Abstract
A less than adequate therapeutic plan for the treatment of anthrax in the 2001 bioterrorism attacks has highlighted the importance of developing alternative or complementary therapeutic approaches for biothreat agents. In these regards passive immunization possesses several important advantages over active vaccination and the use of antibiotics, as it can provide immediate protection against Bacillus anthracis. Herein, we report the selection and characterization of several human monoclonal neutralizing antibodies against the toxin of B. anthracis from a phage displayed human scFv library. In total fifteen clones were selected with distinct sequences and high specificity to protective antigen and thus were the subject of a series of both biophysical and cell-based cytotoxicity assays. From this panel of antibodies a set of neutralizing antibodies were identified, of which clone A8 recognizes the lethal (and/or edema) factor binding domain, and clone F1, G11 and G12 recognize the cellular receptor binding domain within protective antigen. It was noted that all clones distinguish a conformational epitope existing on the protective antigen; this steric relationship was uncovered using a sequential epitope mapping approach. For each neutralizing antibody, the kinetic constants were determined by surface plasmon resonance, while the potency of protection was established using a two-tier macrophage cytotoxicity assay. Among the neutralizing antibodies identified, clone F1 possessed the highest affinity to protective antigen, and provided superior protection from lethal toxin in the cell cytotoxicity assay. The data presented provides to the ever-growing arsenal of immunological and functional analysis of monoclonal antibodies to the exotoxins of anthrax. In addition it grants new candidates for the prophylaxis and therapeutic treatment against this toxin.
Keywords: Bacillus anthracis, protective antigen, human monoclonal antibodies, neutralizing antibodies, phage antibody library
1. Introduction
The use of anthrax spores as an agent for bioterrorism activities in 2001 has led to a renewed interest in the fundamental biology and pathology of the bacterium B. anthracis as well as research initiatives to identify reliable treatments for the inhalation anthrax. Indeed, this terrorist attack has elevated anthrax to the position of one of the six highest-risk threat agents for bioterrorism (category A agents defined by the Center for Disease Control and Prevention (CDC), http://www.bt.cdc.gov/agent/agentlist-category.asp#a). Consequently, the development of vaccines has come under intense public pressure as they are viewed as safe therapeutic countermeasures against anthrax.
Anthrax toxin, one of the two major virulence factors of B. anthracis, is a tripartite toxin composed of protective antigen (PA), lethal factor (LF), and edema factor (EF). Toxic activity is expressed only when receptor-binding moiety PA is combined with distinct enzymatic moieties LF and/or EF, consequently forming the lethal toxin (LeTx) and edema toxin (EdTx), respectively 1–3. Protective antigen, named for its use in vaccines, is central to the action of the LeTx and EdTx, and is singularly the most important antigen required for conferring specific immunity to anthrax; additionally it is the most critical factor presented in all current anthrax vaccine efforts 4–6.
Research using vaccination as a therapeutic for the protection against anthrax exposure has been known for more than a century 5. However, the prolonged vaccination schedules and induction times required in mounting an immune response are considered serious drawbacks, as the therapeutic window for treatment of a deliberate release of B. anthracis is limited. Furthermore, vaccination would not be effective with immunocompromised individuals, and would offer little or no protection for the inhalation of anthrax and thus the mucosal surface. Alternatively, recently developed antibiotic prophylaxis for the treatment of anthrax exposure while important, would also be of lesser value in cases of infection with antibiotic-resistant strains that could be encountered 7–9.
Passive immunization has provided an attractive avenue as a post-exposure and/or pre-exposure treatment. Indeed, passive transfer of antiserum has successfully provided protection from anthrax in a large body of animal studies 10–13. Serum therapy has also been used in the past for the treatment of human anthrax with some success 14. Furthermore, studies with various vaccines indicate a strong correlation between the titer of PA neutralizing antibodies and the potency of the vaccine 15, and suggest that PA neutralizing antibodies are the main mechanism of vaccine-induced protective immunity 16. Overall these findings highlight the importance of PA neutralizing antibodies in conferring protection against anthrax and also demonstrate the ability of such antibodies to be effectively applied as a post-exposure therapy. Finally, passive immunization could have advantages over active vaccination and antibiotic treatments via low toxicity, high specificity, large scale stockpile capabilities, and immediate protection against a biological attack 17.
The molecular mechanisms by which anthrax toxin enters cells 18, structural information on each of the toxin components 19–21, and the action of toxin enzymes are fairly well understood 22, 23. Hence, anthrax presents a very good model for antidote design, and antitoxins that act upon the mechanism of action of the toxin (including toxin binding, assembly, translocation into target cells) have been developed 12, 24–27. This suggests that intrinsic neutralizing epitopes exist within the toxin structural motif. However, it may also require a combination of toxin neutralizing antibodies to simultaneously neutralize several epitopes to provide for full protection.
It has been noted that antiserum can provide protection against the toxicity of anthrax toxin; yet, the development of effective human neutralizing antibodies that can be produced in sufficient amounts will be of great value not only to allow complete assessment of passive immunization, but as a conduit to provide safe and effective clinical applications in humans. In these regards, human monoclonal antibodies, which are derived from vaccinated donors, have been developed that prevent PA binding to its receptor 12, 13, 28, or LF binding to PA63 heptamer 13. However, complete protection is still an important goal.
Herein, we describe the screening and selection of human antibodies against PA83 and PA63 via phage display technology, including assessment of identified antibodies to neutralize distinct epitopes existing on PA. The rationale of our approach is based on the structure and function of PA as displayed in anthrax toxin’s mechanism of action 19, 29; our experimental design is shown in Figure 1, which features a series of selection and characterization criteria with its foundation grounded upon the following potential neutralizing epitopes: 1) The proteolytic cleavage site as found within PA83 domain 1 that is essential for toxin activation. 2) Oligomerization sites as found on the activated PA63 domain 2, which are important for heptamer formation and subsequent LF and EF binding; 3) LF and/or EF binding site(s) on heptameric (or oligomeric) PA63 domain 1’, which are essential for both LeTx and EdTx action. 4) The cellular receptor-binding site on PA domain 4, which is central in strategies for the development vaccines for anthrax.
Figure 1.
