Abstract
Traditional cell culture substrates consist of static, flat surfaces although in vivo, cells exist on various dynamic topographies. We report development of a reconfigurable microtopographical system compatible with cell culture that is comprised of reversible wavy microfeatures on poly(dimethylsiloxane). Robust reversibility of the wavy micropattern is induced on the cell culture customized substrate by first plasma oxidizing the substrate to create a thin, brittle film on the surface and then applying and releasing compressive strain, to introduce and remove the microfeatures, respectively. The reversible topography was able to align, unalign, and realign C2C12 myogenic cell line cells repeatedly on the same substrate within 24 h intervals, and did not inhibit cell differentiation. The flexibility and simplicity of the materials and methods presented here provide a broadly applicable capability by which to investigate and compare dynamic cellular processes not yet easily studied using conventional in vitro culture substrates.
1. Introduction
Surface topographies are useful in the study and control of cell adhesion, orientation, motility, and proliferation and differentiation [1-3]. A materials constraint of most conventional man-made micromachined topographical substrates is that the features are not dynamic [4] as microenvironments in vivo often are [1,5-8]. Reconfigurable surfaces have been developed to a much lesser degree and even then mostly for regulating surface chemistry not surface topography [9-12]. Here, we describe a reconfigurable microtopographical system customized for cell culture and imaging that consists of reversible wavy microfeatures on poly(dimethylsiloxane) (PDMS). The wavy features are created by compression-induced buckling of a brittle thin film on the surface of a PDMS substrate [13]. We observed that by uncompressing such a substrate, the microfeatures would also reversibly disperse, so that by applying or releasing compressive strain, waves could be obtained or removed, respectively. Using these reversibly reconfigurable topographical features, we demonstrate the ability of C2C12 myoblast cells to align and realign on wavy features that are created and recreated on a substrate. Myoblasts were studied because alignment is a critical step in musculoskeletal myogenesis that is required prior to fusion into multinucleated myotubes, eventually leading to the development of the discernable muscular structure [14]. Abnormalities in the muscle structure alignment are involved in some musculoskeletal disorders [15,16]. Although microtopographical features are known to be present in vivo [17] and to be effective for aligning myoblasts in vitro [2,18], much of the mechanisms by which various types of myoblasts align and muscle tissues organize is still unclear [19]. What is needed for studying the adaptive properties of such dynamic living cells is the development of correspondingly adaptive materials with dynamically reconfigurable topographical features.
2. Materials and methods
2.1 Cell source and media
Mouse myogenic cell line C2C12 cells were obtained from American Tissue Type Culture Collection (ATCC, Manassas, VA). Growth medium (GM) consisted of Dulbecco's Modified Eagle Medium (DMEM, Invitrogen, Carlsbad, CA 11965) with 20% fetal bovine serum (FBS, Invitrogen 26140) and 1% antibiotic-antimycotic (Invitrogen 15240). Differentiation medium (DM) consisted of DMEM with 7.5% horse serum (HS, Invitrogen 16050) and 1% antibiotic-antimycotic.
2.2 Substrate fabrication
Wells of poly(dimethylsiloxane) (PDMS, type 184 silicone elastomer, Dow Chemical Corporation, Midland, MI) were made for reversible patterning. Molds were made for fabricating wells. A pre-mold was made by pouring 10:1 (prepolymer to curing agent) PDMS into square Petri dishes and cured overnight at room temperature to remove air bubbles followed by a 2 h oven cure at 60°C. 1.5 cm2 pieces were cut out of the cured PDMS and the holes filled with Epo-Tek polymer (Epoxy Technology, Boston, MA). The Epo-Tek was cured under UV light for 2 h. The PDMS pre-mold was then removed leaving the final Epo-Tek mold consisting of blocks of epoxy sized 1.5 cm × 1.5 cm × 0.3 cm. 15:1 prepolymer to curing agent PDMS was poured into the Epo-Tek molds and substrate wells were cut out once the PDMS was room temperature cured overnight then oven cured at 60°C for 2−3 h.
