Abstract
SpvC is encoded by the Salmonella virulence plasmid. We have investigated the biochemical function of SpvC and the mechanism by which it is secreted by bacteria and translocated into infected macrophages. We constructed a strain carrying a deletion in spvC and showed that the strain is attenuated for systemic virulence in mice. SpvC can be secreted in vitro by either the SPI-1 or SPI-2 type III secretion systems. Cell biological and genetic experiments showed that translocation of the protein into the cytosol of macrophages by intracellular bacteria is dependent on the SPI-2 T3SS. Using antibodies specific to phospho-amino acids and mass spectrometry we demonstrate that SpvC has phosphothreonine lyase activity on full-length phospho-Erk (pErk) and a synthetic 13-amino-acid phospho-peptide containing the TXY motif. A Salmonella strain expressing spvC from a plasmid downregulated cytokine release from infected cells.
Introduction
Salmonella enterica serovar Typhimurium (S. Typhimurium) is a facultative intracellular pathogen that causes a self-limiting gastroenteritis in humans and, less commonly, a non-typhoidal bacteremia (Velge et al., 2005). S. Typhimurium pathogenesis depends upon a large number of virulence proteins. Some of these, called effectors, are injected into the host cell by two specialized protein translocation machineries – type three secretion systems (T3SS). The T3SS encoded within Salmonella pathogenicity island 1 (SPI-1) is produced by extracellular bacteria and delivers effectors across the host cell plasma membrane; these are necessary for bacterial internalization and the early stages of Salmonella-containing vacuole (SCV) formation (Zhou and Galan, 2001; Schlumberger and Hardt, 2006). The SPI-2-encoded T3SS is activated by internalized Salmonella and translocates effectors across the vacuolar membrane. These support bacterial replication in epithelial cells and macrophages, interfere with MHC class II presentation (Mitchell et al., 2004) and also induce macrophage motility (Worley et al., 2006) and cytotoxicity (Rytkonen et al., 2007). Together, SPI-2 T3SS effectors are required for systemic virulence in the murine model of salmonellosis (Shea et al., 1996; Waterman and Holden, 2003).
Some Salmonella serovars carry plasmids which share a highly conserved locus called the spv (salmonella plasmid virulence) operon (Boyd and Hartl, 1998). This operon is primarily responsible for the virulence phenotype associated with these plasmids (Gulig et al., 1993; Gulig and Doyle, 1993; Chu and Chiu, 2006). It has been suggested that spv genes are important for pathogenesis in humans as spv-carrying strains dominate among clinical isolates from patients with non-typhoidal bacteremia (Montenegro et al., 1991; Fierer et al., 1992). spv genes are also necessary for full virulence of S. Typhimurium in mouse models of systemic infection (Gulig and Doyle, 1993), but their functions are poorly understood. The operon consists of five genes, named spvR, A, B, C and D (Krause et al., 1992). The expression of spv genes is strongly induced in intracellular bacteria in tissue culture, in animal infection models and in media mimicking the ion composition and low pH within the SCV (Wilson et al., 1997; Wilson and Gulig, 1998; Ygberg et al., 2006). The expression of the operon depends on spvR, which encodes a positive transcriptional regulator (Fang et al., 1991; Krause et al., 1992; Grob and Guiney, 1996). spvA seems to be dispensable for virulence and its function is unknown (Roudier et al., 1992; Wilson and Gulig, 1998). spvB encodes an actin-ribosyltransferase, which upon delivery to host cells modifies actin and blocks its polymerization into F-actin filaments (Otto et al., 2000; Lesnick et al., 2001; Tezcan-Merdol et al., 2001). During infections of macrophages in vitro, the loss of actin cytoskeleton results in a form of delayed cytotoxicity, manifested by cell detachment and apoptosis of infected cells (Libby et al., 2000). The function of spvD is unknown but it contributes to virulence of S. Typhimurium in mice (Matsui et al., 2001).
SpvC shares 63% identity at the amino acid level with OspF of Shigella flexneri. Two recent reports demonstrated that OspF can remove phosphate groups from host cell mitogen-activated protein kinases (MAPKs) thereby inactivating them (Arbibe et al., 2007; Li et al., 2007). OspF interference with MAPK-pathways blocks the activation of a subset of NF-kB-regulated genes resulting in impaired chemokine signalling and compromised recruitment of polymorphonuclear leukocytes to infected tissues (Arbibe et al., 2007). OspF removes phosphate groups of Erk and p38 MAPKs by either phosphatase (Arbibe et al., 2007) or phosphothreonine lyase (Li et al., 2007) activity. Experiments with recombinant SpvC and cells producing SpvC following transfection suggested that it is also capable of inactivating MAPKs (Li et al., 2007).
In this study we have investigated the function of SpvC in more detail. We show that SpvC is important for Salmonella virulence in mice and that it can be secreted by either the SPI-1 or SPI-2 T3SS. Translocation of SpvC into the cytosol of macrophages is shown to be dependent on the SPI-2 T3SS. Using antibodies specific to phospho-amino acids and mass spectrometry we demonstrate that SpvC has phosphothreonine lyase activity on full-length phospho-Erk (pErk) and a synthetic 13-amino-acid phospho-peptide containing the TXY motif. A Salmonella strain expressing spvC from a plasmid downregulated cytokine release from infected cells.
Results
SpvC is a Salmonella T3SS effector
spvC is essential for full virulence of S. Typhimurium in mice (Matsui et al., 2001) but it has not been established if SpvC functions within the bacterium or is an effector protein translocated into the host cells during or after invasion. To investigate the possibility that SpvC is a secreted effector we used homologous recombination to introduce a double haemagglutinin (2HA) tag at the C-terminus of SpvC on the Salmonella virulence plasmid (Uzzau et al., 2001). Then we checked production of SpvC-2HA by Salmonella grown in Luria–Bertani (LB) or MgM-MES media. SpvC-2HA could only be detected at a very low level in the stationary growth phase in LB medium, but a strong induction was observed during exponential growth phase in MgM-MES (data not shown). Production of SpvC-2HA in MgM-MES medium (which mimics the luminal environment of the SCV and which induces expression of SPI-2 T3SS genes) raised the possibility that SpvC might be translocated by the SPI-2 T3SS. Salmonella strains carrying mutations in SPI-1 or SPI-2 genes and expressing SpvC-2HA from the virulence plasmid were grown in MgM-MES medium. Immunoblotting was used to detect HA-tagged protein in bacterial lysates and on the plastic surface of the culture vessels, which adsorbs secreted proteins (Daefler, 1999). Similar amounts of SpvC-2HA were present in lysates from wild-type bacteria and strains carrying mutations in prgH and ssaV (which inactivate the SPI-1 and SPI-2 T3SSs respectively) (Fig. 1A). SpvC-2HA was also present on the plastic surface of the tubes in which wild-type and prgH mutant bacteria were cultured, but it was absent from the surface of the tube containing the ssaV mutant strain, indicating that SpvC is secreted in a SPI-2 T3SS-dependent manner (Fig. 1A). To test the activity of the SPI-2 T3SS in the strains used, we examined the secretion of SseB, a component of the SPI-2 T3SS translocon (Beuzon et al., 1999). As expected, SseB was detected in secreted fractions from wild-type Salmonella and prgH mutant bacteria but not from the ssaV mutant (Fig. 1A). When SpvC-2HA was overproduced from the constitutive tet promoter of pACYC184 (pspvC), a SPI-1 T3SS-dependent secretion was detected in SPI-1-inducing conditions (Fig. 1B).
Fig. 1.
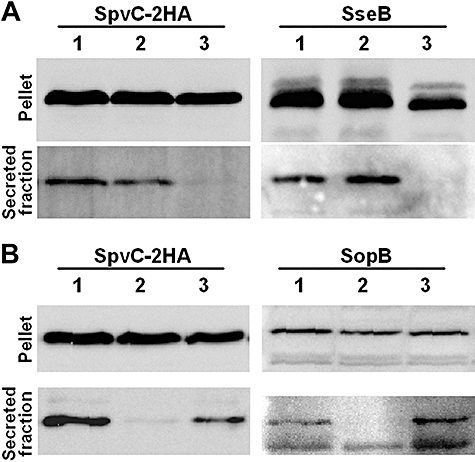
In vitro secretion of SpvC-2HA. A. Secretion of double HA-tagged SpvC produced from the virulence plasmid by Salmonella strains cultured in low magnesium minimal medium (MgM-MES): 1 –spvC-2HA, 2 –prgHspvC-2HA, 3 –ssaVspvC-2HA. B. Constitutively produced SpvC-2HA is secreted in SPI-1 T3SS-dependent manner by Salmonella grown in LB medium: 1 – WT pspvC, 2 –prgHpspvC, 3 –ssaVpspvC. Samples were probed for SseB and SopB to confirm the activity of SPI-2 and SPI-1 T3SSs respectively.