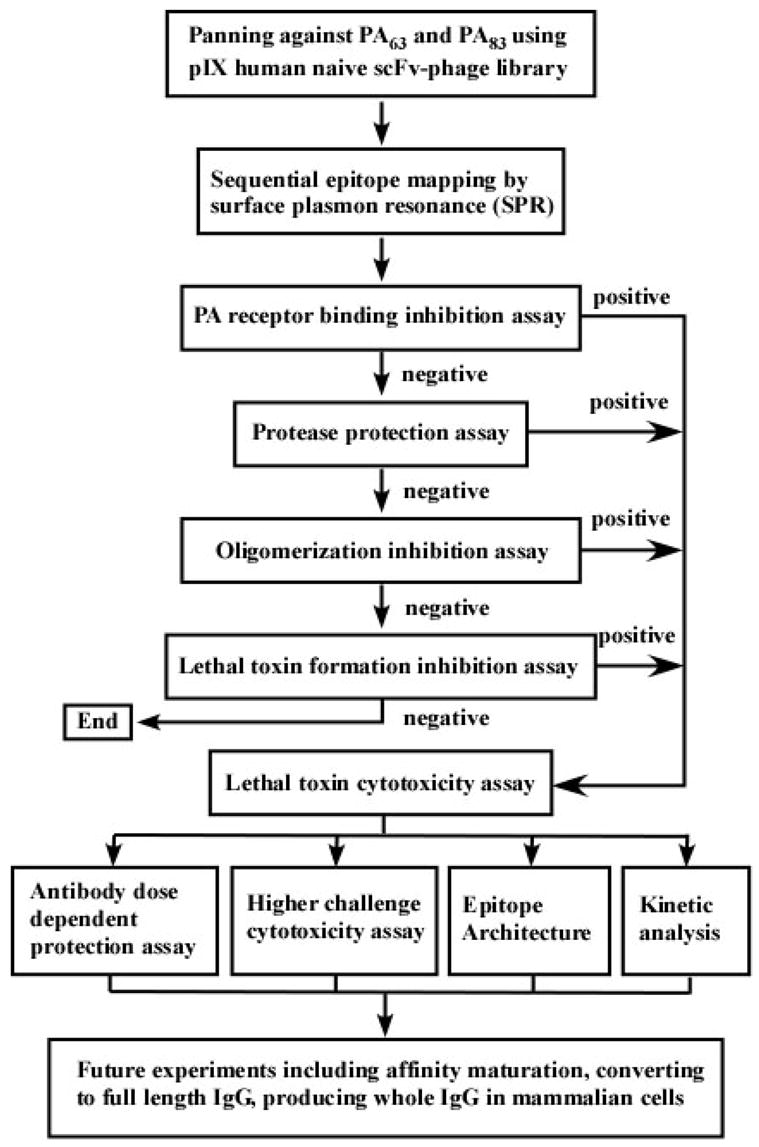
Flowchart detailing the process for the selection and characterization of human monoclonal neutralization antibodies against B. anthracis toxin.
Using this “road map” for selection purposes, fifteen unique human monoclonal antibody scFvs were uncovered, the most important of which, F1, G11, and G12 recognize and obstruct the PA cellular receptor-binding site; while Fab A8 blocked the LF/EF binding site on PA63 heptamer (or oligomer). We also demonstrate that these antibodies can provide protection in a LeTx challenged murine RAW264.7 macrophage cell cytotoxicity assay.
2. Results
2.1. Screening and selection of human anti-PA scFvs
A human naïve pIX display scFv-phage library30 was used to screen for scFvs against PA63 and PA83. The library was subjected to four rounds of panning, and ninety-six clones were randomly picked from each antigen panning experiment, which were subsequently screened for antigen binding by ELISA; the positive rate was 96% for coated PA63 and 80% for coated PA83, respectively. Forty clones for each antigen were selected for sequencing in accordance with the general diversity and the strength of their binding activity. Twelve clones for PA63 and three clones for PA83 with distinct VH and/or VL sequences were employed in specificity analysis by cross-reactive ELISA against PA63, PA83, LF, EF, and BSA. Finally, PCR assembly converted eight selected clones with exquisite specificity against PA63 and/or PA83 to a Fab format. The resulting Fab fragments were subcloned into a pET-His expression vector. While each Fab protein was expressed in Rosetta 2 (DE3) E. coli strain in their corresponding optimum medium, and purified by IMAC and protein G chromatography. The yield of purified Fab was in the range of 2–8 mg/L; ELISA reconfirmed binding activity and specificity. The purity of each Fab was analyzed by SDS-PAGE (reduced and non-reduced) and was found to be greater than 90%.
2.2. Epitope mapping by surface plasmon resonance
Epitope mapping was conducted using surface plasmon resonance (SPR) technology, which granted a means to determine steric relationships between antigenic sites within PA63 as defined by affinity/specificity of each of the selected Fabs. The method used to uncover each epitope was based on a sequential binding assay wherein the antigen was immobilized on a sensory chip surface and different pairs of antibodies were used in an attempt to recognize distinct epitopes. Using this protocol the first probe (antibody) was allowed to bind and reach saturation; this was followed by the addition of a second probe; if distinct epitopes were recognized then a further increase in the resonance signal would be observed with the pair of antibodies. Otherwise, recognition of a same, or a shared overlapping epitope would result in either no signal enhancement or less signal enhancement. The order of probes was also reversed and by examining all selected clone combinations antibody-PA epitopes were confirmed. The results of this sequential epitope mapping technique revealed that there are four clusters of epitopes on PA that can be readily recognized by our defined Fabs. Among them, A8 recognizes a distinct epitope, while F1 and G12, F11 and G3, and G11, H2 and 83H2 recognize same epitope, respectively. The later three clusters also have the similar epitope overlapping between each other. The results of our epitope mapping of these eight defined Fabs are summarized in Table I and illustrated in Figure 3.
Table I.
Reactivity pattern matrix for the panel of eight selected Fabs. The epitope mapping results revealed four different epitopes exist on PA. Fab A8 recognizes a distinct epitope. Fab F1 and G12, F11 and G3, and G11, H2 and 83H2 recognize a similar epitope, respectively. The later three clusters also have epitope overlapping with each other in some extent.
| Second Probe | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| A8 | F1 | F11 | G3 | G11 | G12 | H2 | 83H2 | ||
| First Probe | A8 | ✘ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| F1 | ✓ | ✘ | UC | UC | UC | ✘ | ✓ | ✓ | |
| F11 | ✓ | UC | ✘ | ✘ | UC | UC | ✓ | ✓ | |
| G3 | ✓ | UC | ✘ | ✘ | UC | UC | ✓ | ✓ | |
| G11 | ✓ | UC | UC | UC | ✘ | UC | ✘ | ✘ | |
| G12 | ✓ | ✘ | UC | UC | UC | ✘ | ✓ | ✓ | |
| H2 | ✓ | ✓ | ✓ | ✓ | ✘ | ✓ | ✘ | UC | |
| 83H2 | ✓ | ✓ | ✓ | ✓ | ✘ | ✓ | UC | ✘ | |
Recognition of the same or a very similar epitope;
recognition of a different epitope; UC: uncertain, may recognize an overlapping or slightly shifted epitope.
Figure 3.
Four clusters of epitopes recognized by selected Fabs. The picture depicts a set of potential epitopes (overlapping circles), as defined by the selected Fabs binding to the PA63 heptamer surface (PDB code: 1TZO). Fab A8 recognizes an epitope within LF/EF binding site. Fabs F1 and G12, F11 and G3, and G11, H2 and 83H2 all recognize a similar epitope near the PA cellular receptor binding site. The later three clusters also have epitopes overlapping with each other but with less homology than the other identified epitopes.
2.3. Screening for human PA neutralizing Fabs
An MTT (3-(4,5-dimethylthiazolyl-2)-2, 5-diphenyltetraolium bromide) cell proliferation assay 31 based macrophage cytotoxicity 32 was engaged to identify human PA neutralization antibodies. To begin, a linear range for the MTT assay was determined and found to be 0.8 × 104 – 6 × 104 cells/well; based on this finding we seeded 100 μl of 5 × 105 cells/mL cell suspension for each well in 96-well culture plates. Titration of the cytotoxicity for LeTx on cultured RAW264.7 macrophage cells was determined and the optimal concentration of PA83 and LF used for the experiment was found to be of 1 μg/mL for both components, which resulted in 100% killing of RAW264.7 cells. Using a time course for the LeTx challenge assay, it was discovered that upon exposure of the RAW264.7 cells to LeTx at a concentration of 1μg/mL, 30 min was the threshold time for protection. Hence, 30 min was considered to be the minimum time course needed for LeTx to enter and/or bind to the cell surface and eventually grant 100% killing of the cultured RAW264.7 macrophage cells. This was true even if LeTx was removed from the culture medium in MTT assay that require a longer incubation period.