Wavy micropatterns were generated on elastomeric polymer surfaces by oxidizing the substrate to create a brittle thin film, then compressing it [13]. Upon release of the compressive strain the microfeatures dissipated, thus creating a reversible surface topography. The general method for creating reversible wavy micropatterns into a surface begins with oxidizing a PDMS substrate at 200 mTorr in O2 for an appropriate amount of time to obtain desired wavy feature sizes (5 min was required for the work described in this paper) at 100 W in a Plasma Prep II Plasma Etcher (model 11005, SPI, West Chester, PA) to create a thin brittle oxidized surface layer. Compressive strain was applied with a straining device equipped with a micrometer (Micro Vice, S.T. Japan, Tokyo, Japan) causing the surface to buckle, creating a smooth, sinusoidal-like wave pattern (Fig. 1). These patterns formed on the substrate surface perpendicular to the direction of compression. The micropattern could then be removed from the surface by uncompressing the substrate via removal of the straining device.
Fig. 1.
Micrographs of the compressed substrate with a comparison image. Larger images are of replicas of a compressed substrate from a top view (top left, optical), angled view (top right, SEM), and cross-sectional view (bottom, SEM). Smaller image is of oxidized, not compressed PDMS for comparison (optical).
2.3 Cell culture on reconfigurable microtopography
PDMS wells were incubated in 4 μg natural mouse laminin (Invitrogen 23017) diluted in Dulbecco's Phosphate-Buffered Saline (DPBS, Invitrogen 14190) per cm2 of substrate surface. Wells were placed in a biological hood with the blower on overnight to evaporate the DPBS and deposit laminin onto the surface. Salt crystals left over from the deposition were washed off with additional DPBS. Substrate wells were then oxidized to create a brittle thin film on the well surface, and then UV sterilized for 30 min. UV sterilization did not affect cell attachment, and hence presumably neither affected functionality of the laminin. Strain devices were sterilized with 70% ethanol immediately prior to substrate compression and allowed to air dry in a biological hood. Compressed substrates were incubated in an airtight chamber (Hypoxia Modular Incubator Chamber, Billups-Rothenburg Inc., Del Mar, CA MIC-101) with open petri dishes of sterile distilled water to prevent media evaporation in the small volume wells; adequate gas exchange with the 5% CO2 and 21% O2 incubator conditions was provided by removing clamps on chamber tubing open to the incubator environment.
2.4 Measures
2.4.1 Compressive strain
Strain was calculated by measuring square substrate width before (lo) and after (lf) compression, finding the change in length (Δ l = lo – lf) and inputting the values into the standard strain equation: ε = Δ l / lo.
2.4.2 Wave size
Rough measurements of each substrate were made to ensure that the wave size acquired was around 6 μm, the size previously found optimal for cell alignment [20]. Estimated wave sizes were determined by taking 10× light microscope images of the substrate surface while under compression and after plating cells. The number of waves (N) was counted per a 1 mm distance as seen under 10× magnification. Wavelength (λ), or peak-to-peak wave distance, was then calculated by dividing 1 mm by the number of waves (λ = 1 mm / N).
The distance from the top to the bottom of a wave was defined as the amplitude. Wave amplitude was measured using scanning electron microscopy (SEM) of compressed substrate replicas (Fig. 1). To prepare for SEM, compressed substrates were replicated (compressed substrates were filled with Epo-Tek, UV cured, and PDMS replicas cast against the Epo-Tek patterned piece). PDMS replicas were sputter-coated with a 20 nm layer of Au/Pd at 10 mA, 8V and 70mTorr in Argon gas, and loaded into a Philips XL30 SEM for imaging.
2.4.3 Orientation angle
Orientation angle (OA) reveals the degree of cell alignment along a feature or on a flat surface, and is found by ascertaining the major axis of the cell and finding the angle between this axis and the feature (in this case, the wave). Low OAs denote alignment along the features, with perfect cell alignment resulting in an OA of 0°; randomly oriented cells have an angle of 45° [21].
2.4.4 Statistical analysis
Average orientation angle was calculated as a mean value, that is the sum of the angles divided by the number of values for any given timepoint. Statistical analysis was performed on cell data during compressed and uncompressed states to test if the change in cell orientation angle was statistically significant at these time points. ANVOA with Tukey post-hoc criterion was used to assess statistical significance of orientation angle of cells on flat then compressed then flat substrates. Student's t-test was performed on orientation angle of cells on compressed then flat substrates and also for the repeated compression and uncompression studies at each point the substrate was reversed back to flat from wavy; α level was set at 0.05 in both cases.