To investigate if SpvC is translocated into infected cells, J774 macrophage-like cells were infected with wild-type Salmonella expressing SpvC-2HA from the virulence plasmid. Using antibodies against the 2HA tag we could not detect any translocated SpvC-2HA within infected cells by immunofluorescence microscopy. However, after infection of macrophages with the strain harbouring pspvC it was possible to detect translocated SpvC-2HA 16 h post challenge. The amounts of detectable SpvC-2HA increased when infected cells were incubated for 2 h prior to fixation with MG132, a proteasome inhibitor (Fig. 2A). In the majority of examined cells, translocated SpvC-2HA appeared to be distributed evenly in the cytoplasm. These results suggest that relatively small amounts of SpvC are translocated into the cytoplasm of infected cells, where it is prone to degradation. No colocalization with the SCV membrane or accumulation in the nucleus was noted (Fig. 2B). To confirm that translocation of SpvC-2HA is T3SS-dependent we infected J774 cells with prgH, ssaV and double prgH ssaV mutants overexpressing SpvC-2HA. Only infection with the prgH mutant led to detectable SpvC-2HA in the cytoplasm of infected cells (Fig. 2A). This result is consistent with that of the in vitro secretion experiment, and indicates that a functional SPI-2 T3SS is necessary for translocation of SpvC-2HA.
Fig. 2.
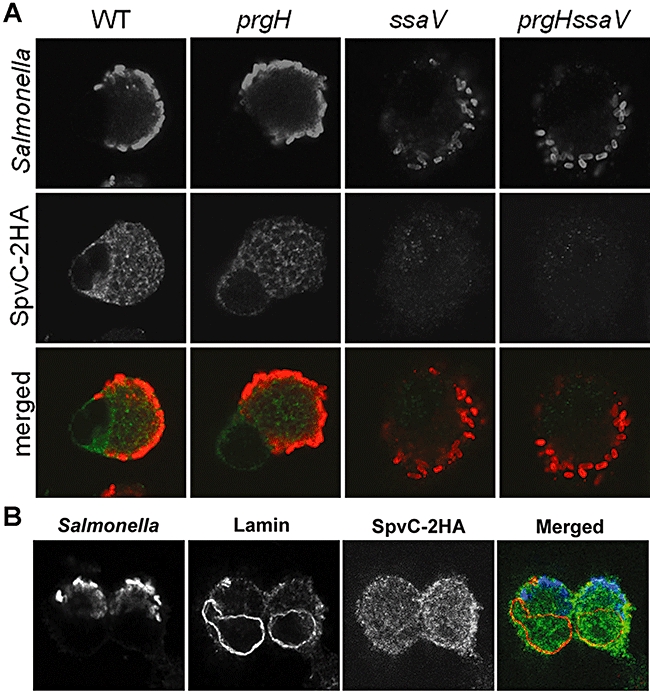
SpvC-2HA is translocated into infected macrophages in SPI-2 T3SS-dependent manner. A. J774 cells were infected for 16 h with Salmonella strains carrying pACYCspvC-2HA; 10 μg ml−1 of a proteasome inhibitor MG132 was added to the culture media for the last 2 h of infection. Samples were fixed and labeled with antibodies to visualize Salmonella and SpvC-2HA (red and green, respectively, on merged images). Similar results were observed in infected HeLa cells (data not shown). B. Translocated SpvC-2HA does not accumulate in the nucleus of infected cells. PFA-fixed samples were labeled with antibodies to visualize Salmonella (blue), SpvC-2HA (green) and lamin, a nuclear membrane protein (red).
Virulence tests
To investigate the role of SpvC in virulence, a clean deletion mutant strain (ΔspvC) was constructed as described in Experimental procedures. Its virulence was tested in mixed infections with a kanamycin-resistant isogenic strain (STM0857) whose virulence level is indistinguishable from that of wild-type S. Typhimurium strain 12023 (J. Poh and D.W. Holden, unpubl. data). BALB/c mice were inoculated intraperitoneally with 5 × 104 cfu ΔspvC and 5 × 104 cfu of STM0857. After 96 h of infection mice were sacrificed and bacteria were recovered from spleens and plated on LB and LB agar plates supplemented with kanamycin to discriminate the strains. The competitive index (CI) of ΔspvC strain was 0.51 (SD ± 0.069) indicating an attenuation in virulence. When mice were coinfected with the ΔspvC strain and ΔspvC carrying pspvC, the CI for ΔspvC was 0.33 (SD ± 0.085), showing that the attenuation of the ΔspvC strain is due to the loss of SpvC, and that SpvC-2HA is a functional protein. Next, we quantified the attenuation caused by the ΔspvC mutation in strains carrying mutations in essential genes for either the SPI-1 (prgH) or SPI-2 (ssaV) T3SS. Mixed infections with prgHΔspvC and prgH mutants showed that the lack of spvC has an additive effect on the attenuation of prgH mutant, indicating that functions of prgH and spvC are unrelated. By contrast, the spvC mutation did not cause any significant additional attenuation to that observed for the ssaV mutant. This provides genetic evidence that the functions of ssaV and spvC are related (Fig. 3). Together with results shown in Figs 1 and 2, these results strongly suggest that SpvC is translocated by the SPI-2 T3SS in vivo.
Fig. 3.
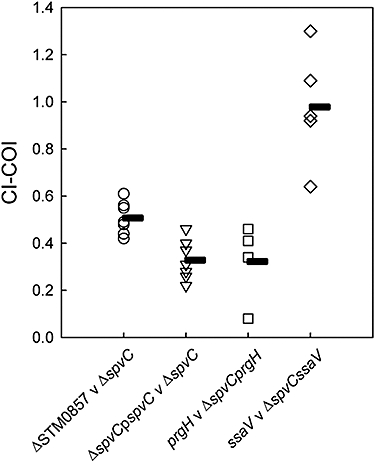
Lack of spvC causes virulence attenuation of Salmonella in the mouse model of systemic infection. Canceled-out index (COI) values show that spvC function can be genetically linked to the SPI-2 T3SS because the COI of ΔssaV versus ΔspvCΔssaV was statistically significant to the CI of the WT versus the ΔspvC strain (P < 0.001). Animals were infected for 96 h; horizontal bars indicate the mean values of CI or COI.
The spvC mutant was also compared with the wild-type strain in terms of its ability to replicate inside host cells. In standard assays involving macrophage-like RAW cells (Ruiz-Albert et al., 2002), there were no significant differences between the two strains (results not shown).
Effect of SpvC on Erk activation in infected cells
OspF inactivates MAP kinases Erk, p38 (Arbibe et al., 2007; Li et al., 2007) and possibly JNK (Li et al., 2007). As SpvC is translocated into host cells and is 63% identical to OspF, we examined its effect on the activation state of host MAP kinase Erk1/2. J774 cells were infected with different strains of S. Typhimurium for 16 h (Fig. 4). Immediately before fixation for immunofluorescence microscopy (Fig. 4A) or cell lysis for Western blotting (Fig. 4B), phorbol-12-myristate 13-acetate (PMA) was added to induce the activation of Erk. This was detected by probing samples with antibodies recognizing double phosphorylated Erk (pErk) on threonine and tyrosine residues of the TXY motif. As shown in Fig. 4, low levels of activated Erk were detected in uninfected, PMA-untreated J774 cells. PMA treatment induced Erk phosphorylation in uninfected control cells, as well as in cells infected with either the ΔspvC or ssaV mutant. However, infection with wild-type S. Typhimurium or the complemented ΔspvC strain strongly reduced Erk activation by PMA treatment. These results show that both SpvC and a functional SPI-2 T3SS are necessary for Erk inactivation.
