Z, Shih M, Shearer TR, Smith JB, Smith DL, David LL. Sequence analysis of betaA3, betaB3, and betaA4 crystallins completes the identification of the major proteins in young human lens. J Biol Chem. 1997;272:2268–75. doi: 10.1074/jbc.272.4.2268. [DOI] [PubMed] [Google Scholar]