3. Results and discussion
Wavelength, λ, can be approximated from the oxidized layer thickness,t, and Young's modulus of the oxidized surface, Es, and the bulk polymer, Ep [13,22]:
| (1) |
As noted by Bowden et al., this equation may not describe the waves exactly because the oxidized PDMS surface layer is not fully characterized and the boundaries between the oxidized surface and bulk polymer may not be discrete, but it does convey the general relation of wavelength to oxidation time [13]. Amplitude, A, can be described by the following approximation,
| (2) |
where Δ/W is the compressive strain (Δ = imposed compressive transverse displacement, W = original width of the substrate) [23]. Hence, wavelength was not dependent on compressive strain, but rather amplitude was. Empirically, substrates exhibited a range of strains within which visible wavy features would form (∼11−15% strain); strains below this range did not produce waves evenly across the entire surface, and strains above it caused the substrate to fold and collapse. The specific resultant wavelength for a given strain within this range varied slightly (λ = 5−7 μm), and depended largely on the oxidation conditions, which are generally reproducible but difficult to control exactly.
For a given strain, the ability to generate waves is determined primarily by oxidation time (hence modulus and SiO2 film thickness on the substrate surface) and the bulk polymer modulus [13,20,22]. Wave feature size was analyzed for dry and cell culture media-wetted plasma oxidized PDMS substrates subjected to 9 intervals of compression (∼11% strain) and uncompression over a period of 9 d (24 h between each compression and uncompression). Wave size did not vary significantly between intervals. Average wavelength during compression (λavg = 7.3 ± 0.8 μm; n = 17) was found to be similar for both dry and wet substrates. Wavelength returned to zero at each point of uncompression. Average amplitude was measured to be 670 ± 120 nm, which coincides with previous findings of similar sized static waves [20].
Practical considerations such as well dimensions, base material thickness, and deformability were also important for successful cell experimentation. For instance, entire substrates would fold under compression if wells were too deep and/or wide. To adapt the reversible waves to cell culture, custom epoxy molds were prepared (see supplementary data for a process diagram) and PDMS substrates cast against it to embody 1.5 × 1.5 cm wells at a depth of 3 mm and an overall substrate thickness of 8 mm. A 15:1 prepolymer to curing agent ratio provided PDMS substrates that were easily and reliably compressed to create wavy microfeatures.
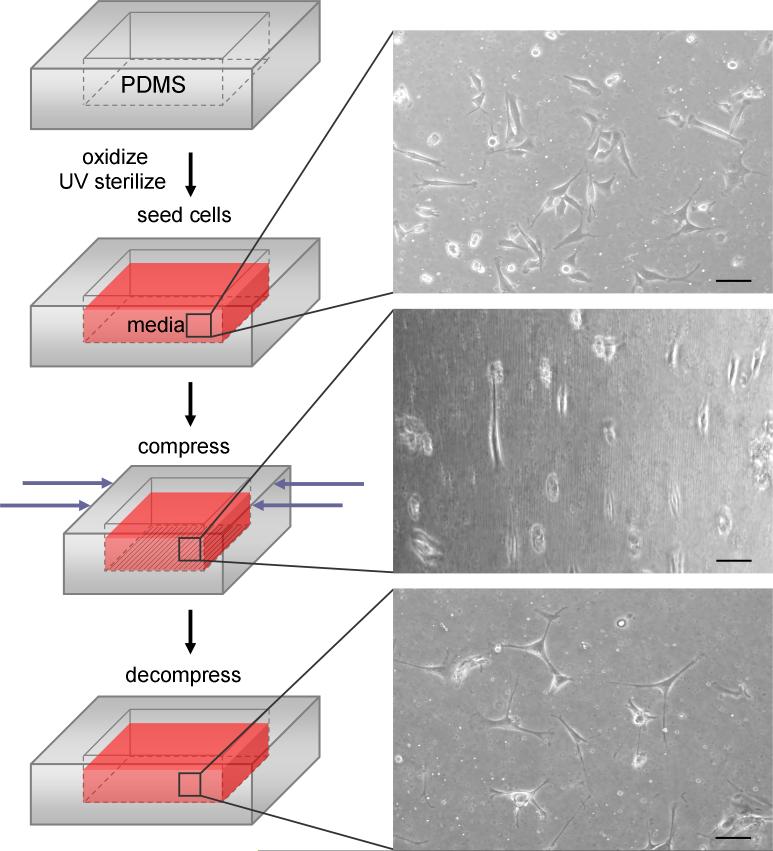
For cell reorientation studies (Fig. 2), PDMS wells were coated with the attachment protein laminin, filled with GM, and UV sterilized for 30 min. C2C12 skeletal muscle cell line cells were plated into the 1.5 cm2 wells at a density of 1.0 × 104 cells/cm2, and cultured in GM. UV sterilized 1 mm thick 15:1 PDMS membranes were cut to size to cover the wells in order to maintain cell culture sterility.