Fig. 4.
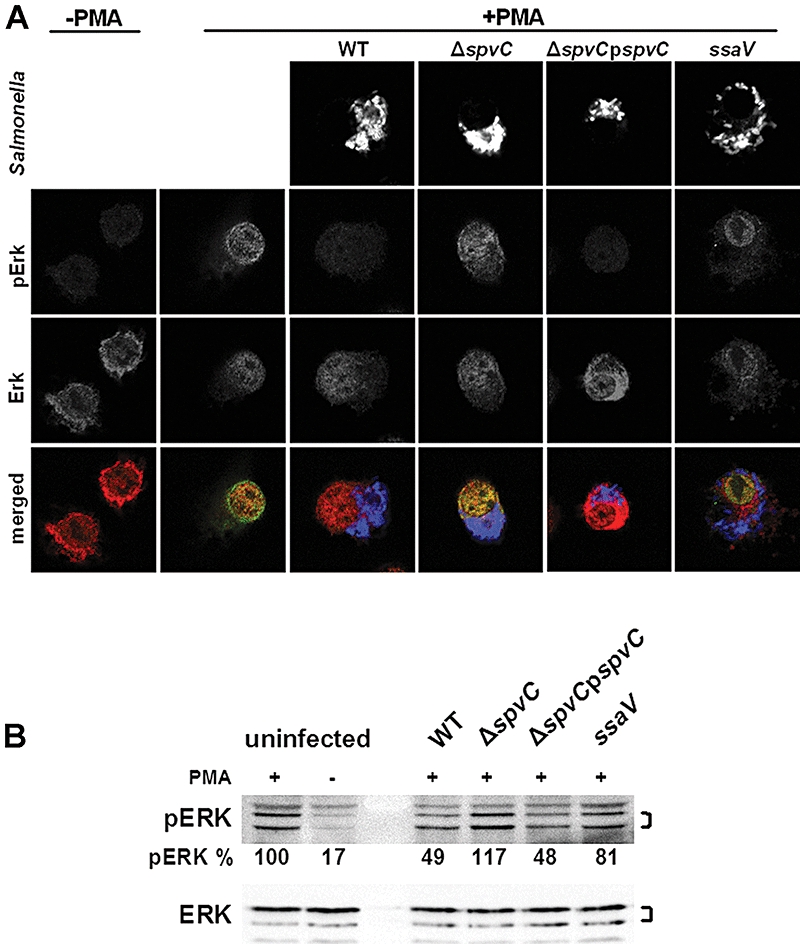
Translocated SpvC prevents Erk phosphorylation upon PMA stimulation. A. J774 cells were infected for 16 h with the indicated Salmonella strains (blue in merged images), then Erk activation was triggered by 15 min of incubation with PMA (1 μg ml−1). To assess Erk activation total Erk and phosphorylated Erk (pErk) were visualized with antibodies (red and green, respectively, in merged images). Similar results were obtained for HeLa cells (data not shown). B. Western blot analysis of the phosphorylation state of Erk in J774 macrophages infected with different Salmonella strains. Lysates of infected cells were probed for pErk and total Erk. Brackets indicate two forms of the kinase detected Erk1 (44 kDa) and Erk2 (42 kDa). pErk bands were quantified using Scion Image software. In both A and B uninfected cells were used as a control for the efficiency of PMA stimulation.
Purified SpvC inactivates Erk and JNK in vitro and is not sensitive to vanadate inhibition
To establish if SpvC is sufficient for Erk inactivation we purified GST–SpvC (Fig. S1), GST and SpvC-His and used these in in vitro dephosphorylation assays with purified, phosphorylated MAP kinases. A dual specificity MAP kinase phosphatase 1 (MKP1) was used as a control for MAP kinase dephosphorylation. Immunoblotting analysis revealed that MKP1 and GST–SpvC can remove phosphate groups from pErk (Fig. 5A). Similar results were obtained when pJNK was incubated with MKP1 and GST–SpvC (Fig. 5B). Next we tested the sensitivity of SpvC to vanadate, a tyrosine phosphatase inhibitor. pErk was incubated with GST–SpvC in the presence or absence of vanadate. As a control, MKP1 (which is susceptible to vanadate inhibition) was used (Charles et al., 1993). As expected, dephosphorylation of pErk by MKP1 was inhibited by addition of vanadate, but the activity of SpvC was not (Fig. 5C), indicating that SpvC might not be a typical dual specificity phosphatase.
Fig. 5.
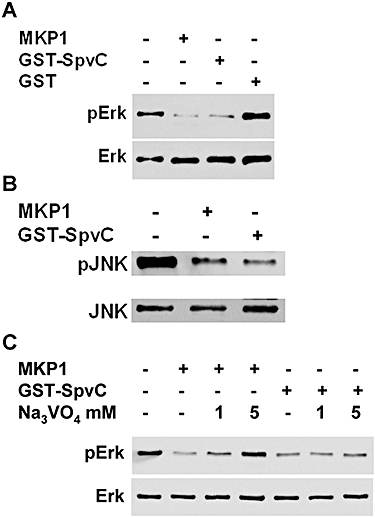
Purified GST–SpvC removes phosphate group(s) from pErk and pJNK in vitro. Phosphorylation status of Erk/JNK was determined using an antibody specific for double (threonine and tyrosine) phosphorylated MAP kinases (upper panels in A, B and C). Antibodies detecting MAP kinases regardless of their phosphorylation state were used to check the amounts of pErk/pJNK used (lower panels in A, B and C). A. One hundred nanograms of pErk were incubated with 30 U (1 μg) of MAP-kinase phosphatase 1 (MKP1), 400 ng of GST–SpvC, 400 ng of GST or left untreated as a positive control for anti-pErk antibody. B. One hundred nanograms of pJNK were incubated with 30 U MKP1, 400 ng of GST–SpvC or left untreated as a positive control for anti-pJNK antibody. C. Effect of orthovanadate on the activity of MKP1 and GST–SpvC. Two hundred nanograms of pErk were incubated with 1 μg of MKP1, 200 ng of GST–SpvC and indicated concentrations of sodium orthovanadate.
SpvC specifically removes phosphate from threonine of the TXY motif
The antibody used above recognizes double-phosphorylated Erk on threonine and tyrosine residues in the conserved activation loop motif TEY. This antibody does not recognize unphosphorylated Erk or Erk phosphorylated on either threonine or tyrosine. Therefore, to establish if SpvC is a dual specificity enzyme or if it acts only on phospho-threonine or phospho-tyrosine residues, we probed MKP1- or SpvC-treated pErk with antibodies recognizing only phospho-threonine or only phospho-tyrosine residues. MKP1-treated pErk lost phosphate groups on both threonine and tyrosine (Fig. 6A). By contrast, SpvC-treated pErk was negative for phospho-threonine labelling but remained positive for phospho-tyrosine labelling. Furthermore, the anti-phosphotyrosine antibody gave a stronger signal from the SpvC-treated pErk sample than from untreated pErk, suggesting that the presence of phosphate group on the threonine residue adjacent to the phospho-tyrosine might reduce binding of anti-phosphotyrosine antibody to double phosphorylated Erk. These results suggest that SpvC is not a dual specificity phosphatase but is either a phosphothreonine-specific phosphatase or, as proposed for OspF (Li et al., 2007), a phosphothreonine lyase.
Fig. 6.