For preliminary screening for potential neutralizing antibodies, the antibody concentration utilized was 400μg/mL. A less stringent protocol (lower challenge assay, LCA) in the cell-based LeTx neutralization assays was initially examined, i.e., an individual antibody was first incubated with PA83 for 1 hr, and then LF was added to cultured RAW264.7 cells.
To identify all possible neutralization Fabs, we designed four different assay protocols (Figure 1) based on the structure of PA and the mechanism of anthrax toxin intoxication, namely: 1) A PA receptor-binding inhibition assay, 2) a protease protection assay, 3) an oligomerization inhibition assay, 4) a LeTx formation inhibition assay. Protocols were also devised for a LeTx cytotoxicity assay for general use. Through the processing of all four protocols, we identified three Fabs (F1, G11, G12) from protocol 1 and one Fab (A8) from protocol 4 that could provide protection from LeTx challenge to various extents. Unfortunately, we did not identify any neutralizing Fabs using protocols 2 and 3. The neutralizing ability of all identified Fabs was further confirmed as positive using the general LeTx cytotoxicity assay.
2.4. Characterization of human PA neutralization Fabs
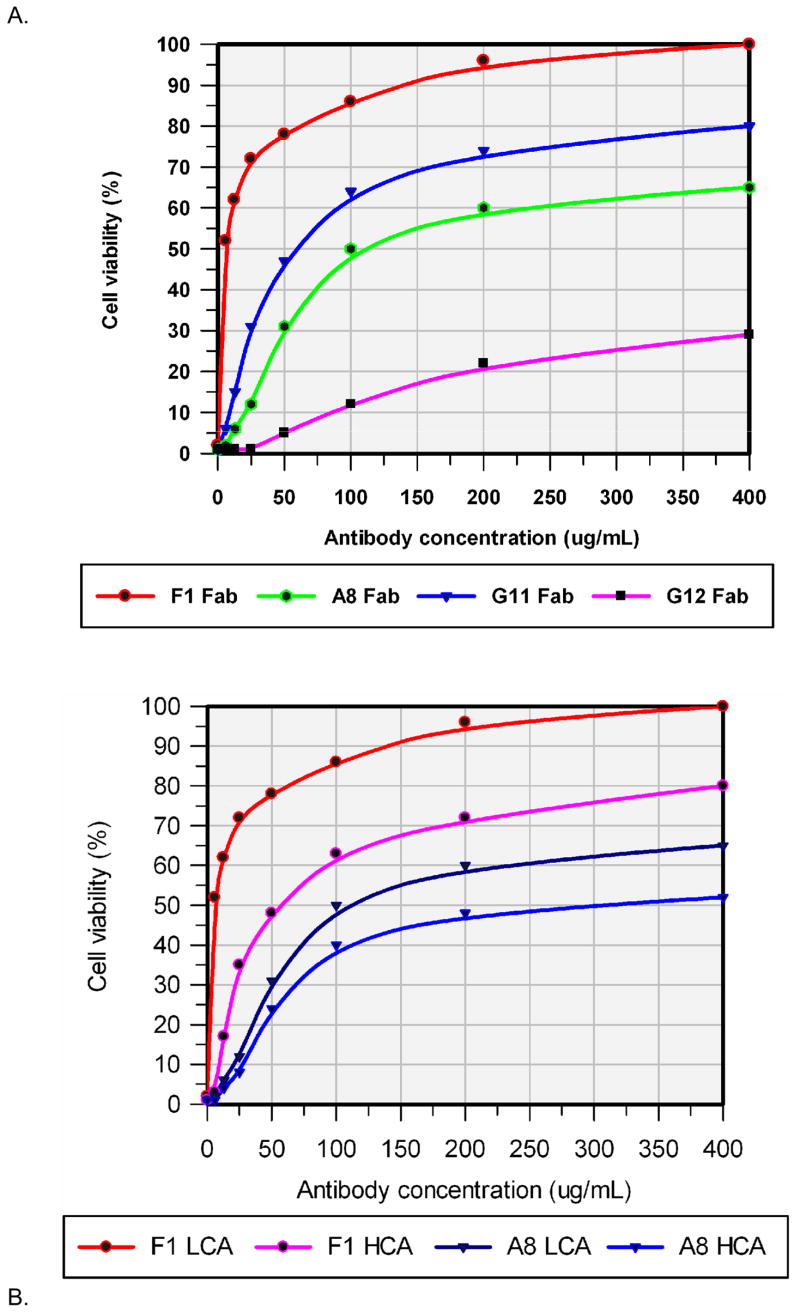
To evaluate the potency of defined neutralizing Fabs, an antibody dose dependent protection assay was conducted. Thus, serially diluted individual antibodies ranging from 400μg/mL to 6.25μg/mL was used in the LeTx cytotoxicity assay; only the assay results are shown in Figure 2A, and are summarized in Table II. Among those defined neutralizing Fabs, F1 provided a superior protection from LeTx in the macrophage cytotoxicity assay, while A8 and G11 provided some protection, while G12 was observed to provide the least protection. Also noted was that Fab F1, could provide up to 50% of protection even at a Fab/LeTx molar ratio as low as ca. 10.
Figure 2.
Antibody dose dependent LeTx protection assay for defined neutralization Fabs. Results represent the mean values from twenty-one wells of a representative experiment. Culture medium with 1μg/mL of PA83 and 1μg/mL of LF was used as a positive control, while culture medium alone was used as a negative control. A. For the lower challenge assay (LCA), individual Fab concentrations ranged from 400μg/mL to 6.25μg/mL; each were incubated with 1μg/mL of PA83 at room temperature for 1 hr, and then added to cultured RAW264.7 cells and incubated at 37 °C for 1 hr. LF (final concentration of 1μg/mL) was added to culture medium and incubated at 37 °C for 4 hr. Cell viability was estimated using an MTT assay. B. For the higher challenge assay (HCA), serial diluted individual Fabs ranging from 400μg/mL to 6.25μg/mL were mixed with 1μg/mL of PA83 and 1μg/mL of LF, the mixture was added to the RAW26.7 cells immediately, and incubated at 37 °C for 4 hr. Cell viability was estimated by the MTT assay.
Table II.