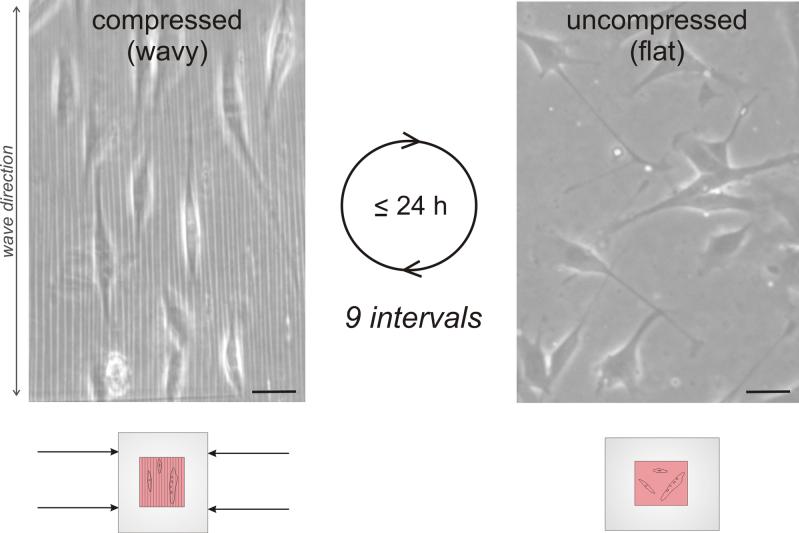
Fig. 2.
Schematic of the process of reversibly introducing microfeatures into the substrate surface (left side); after being prepared for cell culture, substrates are compressed to obtain wavy features and uncompressed to remove the features. Micrographs on the right show cell behavior on the reconfigurable surface before, during and after compression- cells orient from random to aligned and back to random. Scale bars = 100 μm.
Cells attached and spread out on the reversible surfaces within 4 h after plating. Cell proliferation was seen on the reversible topography, and was not inhibited noticeably by the surface changes. ∼13% compressive strain was applied with strain devices to PDMS wells in all the following studies to obtain wavy microfeatures with sufficient amplitudes to align cells. For all cell studies, data was collected from cells cultured on 2 PDMS substrates per experiment.
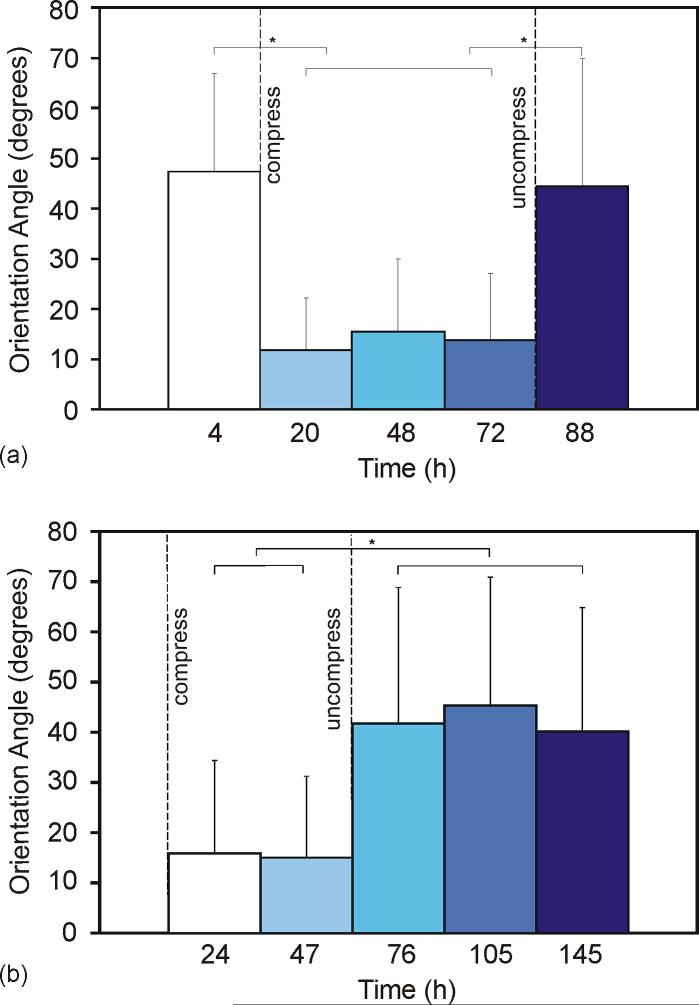
Orientation angles of cells cultured on flat (pre-compression) to wavy (compressed) to flat (uncompressed) surfaces over the course of 4 d verified the reversibility of the system (Figures 3a). The OA data reveal initial random orientation (average OA = 47° ± 20°), followed by alignment (average OA over all compressed time points = 13° ± 12°), and lastly returning to random (average OA = 44° ± 25°). These results show that C2C12 cells that are already spread onto a substrate in an unaligned manner can be aligned by creating wavy topographical features on the surface, and once again unaligned by dissipating the features. Statistical analysis using ANOVA showed a statistically significant difference in orientation angle between groups of cells before substrate compression (cells at 4 h, on the flat surface, group 1), during compression (combined data for cells at 20, 48, and 72 h, on the wavy surface, group 2), and after uncompression (cells at 88h, on flat surface, group 3) (p<0.001). Tukey