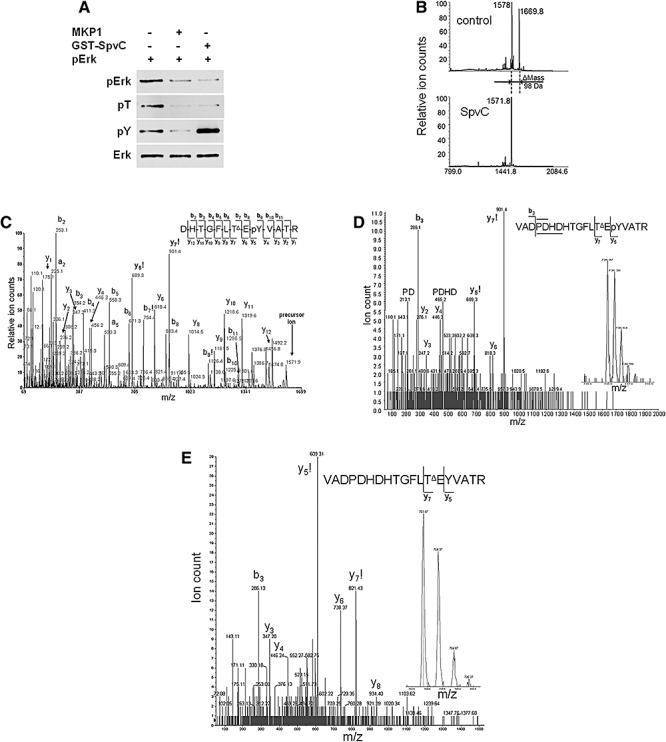
SpvC has phospholyase activity on phospho-threonine but not phospho-tyrosine residues of pErk. A. One hundred nanograms of pErk were incubated with 30 U MKP1, 400 ng of GST–SpvC, or left untreated as a positive control for anti-pErk antibody. Samples were incubated for 1 h at 30°C, and then subjected to SDS-PAGE and blotting. Membranes were probed with antibodies recognizing double phosphorylated Erk (pErk), phosphothreonine residues (pT), phosphotyrosine residues (pY) or Erk regardless of its phosphorylation state (Erk). B. MALDI-TOF spectrum of a synthetic Erk2 diphosphopeptide (DHTGFL-pT-E-pY-VATR) untreated (control) and treated with SpvC. The mass difference of 98 between the diphosphopeptide ([M + H]+ at m/z 1669.8) and the SpvC-treated product ([M + H]+ at m/z 1571.8) is consistent with the loss of phosphoric acid HPO3 and H2O. C. Tandem mass spectrometric (MALDI-CID-MS/MS) analysis of the precursor at m/z 1571.8 from the SpvC-treated fraction. The b- and y-ions are labelled; those with an exclamation mark indicating ions are crucial to assignment of the sites of phosphorylation (tyrosine-9) and dehydration (threonine-7). All ions included in the diagram are visible in the spectrum. TΔ denotes dehydrothreonine and pY phosphotyrosine. D. CID-MS/MS spectrum of the triply charged precursor ([M + 3H]3+) at m/z 736.0 obtained by LC-ES-MS of the tryptic digest of SpvC-treated pErk. The y-ions with an exclamation mark are crucial to assignment of the sites of phosphorylation (tyrosine-15) and dehydration (threonine-13). Double b,y-cleavage ions are indicated by PD and PDHD, and their proposed origins are indicated by horizontal lines above and below the peptide sequence. TΔ denotes dehydrothreonine and pY phosphotyrosine. Inset shows the precursor ion at m/z 736.0 in an LC-MS survey scan recorded at a retention time of 4.96 min. E. CID-MS/MS spectrum of the triply charged precursor ([M + 3H]3+) at m/z 703.9 obtained by LC-ES-MS of the tryptic digest of SpvC-treated pErk. The y-ions with an exclamation mark are crucial to assignment of the site of dehydration (threonine-13). TΔ denotes dehydrothreonine. Inset shows the precursor ion at m/z 703.9 in an LC-MS survey scan recorded at a retention time of 5.08 min.
SpvC is a phosphothreonine lyase
To discriminate between phosphatase and lyase activities we analysed a 13-amino-acid diphosphopeptide [comprising the TEY motif and adjacent amino acids; Erk2(177–189)] after treatment with SpvC, using mass spectrometry. MALDI-TOF mass spectra of Erk2(177–189) before and after treatment with SpvC (Fig. 6B) show that a mass shift of 98 Da occurred as a result of treatment with the enzyme. The monoisotopic [M + H]+ ion at m/z 1669.8 of the diphosphorylated peptide (theoretical m/z 1669.66) was replaced by a product at m/z 1571.8, which was consistent with the loss of phosphoric acid (H3PO4). The loss of phosphoric acid from threonine-7 of the Erk2(177–189) peptide to form dehydrothreonine was confirmed by MALDI-CID-MS/MS of the precursor ion at m/z 1571.8 (Fig. 6C). Product ions crucial to the assignment of the site of dephosphorylation (indicated with an exclamation mark) are the b7 at m/z 754 and y7 at m/z 901. The mass difference of m/z 83 between the y6 and y7 ions is consistent with dehydrothreonine; threonine at this position would result in a mass difference of m/z 101. Phosphorylation at tyrosine-9 was confirmed by the b9 ion at m/z 1126 and y5 at m/z 689. The MS/MS spectrum of the dephosphorylated Erk2(177–189) peptide produced by SpvC treatment (Fig. 6C) is very similar to the spectrum of the OspF product obtained by Li et al. (2007). The compound giving rise to the ion at m/z 1578.0 in the spectrum of the Erk(177–189) peptide preparation could not be identified. Despite being lighter by 91 Da, its MALDI-CID-MS/MS spectrum was almost identical to that of the ion at m/z 1669.8 (data not shown). This compound therefore appears to be a peptide that is very similar to the diphosphorylated Erk(177–189) peptide.
Dephosphorylation of p-Erk2 by SpvC was characterized by mass spectrometry of the tryptic peptides produced by in-gel digestion of the protein after SpvC treatment and SDS-PAGE. The tryptic peptide VADPDHDHTGFLTEYVATR with a single phosphorylation site and a dehydrothreonine residue, which would be produced from the p-Erk2 by SpvC acting as lyase, has an expected [M + H]+ monoisotopic mass of 2206. An ion with this mass was observed in the MALDI-TOF spectrum of the tryptic digest of SpvC-treated p-Erk2 (data not shown), but its intensity was not sufficient for selection as a precursor ion for MS/MS. The same peptide was detected as the triply charged [M + 3H]3+ ion at m/z 736 during LC-ES-MS (Fig. 6D, inset). The CID-MS/MS spectrum of the triply charged ion at m/z 736 contains product ions that are consistent with the sequence VADPDHDHTGFLTΔEpYVATR, where TΔ indicates dehydrothreonine and pY indicates phosphotyrosine in the sequence. Ions crucial to the sequence assignment are present, in particular the y5 and y7 ions at m/z 689 and 901 respectively. The proposed double-cleavage ions at m/z 213 and 465 are entirely consistent with the peptide sequence and the enhanced cleavage normally observed at prolyl and aspartatyl residues (Wysocki et al., 2000). In addition to the tyrosine-phosphorylated tryptic peptide containing dehydrothreonine, the non-phosphorylated version was also observed by LC-ES-MS (Fig. 6E). The presence of this non-phosphorylated peptide product indicates that the p-Erk2 substrate was not completely phosphorylated at tyrosine-412, as well as providing additional evidence for the removal of phosphoric acid from threonine-410 by SpvC.
Effect of SpvC on proinflammatory immune responses
The action of OspF on MAPKs affects histone modification and chromatin accessibility of NF-κB at the IL-8 promoter, and negatively regulates neutrophil recruitment in infected tissues (Arbibe et al., 2007). We therefore measured secretion of IL-8 from HeLa cells at different times after infection with bacterial strains. Infection with wild-type bacteria induced IL-8 production but no increased production could be detected in cells infected with the ΔspvC mutant (Fig. 7A). However, cells infected with the ΔspvCpspvC strain showed significantly less IL-8 production compared with the wild-type strain at all time points tested (P ≤ 0.05 for all time points) (Fig. 7A). As TNF-α production following stimulation of TLR-4 by LPS is MAPK-dependent (van der Bruggen et al., 1999), we also examined production of this cytokine by murine J774 macrophages. Time points for sampling were chosen to coincide with detectable SpvC translocation into host cells. Again, no significant differences were detected between cells infected with the wild-type or spvC mutant, but cells overexpressing SpvC produced less TNF-α at 14, 18 and 21 h post uptake (Fig. 7B). We also infected BALB/c mice with either the wild-type, spvC mutant or ΔspvCpspvC strains. At 4, 24 and 72 h post inoculation, mouse sera were recovered and analysed for IL-6, IL-10, MCP-1, INF-γ, TNF-α and IL12p70 by ELISA. No statistically significant differences were detected between mice infected with these strains (data not shown).
Fig. 7.