Fab neutralization potency and possible PA interaction domains. Antibody PA neutralization potency was determined in an antibody dose dependent protection assay with lower and higher challenge approaches as stated in the text. In a lower challenge assay, antibody is pre-incubated with PA, added to the RAW264.7 cells, and then LF is applied for the MTT assay. In the higher challenge assay, antibody, PA and LF are mixed and immediately applied to the RAW264.7 cells for the MTT assay. The possible epitopes on PA are defined by a neutralizing antibody as deduced via the assay.
| Fab | Neutralization Potency (IC) | Possible Epitope on PA |
|---|---|---|
| A8 | a65% at 400 μg/mL, 50% at 110 μg/mL | Domain 1’, LF binding site |
| b54% at 400 μg/mL, 50% at 300 μg/mL | ||
| F1 | a100% at 400 μg/mL, 50% at .25μg/mL | Domain 4, ATR binding site |
| b80% at 400 μg/mL, 50% at 58 μg/mL | ||
| G11c | a80% at 400 μg/mL, 50% at 60 μg/mL | Domain 4, ATR binding site |
| G12c | a29% at 400 μg/mL | Domain 4, ATR binding site |
Lower challenge assay;
Higher challenge assay;
Lower challenge assay only
To further explore the PA neutralizing activities of defined Fabs, a more challenging antibody dose dependent protection assay (higher challenge assay, HCA) was performed. In this assay, serially diluted individual antibodies ranging from 400μg/mL to 6.25μg/mL were mixed with PA83 and LF, the mixture was immediately added to the RAW26.7 cells, and no pre-incubation was allowed. Only the assay results are shown in Figure 2B, and are also summarized in Table II. As anticipated, in the higher challenge assay, the LeTx neutralization ability for all Fabs declined, and the molar ratio of Fab/LeTx, which could provide 50% protection also increased approximately three-fold for A8 and approximately nine-fold for F1, respectively.
Western-blot and dot-blot investigations were also carried out so as to characterize the epitope architecture, i.e. linear versus conformational epitope. We note that western-blot analysis can only present a linear, hence a contiguous structure to a detecting probe, while dot-blot will provide both linear and a conformational structure to the detecting probe. Here individual Fabs were used as the detecting probe in both blots. The blot results revealed that all neutralizing Fabs recognized a conformational epitope rather than a linear epitope.
To determine the kinetic constants of the defined neutralizing Fabs, kinetic analysis using Biacore 3000 was employed. PA63 or PA83 were immobilized onto a CM5 chip using NHS/EDC chemistry with a target of 500 RU. Various concentrations of each Fab ranging from 800 nM to 3.125 nM were injected over the chip surface, and the interaction between Fab and immobilized PA were recorded within the sensorgram. The kinetic data were evaluated via fitting the sensorgram data by BIAevaluation software using 1:1 (Langmuir) binding model. The kinetic constants, including association and dissociation equilibrium constants (KA and KD) and association and dissociation rate constants (kon and koff), were determined for each Fab and are summarized in Table III. The affinities of the selected neutralizing Fabs to PA were in the range of 50 nM to 470 nM. Among them, F1 possessed the most potent affinity, while G12 had the poorest affinity. The on-rates for all the Fabs were found to be in the lower range i.e. 104 M−1s−1, and the off rates were also in the lower range and found to be 10−3 s−1 except G12, which was determined to at 10−2 s−1.
Table III.
Kinetic constants determined for all neutralizing Fabs by Biacore 3000.
| Fab | ka (1/Ms) | kd (1/s) | KA (1/M) | KD (M) |
|---|---|---|---|---|
| A8 | 3.47 × 104 | 3.78 × 10−3 | 9.17 × 106 | 1.09 × 10−7 |
| F1 | 2.71 × 104 | 1.38 × 10−3 | 1.96 × 107 | 5.09 × 10−8 |
| G11 | 2.84 × 104 | 2.31 × 10−3 | 1.23 × 107 | 8.13 × 10−8 |
| G12 | 4.03 × 104 | 1.96 × 10−2 | 2.13 × 106 | 4.68 × 10−7 |
3. Discussion
The toxin secreted by B. anthracis, i.e., anthrax toxin, possesses the ability to impair innate and adaptive immune responses, which in turn potentates the bacterial infection. Tragically, the use of anthrax spores in several paper envelopes may have exposed as many as 30,000 US citizens, resulting in 11 cases of inhalational anthrax and 5 deaths, which have led to a renewed interest in the fundamental microbiology and pathology of the bacterium B. anthracis. Indeed, it was the inadequate antibiotic therapy for the treatment of anthrax 33 and obstacles confronted by the current anthrax vaccines that has highlighted the importance of, and renewed thrust for the development of alternative or complementary therapeutic approaches, including passive immunization.
Passive immunization has successfully provided protection from anthrax in both animal 10–12 and human studies 14. Factors that have driven these studies have include low toxicity, high specificity, stockpile capabilities, and immediate protection, hence, passive immunization may offer an excellent avenue for either post-exposure and/or pre-exposure treatments for anthrax attack. Accordingly, the goal of our research initiative was to select and characterize human monoclonal neutralizing antibodies directed against the central component of anthrax toxin, the protective antigen. In these regards we have successfully identified several PA neutralizing antibodies; our tact relied upon the use of a high quality human scFv-phage library, coupled with a novel set of panning/selection protocols and conformation of antibody neutralization activity through a reliable cell-based assay.
Since PA binding to its cellular receptor is essential for both LeTx and EdTx activity, prevention and/or inhibition of binding of PA to its receptor are central in current strategies for therapeutic advancement for the treatment of anthrax. In accordance with these tenets we utilized a designed in house pIX human scFv-phage library to directly pan against various forms of PA using a guided approach (Figure 1) based on the molecular mechanism which anthrax enters cells. Thus, using this unique panning strategy we uncovered antibodies against both PA63 and PA83. The results were deemed to be encouraging as four out of fifteen selected human Fabs were identified as PA neutralizing; excitingly some of these antibodies provided excellent protection in an in-vitro macrophage cytotoxicity assay.
A biosensor-based epitope mapping approach was employed that greatly facilitated the efficacy and the accuracy of the mapping process. The most common approach to epitope mapping using Fabs is a two-site binding sandwich assay, where the first probe, (a Fab), is captured or immobilized on the chip followed by injection of antigen and the second probe 34. An alternative approach is sequential epitope mapping, where antigen is first captured or immobilized on a chip, followed by injection of the first probe so as to reach saturation, and finally the injection of the second probe. Because of its adaptability, we used sequential mapping in our experiments that is especially useful, as PA63 tends to form homo-heptamers, which possess problems with two-site mapping. Thus, the first probe can saturate the antigen surface epitope, which in turn can save a significant quantity of antigen, (PA63). More salient is the use of a capture molecule (e.g. antibody) to secure the [PA63]7 or tagged [PA63]7 in the sequential epitope mapping experiment. A significant benefit using this scheme includes the elimination of conformational changes of the antigen that may occur during the immobilization process.
Thus, through sequential epitope mapping, we have identified at least four clusters of epitopes from a panel of eight selected Fabs. The possible cross-relationships of these epitopes between themselves and each other are illustrated in Figure 3. Noteworthy, is that the epitope recognized by Fab A8 is distinct from any other epitope clusters, and we have mapped it’s binding to the LF and/or EF binding domain according to our LF inhibition assay. Whether this antibody interacts with the monomer PA63 or oligomerlized PA63, which would mirror LF/EF 35 interactions is under further investigation.