post-hoc analysis indicated a statistically significant difference between groups 1 and 2, and 2 and 3 (α=0.05). The Tukey post-hoc test (α=0.05) also showed that the difference between groups 1 and 3 (both discontinuous flat conditions) was not statistically significant, though this is to be expected because cells were randomly oriented on a flat surface in both instances.
Fig. 3.
(a) Average orientation angle of cells on reconfigurable surfaces; cells were plated onto flat substrates that were compressed 4 h after initial cell plating and uncompressed at 76 h. 4 h n = 129 (M = 47° ± 20°), 20 h n = 304 (M = 11° ± 10°), 48 h n = 184 (M = 15° ± 14°), 72 h n = 167 (M = 14° ± 13°), 88 h n = 160 (M = 44° ± 25°). (b) Average orientation angle of cells on substrates first compressed (wavy) then released (flat) at 47 h; cells aligned on the wavy surface and unaligned to a random orientation on the flat surface. 24 h n = 266 (M = 16° ± 18°), 47 h n = 413 (M = 15° ± 16°), 76 h n = 278 (M = 42° ± 27°), 105 h n = 290 (M = 45° ± 26°), 145 h n = 449 (M = 40° ± 25°). Bars represent standard deviation; * denotes statistical significance.
Similar reversibility in cell alignment was observed even when cells were initially plated onto wavy features and allowed to spread first in an aligned manner, and then unaligned by removing the wavy features (Fig. 3b). There are no apparent lasting effects from cell adaptive events, such as extracellular matrix (ECM) remodeling or cytoskeleton organization that may arise from the initial cell spreading process that cannot be overridden by topographical reconfiguration of the substrate over the 3−6 d investigated. A t-test was performed on combined data of cells during compression (first group; data from 24 and 47 h) and after uncompression (second group; data from 76, 105, and 145 h). Results indicate the change in orientation angle on the wavy surface compared to the flat surface was statistically significant (p<0.001).