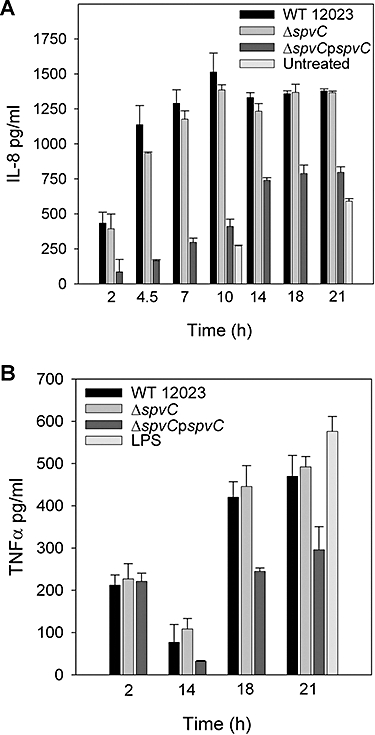
Effect of Salmonella on cytokine release by infected cells. ELISA was used to measure amounts of IL-8 released by infected HeLa cells (A) and TNF-α released by J774 macrophages (B) into the growth media at the indicated time points. Similar infection efficiencies was confirmed by microscopy in every experiment. P-values were calculated for cytokine levels from cells infected with the ΔspvCpspvC strain compared with cells infected with WT bacteria were significantly different (> 0.05) at all time points tested for A, and at 14, 18 and 21 h for B.
Discussion
Phylogenetic analysis suggests that the spv operon is widely distributed throughout the subspecies of Salmonella enterica. It is usually carried on the large Salmonella virulence plasmid, but in some serovars is integrated into the chromosome (Boyd and Hartl, 1998). The best characterized Spv protein is SpvB (which is encoded immediately upstream of spvC in the operon). SpvB ADP-ribosylates actin and is associated with cytotoxic activity in infected host cells (Otto et al., 2000; Tezcan-Merdol et al., 2001; Browne et al., 2002). SpvB must therefore be translocated into host cells from extracellular or intracellular bacteria. Its cytopathic effects seem to be dependent on a functional SPI-2 T3SS (Browne et al., 2002), but paradoxically its secretion into the culture medium in vitro was reported to be independent of either the SPI-1 or SPI-2 T3SSs (Gotoh et al., 2003).
Until recently little has been known about the function of SpvC, apart from its requirement for virulence in mice (Matsui et al., 2001). Using an epitope-tagged version produced from its own promoter in the virulence plasmid, we have shown here that SpvC can be secreted into the culture medium in either a SPI-1- or SPI-2-dependent manner, according to which secretion system is activated by bacterial growth conditions. However, we were unable to detect translocation of the protein by immunofluorescence microscopy following its normal production in infected cells. Only when SpvC was produced from a multicopy plasmid in infected cells that had been treated with a proteasome inhibitor could its SPI-2 T3SS-dependent translocation be detected clearly.
Although SpvC was not observed in infected macrophages following expression from its own promoter, phospho-Erk was detected in the cytoplasm of cells infected with either the spvC and SPI-2 T3SS null mutant, but not in cells infected with the wild-type or spvC-complemented strain (Fig. 4). As SpvC removes phosphate groups from Erk, this provides strong indirect evidence for SPI-2-dependent translocation of SpvC by wild-type bacteria. Furthermore, virulence tests with single and double mutants demonstrated that the attenuation caused by mutation of spvC is SPI-2-dependent, indicating that SpvC uses this secretion system in vivo. This contrasts with the results of an earlier study in which a polar spvA mutation had an additive effect on virulence attenuation of a SPI-2 null mutant (Shea et al., 1996). It is possible that other protein(s) encoded by the operon might be functionally independent of the SPI-2 T3SS, but further work on secretion and translocation of the Spv proteins is necessary to clarify this issue. Taken together, the results of this study provide strong evidence that during intracellular growth of bacteria, SpvC is translocated into the cytosol of macrophages in relatively small amounts through the SPI-2 T3SS. Whether or not it is also translocated along with SPI-1 T3SS effectors during invasion of epithelial cells remains to be determined.
There are approximately 31 genes in SPI-2 that encode the secretion apparatus, translocon, chaperones and at least two effectors. These, together with genes for several other effectors located outside SPI-2, are strongly induced following uptake of bacteria by host cells and require the SPI-2-encoded SsrA-B two-component system for their expression (Cirillo et al., 1998). The spv operon is known to be induced during intracellular growth in macrophages (Rhen et al., 1993; Eriksson et al., 2003), but to our knowledge there is no evidence indicating that its expression is dependent on the SsrA-B two-component system. Indeed, we found that RNA levels for spvA, B, C and D were similar in wild-type and ssrA mutant bacteria that had been grown in conditions that result in strong expression of the SsrA-SsrB regulon (Rytkonen et al., 2007; P. Mazurkiewicz and D.W. Holden, unpubl. results). SlrP and SspH 1 are two other effectors that can be translocated by either the SPI-1 or SPI-2 T3SSs, and neither seems to be regulated by SsrA-B (Miao and Miller, 2000). Therefore, the repertoire of SPI-2 T3SS effectors is not restricted to the SsrA-B regulon, and the total number of SPI-2 effectors could be significantly larger than previously anticipated.
A purified GST–SpvC fusion protein removed phosphate groups from phospho-Erk and phospho-JNK in vitro, and its insensitivity to vanadate suggested that it is not a typical tyrosine phosphatase. Subsequent mass spectrometry analysis of both full-length Erk and a synthetic phosphopeptide established that it acts as a phosphothreonine lysase, as has been shown for OspF of S. flexneri (Li et al., 2007). A potential catalytic mechanism has been proposed recently, based on an analysis of the crystal structure of SpvC (Zhu et al., 2007). This form of phosphate removal appears to be irreversible, which might help to explain why we could detect the effects of SpvC activity on levels of phospho-Erk in infected cells, but not the bacterial effector itself.
The action of OspF on MAP kinases inhibits immune responses by impairing the formation of histone H3-phosphorylated and NF-κB-regulated promoters (Arbibe et al., 2007). This is associated with a pronounced decrease in IL-8 production by infected cells and reduced recruitment of polymorphonuclear leucocytes to infected tissues (Arbibe et al., 2007). It would appear that SpvC performs a similar function in Salmonella. However, while SpvC is clearly a phosphothreonine lyase, it is possible that its substrate specificity and/or overall physiological effects are different to OspF. The proteins share 63% amino acid identity, and this could allow differential specificity towards MAP kinases. In support of this, Arbibe et al. (2007) reported that OspF is active towards Erk and p38, but not JNK, while SpvC acts on Erk, p38 and JNK (Li et al., 2007; and this work). A second possible means by which the proteins could exert different activities results from the spatio-temporal context in which they are produced: OspF is injected by extracellular bacteria shortly after bacterial contact with host cells and localizes to the nucleus, while SpvC is translocated into the host cytoplasm relatively late in the infective process from intracellular bacteria. Furthermore, whereas OspF accounted for strong suppression of IL-8 transcription and secretion between 2 and 5 h following bacterial invasion of Caco-2 cells, we did not observe a significant difference between the wild-type or spvC mutant strains in the levels of IL-8 produced from HeLa cells, TNF-α released from mouse macrophages (despite the clear difference in levels of phospho-Erk in infected cells) or six cytokines that were tested in sera from infected mice. Nevertheless, it seems likely that SpvC does inhibit proinflammatory responses, because the complemented mutant overexpressing SpvC caused a strong reduction of IL-8 and TNF-α production in tissue cultured cells. It is possible that the effects of SpvC produced by wild-type bacteria are masked by other immune-modulating bacterial molecules present in the infection assays. In vivo, SpvC could help suppress localized proinflammatory responses at infection foci in the spleen and liver, and thereby facilitate bacterial growth. This would be consistent with our finding that the spvC mutant does not have a replication defect in tissue cultured macrophages, but is attenuated in the mouse model of systemic infection.