The stoichiometry of Fab A8 with [PA63]7 is unknown, but should be less than seven as judged by size exclusion chromatography analysis (data not shown). The second (Fab F1 and G12), third (F11 and G3), and fourth (G11, H2 and 83H2) cluster of antibodies recognize the same epitope, respectively. However, in the fourth cluster, the epitope recognized by G11, H2 and 83H2 has slightly shifted, and thus does not overlap or share perfect homology as is seen in the second and third cluster. Based on the results of LeTx cytotoxicity assay, the later three clusters should be localized at the cellular receptor-binding domain and cover a broad area of PA domain 4. The second cluster may be the most potent epitope of the four as Fab F1 provides the most protection under the least antibody/PA ratio. Potentially, the reason of why G12, in cluster 2, provided weak protection maybe due to its lower affinity to PA. Interestingly, no neutralizing antibodies were uncovered to cluster 3, which may imply that this cluster is not a protective epitope(s). In cluster 4, only G11 was protective, this may mean a slight shift in the epitope domain by Fabs H2 and 83H2 that dramatically decreased their protection potency. Finally, the stoichiometry of F1 to PA83 should be one, but is unknown to [PA63]7 where the actual stoichiometry of F1 to [PA63]7 may be equal or less than seven, and will need to be determined in future investigations.
Analysis of our mapping results has provided evidence that at the cellular receptor-binding site there is a broad region that can confer protection. We note that the epitopes in these domains may be more accessible than others, and this may explain why the cellular-receptor-binding domain of PA, in contrast to the EF/LF binding site, is more important in generating a protective antibody response to PA 1. Furthermore, in accordance with our epitope mapping findings, we hypothesize that increased protection maybe obtained by combining antibodies that recognize different clusters of epitopes. We believe this approach would greatly enhance the opportunity of blocking the interaction between PA and its cellular receptor and between enzymatic moiety and its PA docking apparatus.
The molecular mechanisms by which anthrax enters cells 18, structural information on each toxin’s components 19–21, and the enzymatic action of the toxin are fairly well understood 22, 23. Based on the knowledge of the reported structure of PA and its role in anthrax toxicity 19, 29, we developed four protocols for selecting antibodies that may recognize/block the following potential neutralizing epitopes: 1) the proteolytic activation site within PA83 domain 1, which is essential for toxin action; 2) the oligomerization sites on activated PA63 domain 2, which is important for heptamer formation and subsequent LF and EF binding; 3) the LF and/or EF binding sites on heptameric (or oligomeric) PA63 domain 1’, which is essential for both LeTx and EdTx action.; 4) the cellular receptor binding site on PA domain 4. Application of these protocols is a central precept in the strategic development of therapeutic vaccines for anthrax. The data we have provided clearly shows that these protocols when used in concert can successfully grant the identification of several anti-PA human neutralizing monoclonal antibodies. Among those defined as neutralizing human Fabs, F1, G11 and G12 appear to recognize and thus block the PA cellular receptor binding domain, while A8 blocks the LF/EF binding site on PA63 heptamer (or oligomer). We also noted that to gain an insight into how these antibodies exactly interact with anthrax toxin, a fine epitope mapping, including fragment library, site-directed mutagenesis, and/or crystallographic analysis should be conducted in the future.
Recently, a novel antibody neutralizing mechanism for anthrax toxin that requires Fc receptor engagement for maximal activity, has been reported 36. The authors discovered that antibodies elicited by anthrax vaccines do not function by inhibiting initial PA binding but via other epitopes that permit an Fc receptor-mediated interaction on the cell surface. They also demonstrated that this is factual not only for a particular monoclonal antibody, but also to be true with CDC human reference antiserum (AVR801) prepared from pooled antisera of AVA-immunized human individuals and other immune serum collected from different animal species that were vaccinated with recombinant PA83. This has largely extended the possibility of discovering all potential neutralizing antibodies that may greatly enhance the efficacy of antibody mediated prophylactic and postsymptomatic treatment of anthrax. This may also imply that antibodies, especially those obtained from non-immunized approaches (such as naïve antibody-phage library), with very high specificity and affinity to anthrax toxin that can not inhibit the initial PA/ATR or PA/LF/EF binding, should be converted to an IgG format with a proper Fc effector fragment and selection for protective ability via an Fc mediated cell cytotoxicity assay or animal studies.
PA possesses one site for EF/LF binding and one site for receptor binding. These two binding sites may well contain those antigenic epitopes most critical to inducing a protective immune response to anthrax. In live attenuated strains, the cellular receptor binding domain of PA, in contrast to the EF/LF binding site, is more important in generating an antibody response to PA 1. It has been established that vaccination with PA induces antibodies to both the cell receptor and the EF/LF binding domains that neutralize the cytotoxicity of lethal toxin in vitro 37. Our results are consistent with those findings and confirmed the importance of epitopes on these two binding sites, especially the cell receptor binding domain, in conferring the protection to anthrax toxin. Among four of our defined neutralizing Fabs, three of them are directly targeted at PA cellular receptor binding site, and only one is targeted at LF/EF binding domains. Interestingly, we failed to identify neutralizing antibodies against other functional domains, including the protease activation domain, and oligomerization domain. This may due to the fact that the protective epitopes in these domains are not readily exposed to antibodies in general, their conformation may be changed when coated to an immunotube, or the affinity of antibodies against these epitopes is not high enough to be selected from the naïve antibody library.
For each of the defined PA neutralizing Fabs, we ranked their neutralization potency via antibody dose dependent protection using two slightly different approaches. The first approach was less stringent wherein the PA-Fab mixture was pre-incubated before addition to the cells or the addition of the LF. As a more rigorous test PA, LF, and Fab were directly mixed into cultured RAW264.7 cells without pre-incubation. Under such conditions, the antibody must compete with PA for cellular receptor and/or LF binding. The protection potency was decreased for all antibodies in this assay, and thus may imply that a higher affinity neutralizing antibody is needed to compete with the strong interaction between PA and its cell receptor (KD ~ 0.2 nM) 38 or LF (KD ~ 1 nM) 39. This later experiment is a more likely scenario for a pre-exposure challenge to anthrax and thus we note a post-exposure cell based assay was not conducted. Our reasoning here were that results encountered for the time course of LeTx toxicity revealed that the time window available for antibody protection in our cell assay could be found only in the initial 30 min, which would have been difficult to correlate in this cell cytotoxcicity assay.
Through kinetic analysis using surface plasma resonance technology (SPR) via Biacore, we have determined the association and dissociation equilibrium constants (KA and KD) and association and dissociation rate constants (kon and koff) for all defined neutralizing antibodies. From such studies, the affinities displayed by the neutralizing Fabs were in a range from 50 to 470 nM. Notably, the association rate constants (ka or kon) of these Fabs were on the order of 104 (M−1s−1) and the dissociation rate constants (kd or koff) were 10−3(s−1), which is at the lower end of naturally occurring antibodies. The PA heptamer can bind to its cellular receptor and a maximum of three molecules of LF and/or EF with high affinity (KD 0.2–1 nM) 35, 38–43. Furthermore, the dissociation rate constant for [PA63]7 itself is even higher (1 × 10–6 s−1, t1/2 ~ 7 days) 44. These pieces of data suggest that neutralizing antibodies with greater affinity to PA warrant investigation and thus could provide greater protection. With this thought in mind we plan to investigate in-vitro affinity maturation approaches to improve not only the dissociation rate constant (koff), but also the association rate constant (kon) for all neutralizing antibodies to enhance their affinity and hopefully their protection potency.
At this time, more than ever, there is a need for additional countermeasures against anthrax. As evidenced by the comparatively small-scale events after September 11, 2001, there are weaknesses in our capabilities to deal with an anthrax attack. Vaccines and antibiotics must continue to undergo development, as well as new approaches based on the inhibition of toxin-receptor interactions as well as bacteriolytic phage 45–47. We believe that continued investigations into human PA neutralizing monoclonal antibodies for prophylaxis and/or antibody therapeutics are attractive paths that eventually will be effective in curtailing future acts of bioterrorism.