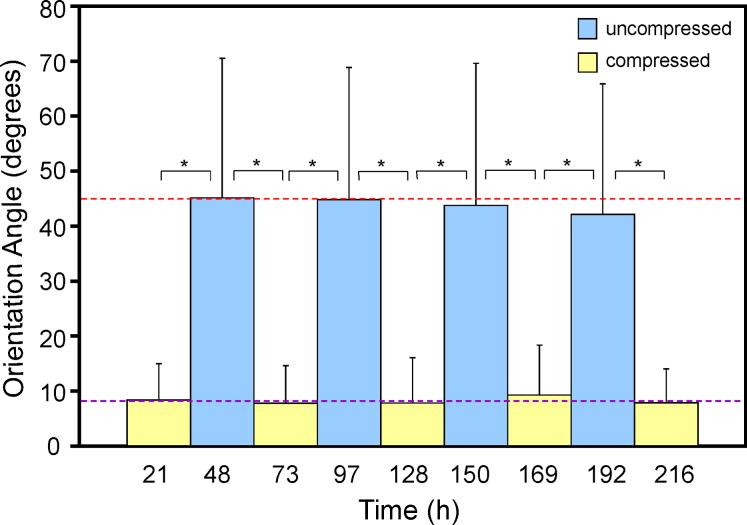
To test robustness of the reconfigurable topographical system as well as the plasticity of cells, substrates with cells were compressed and uncompressed repeatedly at faster rates (24 h intervals) than in previous experiments (2−3 d intervals). Cells were able to switch from random orientation to a highly aligned arrangement with each compression interval (representative micrographs in Fig. 4a). Cells were observed periodically up to 24 h after substrate compression or uncompression. Noticeable cell reorientation occurred between 19−24 h post-topography switching. OA for cells on wavy surfaces across all time points averaged at 8° ± 7° and is represented by the purple dashed line (Fig. 4b). Average OAs on flat, uncompressed surfaces were close to that of random (i.e. 45°, indicated by the dashed red line). Results in OAs of cells on waves in the aforementioned experiments (Fig. 3) were slightly higher than during repeated compression and uncompression, suggesting that oxidation times alter materials properties to produce some variability in actual waves formed by compression. However, for a given material, wave size is consistent across repeated compressions. The graph in Fig. 4b reveals a robust reversibility of cell behavior in response to dynamically altered surface microfeatures (corresponding cell images in Fig. 4c). A minute statistically insignificant decline in cell reversion back to random during uncompression is noticed. This may suggest decreasing reversibility over longer durations and over repeated cell realignment due to phenomena such as increased extracellular matrix deposition (thus limiting cell movement on the substrate) or unseen material deformations remaining after uncompression, or a combination of factors. After ∼9 compression-uncompression intervals, the substrate started to show visible signs of plastic deformation, with slight wavy features remaining in the surface even after uncompression and cells not returning to random orientation. In addition, at this point cells began to die due to repeated handling outside the biological safety cabinet and incubator during unavoidable unsterile optical microscope imaging conditions.
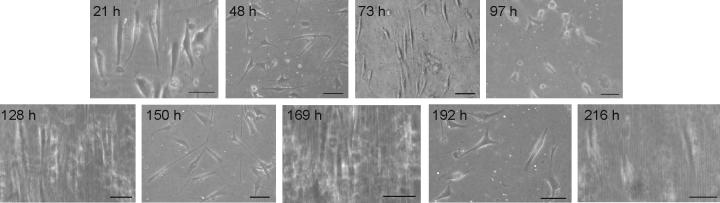
Fig. 4.
(a) Optical microscopy images of muscle cells on the reversible wavy surfaces; cell aligned along the waves and returned to random orientation on the uncompressed, flat surface within 24 h. Scale bars = 50 μm. (b) Average orientation angles for cells during repeated compression (wavy) and uncompression (flat), and (c) corresponding cell images (scale bars = 100 μm); some images are slightly blurred due to imaging during compression through a curved surface. In (b), upper, red dashed line indicates the angle for random orientation (45°); cells on flat surfaces exhibited angles close to random; lower, purple dashed line indicates average angle for cells over all compressed time points (8° ± 7°). 21 h n = 153, 48 h n = 283, 73 h n = 188, 97 h n = 208, 128 h n = 125, 150 h n = 149, 169 h n = 109, 192 h n = 150, 216 h n = 83. Bars represent standard deviation; * denotes statistical significance.
Cells were also able to differentiate, even through repeated substrate compression and uncompression, using the loading regime illustrated in Figure 4a. Despite use of a growth-promoting media, some differentiated muscle cells, or myotubes, were seen as early as 20 h following cell plating (which is not unusual for C2C12 cell line cells); more myotubes appeared by 88 h in culture, demonstrating that the actively changing topography of the culture surface did not preclude cellular differentiation. Once a myotube formed, it did not reorient and remained in its last assumed position, perhaps due to increased ECM deposited and reduced cell motility over time as the cell matured.