Experimental procedures
Bacterial strains and growth conditions
All bacterial strains used in this study are listed in Table S1. Strains were grown in LB medium or a medium based on magnesium minimal medium MES (MgM-MES), containing 170 mM 2-[N-morpholino]ethane-sulphonic acid at pH 5.0, 5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 1 mM KH2PO4, 8 μM MgCl2, 38 mM glycerol and 0.1% casamino acids supplemented with appropriate antibiotics when necessary (For E. coli: ampicilin 100 μg ml−1, kanamycin 50 μg ml−1, chloramphenicol 50 μg ml−1; for Salmonella: ampicilin 50 μg ml−1, kanamycin 50 μg ml−1, chloramphenicol 50 μg ml−1 or tetracycline 12.5 μg ml−1), at 37°C with aeration if not stated differently.
Construction of the spvC mutant strains and HA-tagging of spvC on the virulence plasmid
Deletion of the spvC gene was performed by the method of Datsenko and Wanner (2000), using primers Del-spvC-F and Del-spvC-R (Table S2) to amplify the Cmr and Knr cassettes from pSU314 or pSU315 respectively. The resulting PCR product was integrated into the chromosome of S. Typhimurium expressing the λ red recombinase from pKD46. To excise the resistance marker the ΔspvCKn strain was transformed with pCP20 helper plasmid expressing the FLP recombinase (Cherepanov and Wackernagel, 1995). Deletion of the spvC gene was confirmed by PCR using primers Del-F and Del-R which flank the deleted region. Virulence plasmid-encoded spvC was double HA-tagged using the procedure described by Uzzau et al. (2001). The Cmr and Knr cassettes were amplified from pSU314 or pSU315, respectively, using primers spvC-F and spvC-R and the resulting PCR products were integrated into virulence plasmid of various S. Typhimurium strains (Table S1).
Construction of plasmids
The bacterial plasmids used in this study are listed in Table S1. To obtain pACYCspvC-2HA the double-HA-tagged spvC was amplified from the virulence plasmid of spvC-2HA strain using pacyc-F and pacycR2HA-C primers; the PCR product was cloned into pACYC184 EcoRV and SalI sites. To amplify spvC for cloning into BamHI and EcoRI sites of pGEX4T2, primers pGEXspvC-F and pGEXspvC-R were used. To amplify spvC for cloning into NdeI and XhoI sites of pET22b, primers spvC-his-F and spvC-his-R were used. Obtained plasmids were verified by sequencing.
Cell culture
HeLa (93021013) and J774 (91051511) cells were obtained from the European Collection of Cell Cultures, Salisbury, UK. Cells were grown in DMEM (Gibco, Carlsbad, CA) supplemented with 10% FCS at 37°C in 5% CO2.
Antibodies
Anti-Salmonella goat polyclonal antibody CSA-1 (Kirkegaard and Perry Laboratories, Gaithersburg, MD) was used at a dilution of 1:200. Anti-HA mouse monoclonal antibody (HA.11; Covance) was used at a dilution of 1:200 (immunofluorescence microscopy) and 1:1000 (Western blot). Anti-SseB rabbit serum (Beuzon et al., 1999) was used at a dilution of 1:5000 for Western blot. Mouse monoclonal anti-pErk antibody (M8159, Sigma) was used at a dilution of 1:200 (immunofluorescence) and 1:1000 (Western blot). Rabbit polyclonal anti-Erk antibody (06–182, Upstate) was used at a dilution of 1:200 (immunofluorescence) and 1:1000 (Western blot). Cy2- and Rhodamine red X (RRX)-conjugated donkey anti-goat, anti-mouse or anti-rabbit antibodies (Jackson Immunoresearch Laboratories) were used for immunofluorescence at a dilution of 1:200 and 1:400 for Cy5-conjugated antibody. Anti-mouse and anti-rabbit HRP (Amersham Pharmacia Biosciences) were used at a dilution of 1:10 000 for Western blot analysis.
Mice infections
Female BALB/c mice (B and K Universal, UK) that were 7–12 weeks old were used in accordance with UK Home Office regulations. To prepare the inocula, bacteria were first grown overnight in LB broth and then subcultured at a dilution of 1:100 for a further 2 h. Cultures were diluted to a concentration of 2.5 × 104 cfu ml−1 in physiological saline and mixed for intraperitoneal inoculation (0.2 ml per mouse). Viable bacteria in inocula were quantified by dilution and plating onto LB agar plates with appropriate antibiotics to distinguish between strains. Mice were sacrificed at various times post inoculation. The spleens were removed aseptically and homogenized in distilled water by mechanical disruption in a Colworth stomacher. Serial dilutions were plated on LB agar for cfu enumeration. Strains were distinguished by differential counting or replica plating on antibiotic-supplemented plates.
For each mouse, the CI or cancelled-out index (COI) was calculated by dividing the output ratio (i.e. wild-type versus mutant a, or mutant a versus mutant ab) divided by the input ratio. The CI values were used to calculate means and in statistical analyses. Student's t-test was used to analyse the COI of the single-mutant versus double-mutant strains (i.e. mutant b versus mutant ab) with the null hypothesis that the mean COI was equal to the CI of the wild-type versus the single mutant (i.e. wild-type versus mutant a).
Infection of epithelial cells and macrophages
Infection of HeLa cells and macrophages was performed as described (Beuzon et al., 2000; Ruiz-Albert et al., 2002). Cells were fixed with paraformaldehyde, permeabilized and incubated with antibodies as described (Beuzon et al., 2000). Labelled cells were analysed by using a fluorescence microscope (BX50; Olympus) or a confocal laser scanning microscope (LSM510; Zeiss).
ELISA
HeLa cells or J774 macrophages were seeded at a density of 105 cells per well in 24-well Plate 24 h before challenge with various Salmonella strains. At different time points after challenge plates were centrifuged at 400 g to settle down non-adherent cells. Supernatants were transferred into clean tubes and stored at −80°C until the assay was performed. Purified LPS (Sigma, St Louis, MO) at 1 μg ml−1 was used as a positive control stimulus for TNF-α production by J774 macrophages. TNF-α and IL-8 levels were measured using sandwich ELISA according to manufacturer's recommendations (Diaclone, Stamford, CT).
Measurement of cytokines in mouse sera
Blood samples were collected at various time points after bacterial challenge from intraperitoneally infected mice. Animals were sacrificed and spleens were isolated for enumeration of bacteria. Blood was allowed to clot for 4 h at room temperature and then overnight at 4°C. The clot was separated by centrifugation and sera were stored at −80°C until used for measurements. BD™ Cytometric Bead Array (BD™ CBA, BD Biosciences, San Diego, CA) specific for IL-6, IL-10, IL-12p70, MCP-1, IFN-γ and TNF-α was used to test the sera. Measurements were performed according to manufacturer's recommendations using BD FACSCalibur™ flow cytometer.
Protein secretion assay
To analyse protein production and secretion in SPI-2 T3SS inducing conditions, Salmonella strains were grown overnight in MgM-MES medium. Overnight cultures were subcultured 1:33 into a tube containing 5 ml of fresh growth medium, and bacteria were allowed to grow for a further 6 h. To ensure that protein from equal numbers of cells was analysed, in all experiments protein samples were adjusted to OD600 values such that a volume corresponding to 1 ml of a culture of OD600 0.5 was taken up in 100 μl of protein-denaturing buffer for gel electrophoresis. The secreted and total bacterial fractions were prepared as described before (Yu et al., 2002).
Protein purification
Plasmids for production of GST and GST fusion SpvC (GST–SpvC) were transformed into E. coli TOP10 cells. Proteins were induced by using IPTG (0.5 mM), and cells were lysed using a French press. Samples were centrifuged at 38 000 g for 45 min, and supernatants were incubated with Glutathione Sepharose 4B beads (Amersham Pharmacia Biosciences) at 4°C. The beads were washed extensively with wash buffer (50 mM Tris-Cl, pH 8.0, 100 mM NaCl, 1 mM DTT) and eluted in the same buffer containing 10 mM glutathione. Samples were dialysed against 50 mM Tris-Cl, pH 7.4, 50 mM NaCl. To dialysed samples, 20% glycerol was added and aliquots were stored at −20°C. For purification of six histidine-tagged SpvC, pET22bspvC-His was introduced to E. coli BL21. Proteins were induced by using IPTG (0.5 mM), and cells were lysed using a French press. Samples were centrifuged at 38 000 g for 45 min, and supernatant was incubated with Ni-NTA Agarose (Qiagen). The beads were washed with wash buffer (20 mM Tris-Cl, 300 mM NaCl, 20 mM imidazole, 10% glycerol, pH 8.0) and SpvC-His was eluted with elution buffer (20 mM Tris-Cl, 150 mM NaCl, 250 mM imidazole, 10% glycerol, pH 7.0). Samples were dialysed in 50 mM Tris-Cl, pH 7.4, 50 mM NaCl, 10% glycerol and stored at −20°C. Protein concentration was determined using Bio-Rad RC-DC assay with bovine serum albumin as a standard. Protein purity was confirmed through analysis of samples by SDS-PAGE (Fig. S1).