4. Experimental
4.1. Screening and selection of human anti-PA scFvs
A naïve human pIX display scFv-phage library 30 was used to screen human scFvs against PA63 and PA83. The panning was performed as described elsewhere 30. In brief, the library was amplified and the scFv-phage was rescued. The purified scFv-phage library was applied to PA63 or PA83 (List Biological Laboratories, Inc.) coated onto immunotubes and incubated for 2 hr at room temperature. The tubes were thoroughly washed with PBST (PBS with 0.05% Tween-20), followed by the addition of freshly prepared TG1 competent cells for infection at 37 °C for 20 min. The cells were placed onto gcLB agar plates (LB agar supplemented with 2% glucose and 100 μg/mL carbenicillin), and incubated at 30 °C overnight. The second day, the cells were scraped off the plate, amplified and the scFv-phage was rescued for the next round of panning. This procedure was repeated three to four times until a desired enrichment was attained. After the final round of panning, ninety-six clones were randomly picked from each antigen panning experiment. The monoclonal scFv-phage was amplified and rescued in 96-well plates, and subjected to positive clone selection via standard ELISA. Forty positive clones for each antigen were selected for sequencing in accordance with the general diversity and the strength of their binding activity. All sequences were analyzed and certain numbers of clones were picked for both PA63 and PA83 with distinct VH and/or VL sequences based on a specificity analysis by cross-reactive ELISA against PA63, PA83, LF, EF, and BSA. Finally, clones with high specificity against PA63 and/or PA83 were selected.
To further characterize these defined scFvs, they were all converted into a Fab format, which is more stable than scFv format. The conversion of scFv to a Fab format was carried out by PCR assembly 48; in brief, the variable region gene of each antibody heavy chain (VH) and light chain (VL representing either Vκ or Vλ) were amplified directly from the defined scFv gene by PCR. The constant region gene of the antibody heavy chain (Cλ1) and light chain (Cκ or Cλ) were amplified from our Fab expression vector (pETFabHis) by PCR. The heavy chain gene (VH-Cλ1) and light chain gene Vκ-Cκ (or Vλ-Cλ) were obtained by PCR assembly. The Fab gene was eventually created by PCR assembling of heavy and light chain genes as one 1.5 kb fragment, and cloned into an expression vector, pET-His 49, by two flanked asymmetric Sfi I site. The antibody gene sequences were further analyzed and confirmed by DNA sequencing. To obtain soluble Fab protein, each Fab construct was transformed and expressed in Rosetta 2 (DE3) E. coli strain (Novagen, Inc.) in their defined optimum expression medium (Media Optimization Kit, United States Biological, Inc.). Fab protein from periplasmic space was purified by IMAC and then refined by protein G chromatography (GE Healthcare). The binding activity and specificity of each converted Fab was characterized by standard ELISA. The purity was analyzed by SDS-PAGE (reduced and non-reduced).
4.2. Epitope mapping by surface plasmon resonance
To reveal the steric relationships between antigenic sites on PA that were recognized by selected Fabs, we performed a sequential epitope mapping experiment using surface plasmon resonance (SPR) technology on a Biacore 3000 instrument (Biacore AB). In brief, PA63 was immobilized onto CM5 chip by NHS/EDC amine-coupling method as specified by the manufacture’s instructions (Biacore AB). Thus, we injected the first Fab and observed the binding of analyte to ligand via the sensorgram. When the surface was saturated, we immediately injected the second Fab and observed the binding activity. The order of injection of the Fabs was reversed to perform additional mapping. This procedure was repeated for all Fabs with all possible binary combinations undertaken, and the binding signal for each pair of Fabs was recorded in a reactivity pattern matrix for further analysis. For all Biacore analysis, we always included double-reference as controls, i.e., a reference flow cell and blank buffer run.
4.3. Screening of human PA neutralization Fabs
The murine macrophage cell line RAW264.7 was used in all cell based assay, and the cells were cultured as described elsewhere 32, 48. An MTT cell proliferation assay31 (ATCC) was engaged for identification of human PA neutralization antibodies. For all cell assays, we used medium alone and cell alone as negative controls, and excessive LeTx as positive control. To obtain an accurate quantification of changes in the rate of cell proliferation, the linear relationship between cell number and signal produced for RAW264.7 cells was first determined according to manufacture’s instructions. The final concentrations of LeTx used for the experiments were also determined by LeTx serial titrations so as to result in a reliable 100% killing end point for the RAW264.7 cells.
Evaluation of PA and LeTx cell entry into macrophages was accomplished using a time course of LeTx challenge on cultured RAW264.7 cells. In brief, PA was added to cultured RAW264.7 cells and incubated for a period of 5, 15, 25, 30, 45 and 60 min at 37 °C. PA was removed and the cells were washed by fresh culture medium, followed by the addition of LF and incubation at 37 °C for 4 hr before conducting the MTT assay. Alternatively, PA and LF were simultaneously added to cultured cells and incubated for selected periods of time as detailed (vide supra). Finally, the PA and LF were removed, and the cells were continuously incubated at 37 °C for 4 hr before conducting MTT assay.
To identify human PA neutralizing Fabs, four different protocols (Figure 1) were performed sequentially as follows: 1). PA receptor-binding inhibition assay; individual Fabs were first incubated with PA83 at room temperature for 1 hr; here PA83 alone was the positive control. The Fab-PA83 mixture or PA83 control was added to cultured RAW264.7 cells and incubated at 4 °C for 1 hr. The cells are thoroughly washed with cold PBS, harvested and lysed with SDS buffer. The lysate was applied and separated by SDS-PAGE gel, which was electronically transferred to a NC membrane, for Western-blot analysis using mouse anti-PA IgG (C86501M, Biodesign Intl.). The antibody was ranked as positive, if it could inhibit the binding and endocytosis of PA83 into the cells, as evidenced by the absence of the 83 kDa (PA83) and/or 63 kDa (PA63) band as visualized by Western-blot analysis. Those antibodies that were positive were examined directly in the LeTx cytotoxicity neutralization assay (vide infra); otherwise, antibodies were advanced to the protease protection assay. 2) Protease protection assay; here individual Fabs were first incubated with PA83 at room temperature for 1 hr. Trypsin (Sigma) was added to the Fab-PA83 complex to a final concentration of trypsin/PA83 ratio of 1:1000 (w/w), and the mixture was incubated at room temperature for 30 min. A trypsin/PA83 reaction without antibody was used as a positive control, while PA83 and antibody were the negative controls. The reaction was stopped by the addition of a 10-fold molar excess of soybean trypsin inhibitor (Sigma). The sample was separated by SDS-PAGE gel, and Western-blot analysis was performed as shown in protocol 1. The antibody was ranked as positive, if it was able to protect PA83 from trypsin cleavage, as evidenced by the presence of an 83 kDa (PA83) band on the Western-blot. Those antibodies that were positive were examined directly in the LeTx cytotoxicity neutralization assay; otherwise, the antibodies were put forth into the oligomerization inhibition assay. 3) Oligomerization inhibition assay; in this assay individual Fabs were first incubated with PA83 at room temperature for 1 hr. PA83 and antibody were viewed as controls while all samples were activated by trypsin as described in protocol 2 and the trypsin activated samples were applied and separated by Superdex-200 (GE Healthcare). The antibodies were ranked as positive, if they could inhibit PA from forming an oligomer, as evidenced by the presence of low molecular weight peaks (e.g., Fab and Fab-PA63), but without and/or with reduced concentrations of high molecular weight peaks (e.g., [PA63]7 ~ 441 kDa). Those antibodies that were positive were examined directly in the LeTx cytotoxicity neutralization assay; otherwise, the antibodies were moved forward into the PA LF-binding inhibition assay. 4) LeTx formation inhibition assay; PA83 was preincubated with RAW264.7 cells at 4°C for 2 hr followed by the addition of individual Fabs and incubation at 4 °C for another hour. In this experiment PA83 was the positive control and cultured medium alone a negative control. Next, LF was added to the Fab-PA63-cell mixture and incubated at 37 °C for 4 hr. Cell viability was determined by the MTT assay and the antibody was ranked as positive, if it could inhibit LeTx cytotoxicity, while those antibodies that were negative were disregarded for further studies.