Cells have been known to reorient in vitro in response to stimuli such as fluid shear stress, multi-directional cyclical stretch, and electrical fields [24-26]. Cell reorientation due to rapid topographical changes, to our knowledge, has not been previously shown. We observed that cells are able to reorient repeatedly when given appropriate topographical cues, and that realignment of initially randomly spread cells produce equally well aligned myoblasts as cells allowed to align during initial spreading. Cells and their intracellular components are anchored to substrates indirectly through their ECM [27-30]. The reversibility of cell alignment we observed in our system suggests that cell alignment does not, within the time frame investigated, create irreversible changes to the intracellular structures or ECM environment. In fact, topographical cues appear capable of triggering relatively rapid reorganization of the intracellular machinery and any small variations in adhesive cues that may be generated by cellular matrix remodeling. Although the system described in this paper is synthetic, there are wavy topographical features that arise due to buckling in nature, such as embryonic buckling resulting in congenital disease [31], bucking of extracellular matrix components during contraction [32], and buckling during growth of anisotropic tissues [33]. Thus, material-induced cellular reorientation may be a physiologically relevant mechanism of cellular organization.
Advantages of this adaptable materials system include: (i) convenient creation, dissipation, and recreation of the wavy microfeatures using an inexpensive cell culture compatible device; (ii) readily usable fabrication protocols for biological labs without access to clean rooms or other expensive microfabrication equipment; (iii) use of a non-toxic material that is optically transparent to allow convenient analysis of cells using inverted microscopes common to biological labs, and (iv) the ability to align, unalign, and realign the same cells repeatedly on the same substrate. Some weaknesses of the system are: (i) the use of a relatively thick (∼8 mm) bottom substrate to provide mechanical support reduces the quality of optical micrographs and excludes use of short working distance, high numerical aperture objectives with inverted microscopes, (ii) difficulty in precisely reproducing the oxidation conditions leads to some variability in the wave sizes obtained from batch to batch (although the ability to create and recreate very similar features on the same substrate is excellent); and (iii) the use of a material, PDMS, that may adsorb lipophilic chemicals and alter the effective concentration of reagents for cell biological studies [34] (though the relatively large volumes of liquid to surface area in this device, compared to microfluidics systems, should mitigate this effect). Even though this paper focused exclusively on alignment of C2C12 myoblasts, many other cell types respond to topographical features, including other muscle cells [35,36], osteoblasts [37], endothelial cells [38], HeLa cells, hepatocytes and fibroblasts [39,40].
4. Conclusions
In summary, we present a reconfigurable microtopography system for reversible on-demand cell alignment. Using the simple material manipulations of oxidation and compression, wavy microfeatures can be introduced into and removed from silicone substrate surfaces. Myoblasts were shown to rapidly reorient within 24 h on the reversible waves. Our described system provides an easily applicable means by which to examine, for a wide range of cell types, dynamic cellular processes that traditional in vitro culture substrates are not capable of studying.
Acknowledgements
This work has been funded by NIH (EB003793-01) and NSF (BES-0238625). M.T.L. would like to thank the National Institute on Aging T32 AG00114 Multidisciplinary Research Training in Aging Grant for support. We also thank Wei Gu for critical review of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
Fig. S1. Diagram of the process of molding substrate wells. (a) PDMS was poured into a square Petri dish, allowed to cure, and material was cut out (b). (c) Epoxy was poured into the cut-out regions, UV cured, and the surrounding PDMS removed, leaving an epoxy mold. (d) PDMS was poured over the epoxy, cured, and regions cut out around the epoxy, resulting in customized-sized PDMS wells.