Dephosphorylation assay
For in vitro dephosphorylation reactions 100 ng of GST-pErk2 (pErk) (14–173, Upstate) was incubated with 30 U (1 μg) of MAP-kinase phosphatase 1 (MKP1) 14–391, Upstate, 400 ng of GST–SpvC, 400 ng of GST or left untreated as a positive control for Erk phosphorylation in 100 mM TRIS-Cl pH 7.0, 50 mM NaCl (DB buffer) for 30 min at 30°C. The reactions were stopped by boiling samples in SDS-PAGE sample buffer. For orthovanadate inhibition, an appropriate amount of activated Na3VO4 was added to the DB buffer. The phosphorylation state of Erk was assessed by Western blot analysis. For mass spectrometry pErk or pErk-derived phosphopeptide (1 μg) were incubated with 400 ng of SpvC-His for 1 h at 30°C in DB buffer. The mixture of pErk and SpvC was separated on a precast Bio-Rad Tris-Glycine acrylamide gel and the band corresponding to pErk was then excised and used for in-gel tryptic digestion. The pErk-derived peptide was used directly for mass spectrometry analysis.
In-gel tryptic digestion
In-gel tryptic digestion of SDS-PAGE bands was performed after reduction with DTT and S-carbamidomethylation with iodoacetamide. Gel pieces were washed three times with 50% (v/v) aqueous acetonitrile containing 25 mM ammonium bicarbonate and dried in a vacuum concentrator for 30 min. Sequencing-grade, modified porcine trypsin (Promega) was dissolved in the 50 mM acetic acid supplied by the manufacturer, and then diluted fivefold by adding 25 mM ammonium bicarbonate to give a final trypsin concentration of 0.02 μg μl−1. Gel pieces were rehydrated by adding 10 μl of trypsin solution, and after 30 min enough trypsin solution was added to cover the gel pieces. Digests were incubated overnight at 37°C.
MALDI-TOF mass spectrometry
A 0.5 μl aliquot of peptide or tryptic digest was applied directly to a MALDI target plate, followed immediately by an equal volume of a freshly prepared 5 mg ml−1 solution of α-cyano-4-hydroxycinnamic acid (Sigma, Poole, UK) in 50% aqueous (v/v) acetonitrile containing 0.1% TFA (v/v). Positive ion mass spectra were obtained using an Applied Biosystems 4700 Proteomics Analyzer (Applied Biosystems, Foster City, CA, USA) in reflectron mode. MS spectra were acquired with a total of 1000 laser pulses over a mass range of m/z 800–4000 with a focus mass of m/z 1500. CID-MS/MS spectra were acquired using a collision energy of 1 kV with air as the gas at a Source 2 pressure of about 1 × 10−6 torr. The precursor mass window was set to a relative resolution of 50, and the metastable suppressor was enabled. MS/MS spectra were baseline-subtracted (peak width 50) and smoothed (Savitsky-Golay with three points across a peak and polynomial order 4) Peak detection used a minimum S/N of 5, local noise window of 50 m/z, and minimum peak width of 2.9 bins.
LC-electrospray (ES) mass spectrometry
Peptide separations were achieved using a 5 cm polystyrene-divinlybenzene (PS-DVB) monolithic column (100 μm inside diameter) and UltiMate capillary HPLC system (Dionex, Sunnyvale, CA, USA) with the column at 60°C. Samples (3 μl) were injected directly onto the column and eluted with a linear gradient of acetonitrile in 0.1% formic acid from 2 to 50% in 10 min at a flow rate of 1.1 μl min−1. Peptides eluting from the column were analysed using a QSTAR Pulsar i hybrid mass spectrometer (Applied Biosystems, Foster City, CA, USA) with Analyst QS software and fitted with a modified MicroIonSpray source. The mass spectrometer was operated in information-dependent acquisition mode with an MS survey scan of 1 s and two 0.5 s MS/MS spectra acquired per experiment. Precursor masses were excluded for 120 s after selection, and the collision offset was selected automatically.
Acknowledgments
This work was supported by grants from the MRC and Wellcome Trust (UK) to D.H. The author J.T. was supported by a Biotechnology and Biological Sciences Research Council (BBSRC) studentship. We thank Dr David Ashford for LC-ES-MS and Dr Jaime Mota for helpful suggestions.
Supplementary material
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1365-2958.2008.06134.x
(This link will take you to the article abstract).
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Arbibe L, Kim DW, Batsche E, Pedron T, Mateescu B, Muchardt C, et al. An injected bacterial effector targets chromatin access for transcription factor NF-kappaB to alter transcription of host genes involved in immune responses. Nat Immunol. 2007;8:47–56. doi: 10.1038/ni1423. [DOI] [PubMed] [Google Scholar]
- Beuzon CR, Banks G, Deiwick J, Hensel M, Holden DW. pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol Microbiol. 1999;33:806–816. doi: 10.1046/j.1365-2958.1999.01527.x. [DOI] [PubMed] [Google Scholar]
- Beuzon CR, Meresse S, Unsworth KE, Ruiz-Albert J, Garvis S, Waterman SR, et al. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 2000;19:3235–3249. doi: 10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd EF, Hartl DL. Salmonella virulence plasmid. Modular acquisition of the spv virulence region by an F-plasmid in Salmonella enterica subspecies I and insertion into the chromosome of subspecies II IIIa, IV and VII isolates. Genetics. 1998;149:1183–1190. doi: 10.1093/genetics/149.3.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne SH, Lesnick ML, Guiney DG. Genetic requirements for salmonella-induced cytopathology in human monocyte-derived macrophages. Infect Immun. 2002;70:7126–7135. doi: 10.1128/IAI.70.12.7126-7135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bruggen T, Nijenhuis S, van Raaij E, Verhoef J, van Asbeck BS. Lipopolysaccharide-induced tumor necrosis factor alpha production by human monocytes involves the raf-1/MEK1-MEK2/ERK1-ERK2 pathway. Infect Immun. 1999;67:3824–3829. doi: 10.1128/iai.67.8.3824-3829.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles CH, Sun H, Lau LF, Tonks NK. The growth factor-inducible immediate-early gene 3CH134 encodes a protein-tyrosine-phosphatase. Proc Natl Acad Sci USA. 1993;90:5292–5296. doi: 10.1073/pnas.90.11.5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- Chu C, Chiu CH. Evolution of the virulence plasmids of non-typhoid Salmonella and its association with antimicrobial resistance. Microbes Infect. 2006;8:1931–1936. doi: 10.1016/j.micinf.2005.12.026. [DOI] [PubMed] [Google Scholar]
- Cirillo DM, Valdivia RH, Monack DM, Falkow S. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol. 1998;30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- Daefler S. Type III secretion by Salmonella typhimurium does not require contact with a eukaryotic host. Mol Microbiol. 1999;31:45–51. doi: 10.1046/j.1365-2958.1999.01141.x. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol. 2003;47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- Fang FC, Krause M, Roudier C, Fierer J, Guiney DG. Growth regulation of a Salmonella plasmid gene essential for virulence. J Bacteriol. 1991;173:6783–6789. doi: 10.1128/jb.173.21.6783-6789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer J, Krause M, Tauxe R, Guiney D. Salmonella typhimurium bacteremia: association with the virulence plasmid. J Infect Dis. 1992;166:639–642. doi: 10.1093/infdis/166.3.639. [DOI] [PubMed] [Google Scholar]
- Gotoh H, Okada N, Kim YG, Shiraishi K, Hirami N, Haneda T, et al. Extracellular secretion of the virulence plasmid-encoded ADP-ribosyltransferase SpvB in Salmonella. Microb Pathog. 2003;34:227–238. doi: 10.1016/s0882-4010(03)00034-2. [DOI] [PubMed] [Google Scholar]