For the general LeTx cytotoxicity assay, individual Fabs were incubated with PA83 at room temperature for 1 hr, and then added to the cultured RAW264.7 cells and incubated at 37 °C for 1 hr. For all LeTx cytotoxicity assay, we used medium alone, cell alone, and cell with defined Fab as negative controls, and cell with 1μg/mL of PA83, cell with excessive LeTx, and cell with 1μg/mL of LeTx with defined amount of mouse PA-neutralizing IgG 106 (RDI-TRK3BA16-106, Research Diagnostics, Inc.) as positive controls. LF was then added to the culture and incubated at 37 °C for 4 hr. Cell viability was estimated by the MTT assay and the antibody was confirmed as positive, if it could inhibit the LeTx cytotoxicity, as evidenced by a higher macrophage cell viability compared to the control.
4.4. Characterization of human PA neutralization Fabs
To evaluate the potency of all defined neutralizing Fabs, an antibody dose dependent protection assay was conducted as follows. Serial diluted individual antibody ranging from 400μg/mL to 6.25μg/mL was used to perform the LeTx cytotoxicity assay as described (vide supra). For each antibody concentration, the assay was repeated within three columns (twenty-one wells in total) of cultured macrophage cells. Antibody dose dependent protection curves were created from mean values by Microsoft Excel.
To further explore the PA neutralizing activity of defined Fabs, a more stringent antibody dose dependent protection assay was examined. Thus, serial diluted individual antibody ranging from 400μg/mL to 6.25μg/mL were mixed with PA83 and LF, the mixture was added to the RAW26.7 cells immediately, and incubated at 37 °C for 4 hr before the MTT assay.
To determine whether the defined Fabs recognized a linear or conformational epitope, Western and dot-blot analysis was performed. PA63 or PA83 was separated on SDS-PAGE gel, and blotted to NC membrane. PA63 or PA83 was also directly dotted onto the NC membrane for the assay. Both membranes were blocked, incubated with defined Fabs, and detected by goat anti-human IgG (Fab specific) HRP conjugate. Fab detected on both blots recognized a linear epitope, while Fab only detected on dot-blot was considered to recognize only a conformational epitope.
To determine the kinetic constants of those defined neutralizing Fabs, Biacore 3000 was engaged for kinetic analysis according to manufacture’s instructions. In brief, PA63 or PA83 was immobilized onto CM5 chip targeting at 500 RU using NHS/EDC coupling chemistry, while an in-line reference with BSA immobilized at the same level was also setup. Various concentrations of each Fab ranging from 800 nM to 3.125 nM were injected onto the chip surface, and the interaction between Fab and immobilized PA were recorded in the sensorgram. The kinetic data were evaluated via fitting the sensorgram data by BIAevaluation software using 1:1 (Langmuir) binding model. The kinetic constants, including association and dissociation equilibrium rate constants (KA and KD) and association and dissociation rate constants (kon and koff), were determined for each Fab.
Acknowledgments
We thank Dr. Gao Changshou for preparing and providing the human naïve pIX display scFv-phage library, and Dr. Han Jiahuai for providing the RAW264.7 cell line. This work was supported by grant from the National Institute of Allergy and Infectious Diseases (Grant Number: AI061271-03).
Abbreviations
- B. anthracis
Bacillus anthracis
- scFv
single chain Fv
- PA
protective antigen
- PA83
mature protein of protective antigen
- PA63
protective antigen 63-kDa fragment
- [PA63]7
PA63 heptamer
- LF
lethal factor
- LeTx
lethal toxin
- EF
edema factor
- EdTx
edema toxin
- ELISA
enzyme-linked immunosorbent assay
- SPR
surface plasmon resonance
- PCR
polymerase chain reaction
- HRP
horse radish peroxidase
- BSA
bovine serum albumin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brossier F, Weber-Levy M, Mock M, Sirard JC. Infect Immun. 2000;68:1781. doi: 10.1128/iai.68.4.1781-1786.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abrami L, Reig N, van der Goot FG. Trends Microbiol. 2005;13:72. doi: 10.1016/j.tim.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Barth H, Aktories K, Popoff MR, Stiles BG. Microbiol Mol Biol Rev. 2004;68:373. doi: 10.1128/MMBR.68.3.373-402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brey RN. Adv Drug Deliv Rev. 2005;57:1266. doi: 10.1016/j.addr.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 5.Friedlander AM, Welkos SL, Ivins BE. Curr Top Microbiol Immunol. 2002;271:33. doi: 10.1007/978-3-662-05767-4_3. [DOI] [PubMed] [Google Scholar]
- 6.Leppla SH, Robbins JB, Schneerson R, Shiloach J. J Clin Invest. 2002;110:141. doi: 10.1172/JCI16204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Price LB, Vogler A, Pearson T, Busch JD, Schupp JM, Keim P. Antimicrob Agents Chemother. 2003;47:2362. doi: 10.1128/AAC.47.7.2362-2365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brook I, Elliott TB, Pryor HI, 2nd, Sautter TE, Gnade BT, Thakar JH, Knudson GB. Int J Antimicrob Agents. 2001;18:559. doi: 10.1016/s0924-8579(01)00464-2. [DOI] [PubMed] [Google Scholar]
- 9.Choe CH, Bouhaouala SS, Brook I, Elliot TB, Knudson GB. Antimicrob Agents Chemother. 2000;44:1766. doi: 10.1128/aac.44.6.1766-1766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobiler D, Gozes Y, Rosenberg H, Marcus D, Reuveny S, Altboum Z. Infect Immun. 2002;70:544. doi: 10.1128/IAI.70.2.544-550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maynard JA, Maassen CB, Leppla SH, Brasky K, Patterson JL, Iverson BL, Georgiou G. Nat Biotechnol. 2002;20:597. doi: 10.1038/nbt0602-597. [DOI] [PubMed] [Google Scholar]
- 12.Sawada-Hirai R, Jiang I, Wang F, Sun SM, Nedellec R, Ruther P, Alvarez A, Millis D, Morrow PR, Kang AS. J Immune Based Ther Vaccines. 2004;2:5. doi: 10.1186/1476-8518-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wild MA, Xin H, Maruyama T, Nolan MJ, Calveley PM, Malone JD, Wallace MR, Bowdish KS. Nature biotechnology. 2003;21:1305. doi: 10.1038/nbt891. [DOI] [PubMed] [Google Scholar]
- 14.Knudson GB. Mil Med. 1986;151:71. [PubMed] [Google Scholar]
- 15.Cohen S, Mendelson I, Altboum Z, Kobiler D, Elhanany E, Bino T, Leitner M, Inbar I, Rosenberg H, Gozes Y, Barak R, Fisher M, Kronman C, Velan B, Shafferman A. Infect Immun. 2000;68:4549. doi: 10.1128/iai.68.8.4549-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Little SF, Ivins BE, Fellows PF, Pitt ML, Norris SL, Andrews GP. Vaccine. 2004;22:422. doi: 10.1016/j.vaccine.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Casadevall A. Emerg Infect Dis. 2002;8:833. doi: 10.3201/eid0808.010516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abrami L, Liu S, Cosson P, Leppla SH, van der Goot FG. J Cell Biol. 2003;160:321. doi: 10.1083/jcb.200211018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petosa C, Collier RJ, Klimpel KR, Leppla SH, Liddington RC. Nature. 1997;385:833. doi: 10.1038/385833a0. [DOI] [PubMed] [Google Scholar]
- 20.Pannifer AD, Wong TY, Schwarzenbacher R, Renatus M, Petosa C, Bienkowska J, Lacy DB, Collier RJ, Park S, Leppla SH, Hanna P, Liddington RC. Nature. 2001;414:229. doi: 10.1038/n35101998. [DOI] [PubMed] [Google Scholar]
- 21.Shen Y, Guo Q, Zhukovskaya NL, Drum CL, Bohm A, Tang WJ. Biochem Biophys Res Commun. 2004;317:309. doi: 10.1016/j.bbrc.2004.03.046. [DOI] [PubMed] [Google Scholar]
- 22.Mourez M. Rev Physiol Biochem Pharmacol. 2004;152:135. doi: 10.1007/s10254-004-0028-2. [DOI] [PubMed] [Google Scholar]
- 23.Moayeri M, Leppla SH. Curr Opin Microbiol. 2004;7:19. doi: 10.1016/j.mib.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Xu J, Li G, Dong D, Song X, Guo Q, Zhao J, Fu L, Chen W. Biochem Biophys Res Commun. 2006;341:1164. doi: 10.1016/j.bbrc.2006.01.080. [DOI] [PubMed] [Google Scholar]
- 25.Zhao P, Liang X, Kalbfleisch J, Koo HM, Cao B. Hum Antibodies. 2003;12:129. [PubMed] [Google Scholar]
- 26.Brossier F, Levy M, Landier A, Lafaye P, Mock M. Infect Immun. 2004;72:6313. doi: 10.1128/IAI.72.11.6313-6317.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang F, Ruther P, Jiang I, Sawada-Hirai R, Sun SM, Nedellec R, Morrow PR, Kang AS. Human antibodies. 2004;13:105. [PubMed] [Google Scholar]
- 28.Cirino NM, Sblattero D, Allen D, Peterson SR, Marks JD, Jackson PJ, Bradbury A, Lehnert BE. Infection and immunity. 1999;67:2957. doi: 10.1128/iai.67.6.2957-2963.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lacy DB, Collier RJ. Curr Top Microbiol Immunol. 2002;271:61. doi: 10.1007/978-3-662-05767-4_4. [DOI] [PubMed] [Google Scholar]
- 30.Gao C, Mao S, Kaufmann G, Wirsching P, Lerner RA, Janda KD. Proc Natl Acad Sci U S A. 2002;99:12612. doi: 10.1073/pnas.192467999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen MB, Nielsen SE, Berg K. J Immunol Methods. 1989;119:203. doi: 10.1016/0022-1759(89)90397-9. [DOI] [PubMed] [Google Scholar]
- 32.Mohamed N, Li J, Ferreira CS, Little SF, Friedlander AM, Spitalny GL, Casey LS. Infect Immun. 2004;72:3276. doi: 10.1128/IAI.72.6.3276-3283.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inglesby TV, O’Toole T, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Friedlander AM, Gerberding J, Hauer J, Hughes J, McDade J, Osterholm MT, Parker G, Perl TM, Russell PK, Tonat K. JAMA. 2002;287:2236. doi: 10.1001/jama.287.17.2236. [DOI] [PubMed] [Google Scholar]
- 34.Mao S, Gao C, Lo CH, Wirsching P, Wong CH, Janda KD. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:6953. doi: 10.1073/pnas.96.12.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mogridge J, Cunningham K, Lacy DB, Mourez M, Collier RJ. Proc Natl Acad Sci U S A. 2002;99:7045. doi: 10.1073/pnas.052160199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vitale L, Blanset D, Lowy I, O’Neill T, Goldstein J, Little SF, Andrews GP, Dorough G, Taylor RK, Keler T. Infection and immunity. 2006;74:5840. doi: 10.1128/IAI.00712-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Little SF, Ivins BE, Fellows PF, Friedlander AM. Infect Immun. 1997;65:5171. doi: 10.1128/iai.65.12.5171-5175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wigelsworth DJ, Krantz BA, Christensen KA, Lacy DB, Juris SJ, Collier RJ. The Journal of biological chemistry. 2004;279:23349. doi: 10.1074/jbc.M401292200. [DOI] [PubMed] [Google Scholar]
- 39.Elliott JL, Mogridge J, Collier RJ. Biochemistry. 2000;39:6706. doi: 10.1021/bi000310u. [DOI] [PubMed] [Google Scholar]
- 40.Cunningham K, Lacy DB, Mogridge J, Collier RJ. Proc Natl Acad Sci U S A. 2002;99:7049. doi: 10.1073/pnas.062160399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mogridge J, Cunningham K, Collier RJ. Biochemistry (Mosc) 2002;41:1079. doi: 10.1021/bi015860m. [DOI] [PubMed] [Google Scholar]
- 42.Bradley KA, Mogridge J, Mourez M, Collier RJ, Young JA. Nature. 2001;414:225. doi: 10.1038/n35101999. [DOI] [PubMed] [Google Scholar]
- 43.Escuyer V, Collier RJ. Infect Immun. 1991;59:3381. doi: 10.1128/iai.59.10.3381-3386.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christensen KA, Krantz BA, Collier RJ. Biochemistry (Mosc) 2006;45:2380. doi: 10.1021/bi051830y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mourez M, Lacy DB, Cunningham K, Legmann R, Sellman BR, Mogridge J, Collier RJ. Trends Microbiol. 2002;10:287. doi: 10.1016/s0966-842x(02)02369-7. [DOI] [PubMed] [Google Scholar]
- 46.Sellman BR, Mourez M, Collier RJ. Science. 2001;292:695. doi: 10.1126/science.109563. [DOI] [PubMed] [Google Scholar]
- 47.Schuch R, Nelson D, Fischetti VA. Nature. 2002;418:884. doi: 10.1038/nature01026. [DOI] [PubMed] [Google Scholar]
- 48.Steiniger SC, Altobell LJ, 3rd, Zhou B, Janda KD. Molecular immunology. 2007;44:2749. doi: 10.1016/j.molimm.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 49.Gao C, Lin CH, Lo CH, Mao S, Wirsching P, Lerner RA, Janda KD. Proc Natl Acad Sci U S A. 1997;94:11777. doi: 10.1073/pnas.94.22.11777. [DOI] [PMC free article] [PubMed] [Google Scholar]