- Grob P, Guiney DG. In vitro binding of the Salmonella dublin virulence plasmid regulatory protein SpvR to the promoter regions of spvA and spvR. J Bacteriol. 1996;178:1813–1820. doi: 10.1128/jb.178.7.1813-1820.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulig PA, Doyle TJ. The Salmonella typhimurium virulence plasmid increases the growth rate of salmonellae in mice. Infect Immun. 1993;61:504–511. doi: 10.1128/iai.61.2.504-511.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulig PA, Danbara H, Guiney DG, Lax AJ, Norel F, Rhen M. Molecular analysis of spv virulence genes of the Salmonella virulence plasmids. Mol Microbiol. 1993;7:825–830. doi: 10.1111/j.1365-2958.1993.tb01172.x. [DOI] [PubMed] [Google Scholar]
- Krause M, Fang FC, Guiney DG. Regulation of plasmid virulence gene expression in Salmonella dublin involves an unusual operon structure. J Bacteriol. 1992;174:4482–4489. doi: 10.1128/jb.174.13.4482-4489.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesnick ML, Reiner NE, Fierer J, Guiney DG. The Salmonella spvB virulence gene encodes an enzyme that ADP-ribosylates actin and destabilizes the cytoskeleton of eukaryotic cells. Mol Microbiol. 2001;39:1464–1470. doi: 10.1046/j.1365-2958.2001.02360.x. [DOI] [PubMed] [Google Scholar]
- Li H, Xu H, Zhou Y, Zhang J, Long C, Li S, et al. The phosphothreonine lyase activity of a bacterial type III effector family. Science. 2007;315:1000–1003. doi: 10.1126/science.1138960. [DOI] [PubMed] [Google Scholar]
- Libby SJ, Lesnick M, Hasegawa P, Weidenhammer E, Guiney DG. The Salmonella virulence plasmid spv genes are required for cytopathology in human monocyte-derived macrophages. Cell Microbiol. 2000;2:49–58. doi: 10.1046/j.1462-5822.2000.00030.x. [DOI] [PubMed] [Google Scholar]
- Matsui H, Bacot CM, Garlington WA, Doyle TJ, Roberts S, Gulig PA. Virulence plasmid-borne spvB and spvC genes can replace the 90-kilobase plasmid in conferring virulence to Salmonella enterica serovar Typhimurium in subcutaneously inoculated mice. J Bacteriol. 2001;183:4652–4658. doi: 10.1128/JB.183.15.4652-4658.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao EA, Miller SI. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc Natl Acad Sci USA. 2000;97:7539–7544. doi: 10.1073/pnas.97.13.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell EK, Mastroeni P, Kelly AP, Trowsdale J. Inhibition of cell surface MHC class II expression by Salmonella. Eur J Immunol. 2004;34:2559–2567. doi: 10.1002/eji.200425314. [DOI] [PubMed] [Google Scholar]
- Montenegro MA, Morelli G, Helmuth R. Heteroduplex analysis of Salmonella virulence plasmids and their prevalence in isolates of defined sources. Microb Pathog. 1991;11:391–397. doi: 10.1016/0882-4010(91)90035-9. [DOI] [PubMed] [Google Scholar]
- Otto H, Tezcan-Merdol D, Girisch R, Haag F, Rhen M, Koch-Nolte F. The spvB gene-product of the Salmonella enterica virulence plasmid is a mono (ADP-ribosyl) transferase. Mol Microbiol. 2000;37:1106–1115. doi: 10.1046/j.1365-2958.2000.02064.x. [DOI] [PubMed] [Google Scholar]
- Rhen M, Riikonen P, Taira S. Transcriptional regulation of Salmonella enterica virulence plasmid genes in cultured macrophages. Mol Microbiol. 1993;10:45–56. doi: 10.1111/j.1365-2958.1993.tb00902.x. [DOI] [PubMed] [Google Scholar]
- Roudier C, Fierer J, Guiney DG. Characterization of translation termination mutations in the spv operon of the Salmonella virulence plasmid pSDL2. J Bacteriol. 1992;174:6418–6423. doi: 10.1128/jb.174.20.6418-6423.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Albert J, Yu XJ, Beuzon CR, Blakey AN, Galyov EE, Holden DW. Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol Microbiol. 2002;44:645–661. doi: 10.1046/j.1365-2958.2002.02912.x. [DOI] [PubMed] [Google Scholar]
- Rytkonen A, Poh J, Garmendia J, Boyle C, Thompson A, Liu M, et al. SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc Natl Acad Sci USA. 2007;104:3502–3507. doi: 10.1073/pnas.0610095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlumberger MC, Hardt WD. Salmonella type III secretion effectors: pulling the host cell's strings. Curr Opin Microbiol. 2006;9:46–54. doi: 10.1016/j.mib.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Shea JE, Hensel M, Gleeson C, Holden DW. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezcan-Merdol D, Nyman T, Lindberg U, Haag F, Koch-Nolte F, Rhen M. Actin is ADP-ribosylated by the Salmonella enterica virulence-associated protein SpvB. Mol Microbiol. 2001;39:606–619. doi: 10.1046/j.1365-2958.2001.02258.x. [DOI] [PubMed] [Google Scholar]
- Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L. Epitope tagging of chromosomal genes in Salmonella. Proc Natl Acad Sci USA. 2001;98:15264–15269. doi: 10.1073/pnas.261348198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velge P, Cloeckaert A, Barrow P. Emergence of Salmonella epidemics: the problems related to Salmonella enterica serotype enteritidis and multiple antibiotic resistance in other major serotypes. Vet Res. 2005;36:267–288. doi: 10.1051/vetres:2005005. [DOI] [PubMed] [Google Scholar]
- Waterman SR, Holden DW. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell Microbiol. 2003;5:501–511. doi: 10.1046/j.1462-5822.2003.00294.x. [DOI] [PubMed] [Google Scholar]
- Wilson JA, Doyle TJ, Gulig PA. Exponential-phase expression of spvA of the Salmonella typhimurium virulence plasmid: induction in intracellular salts medium and intracellularly in mice and cultured mammalian cells. Microbiology. 1997;143(Part 12):3827–3839. doi: 10.1099/00221287-143-12-3827. [DOI] [PubMed] [Google Scholar]
- Wilson JA, Gulig PA. Regulation of the spvR gene of the Salmonella typhimurium virulence plasmid during exponential-phase growth in intracellular salts medium and at stationary phase in L broth. Microbiology. 1998;144(Part 7):1823–1833. doi: 10.1099/00221287-144-7-1823. [DOI] [PubMed] [Google Scholar]
- Worley MJ, Nieman GS, Geddes K, Heffron F. Salmonella typhimurium disseminates within its host by manipulating the motility of infected cells. Proc Natl Acad Sci USA. 2006;103:17915–17920. doi: 10.1073/pnas.0604054103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocki VH, Tsaprailis G, Smith LL, Breci LA. Mobile and localized protons: a framework for understanding peptide dissociation. J Mass Spectrom. 2000;35:1399–1406. doi: 10.1002/1096-9888(200012)35:12<1399::AID-JMS86>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Ygberg SE, Clements MO, Rytkonen A, Thompson A, Holden DW, Hinton JC, Rhen M. Polynucleotide phosphorylase negatively controls spv virulence gene expression in Salmonella enterica. Infect Immun. 2006;74:1243–1254. doi: 10.1128/IAI.74.2.1243-1254.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XJ, Ruiz-Albert J, Unsworth KE, Garvis S, Liu M, Holden DW. SpiC is required for secretion of Salmonella pathogenicity island 2 type III secretion system proteins. Cell Microbiol. 2002;4:531–540. doi: 10.1046/j.1462-5822.2002.00211.x. [DOI] [PubMed] [Google Scholar]
- Zhou D, Galan J. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect. 2001;3:1293–1298. doi: 10.1016/s1286-4579(01)01489-7. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Li H, Long C, Hu L, Xu H, Liu L, et al. Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphotheonine lyase. Mol Cell. 2007;28:899–913. doi: 10.1016/j.molcel.2007.11.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