References
- 1.Stevens MM, George JH. Exploring and engineering the cell surface interface. Science. 2005;310:1135–1138. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- 2.Curtis A, Wilkinson C. Topographical control of cells. Biomaterials. 1997;18:1573–1583. doi: 10.1016/s0142-9612(97)00144-0. [DOI] [PubMed] [Google Scholar]
- 3.Curtis AS, Wilkinson CD. Reactions of cells to topography. J Biomater Sci Polym Ed. 1998;9:1313–1329. doi: 10.1163/156856298x00415. [DOI] [PubMed] [Google Scholar]
- 4.Flemming RG, Murphy CJ, Abrams GA, Goodman SL, Nealey PF. Effects of synthetic micro- and nano-structured surfaces on cell behavior. Biomaterials. 1999;20:573–588. doi: 10.1016/s0142-9612(98)00209-9. [DOI] [PubMed] [Google Scholar]
- 5.Stockdale FE. Myogenic cell lineages. Dev Biol. 1992;154:284–298. doi: 10.1016/0012-1606(92)90068-r. [DOI] [PubMed] [Google Scholar]
- 6.Cowin SC. Structural changes in living tissues. Meccanica. 1999;34:379–398. doi: 10.1023/a:1004777501507. [DOI] [PubMed] [Google Scholar]
- 7.Gheusi G, Lledo P. Control of early events in olfactory processing by adult neurogenesis. Chem Senses. 2007;32:397–409. doi: 10.1093/chemse/bjm012. [DOI] [PubMed] [Google Scholar]
- 8.Rizzolo LJ, Chen X, Weitzman M, Sun R, Zhang H. Analysis of the RPE transcriptome reveals dynamic changes during the development of the outer blood-retinal barrier. Mol Vis. 2007;13:1259–1273. [PubMed] [Google Scholar]
- 9.Ebara M, Hoffman JM, Hoffman AS, Stayton PS. Switchable surface traps for injectable bead-based chromatography in PDMS microfluidic channels. Lab Chip. 2006;6:843–848. doi: 10.1039/b515128g. [DOI] [PubMed] [Google Scholar]
- 10.Lahann J, Mitragotri S, Tran TN, Kaido H, Sundaram J, Choi IS, et al. A reversibly switching surface. Science. 2003;299:371–374. doi: 10.1126/science.1078933. [DOI] [PubMed] [Google Scholar]
- 11.Zhu X, Mills KL, Peters PR, Bahng JH, Liu EH, Shim J, et al. Fabrication of reconfigurable protein matrices by cracking. Nat Mater. 2005;4:403–406. doi: 10.1038/nmat1365. [DOI] [PubMed] [Google Scholar]
- 12.Kwon OH, Kikuchi A, Yamato M, Sakurai Y, Okano T. Rapid cell sheet detachment from poly(N-isopropylacrylamide)-grafted porous cell culture membranes. J Biomed Mater Res. 2000;50:82–89. doi: 10.1002/(sici)1097-4636(200004)50:1<82::aid-jbm12>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 13.Bowden N, Huck W, Paul K, Whitesides G. The controlled formation of ordered, sinusoidal structures by plasma oxidation of an elastomeric polymer. Appl Phys Lett. 1999;75:2557–2559. [Google Scholar]
- 14.Hauschka S. In: Myology. Engel A, Franzini-Armstrong C, editors. McGraw-Hill; New York: 1994. pp. 1–373. [Google Scholar]
- 15.Kawai H, Nishino H, Nishida Y, Yoneda K, Yoshida Y, Inui T, et al. Skeletal muscle pathology of mannosidosis in two siblings with spastic paraplegia. Acta Neuropathol (Berl) 1985;68:201–204. doi: 10.1007/BF00690195. [DOI] [PubMed] [Google Scholar]
- 16.Ferreiro A, Estournet B, Chateau D, Romero NB, Laroche C, Odent S, et al. Multi-minicore disease--searching for boundaries: phenotype analysis of 38 cases. Ann Neurol. 2000;48:745–757. [PubMed] [Google Scholar]
- 17.Wigmore PM, Maleki F, Evans DJ, McErlain M. After embryonic day 17, distribution of cells on surface of primary muscle fibres in mouse is non-random. Dev Dyn. 1996;207:215–221. doi: 10.1002/(SICI)1097-0177(199610)207:2<215::AID-AJA9>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 18.Evans DJ, Britland S, Wigmore PM. Differential response of fetal and neonatal myoblasts to topographical guidance cues in vitro. Dev Genes Evol. 1999;209:438–442. doi: 10.1007/s004270050275. [DOI] [PubMed] [Google Scholar]
- 19.Sassoon DA. Stem cells and cell signaling in skeletal myogenesis. Elsevier Science B.V.; Amsterdam, Netherlands: 2002. pp. 127–142. [Google Scholar]
- 20.Lam MT, Sim S, Zhu X, Takayama S. The effect of continuous wavy micropatterns on silicone substrates on the alignment of skeletal muscle myoblasts and myotubes. Biomaterials. 2006;27:4340–4347. doi: 10.1016/j.biomaterials.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 21.Jiang X, Takayama S, Qian X, Ostuni E, Wu H, Bowden N, et al. Controlling mammalian cell spreading and cytoskeletal arrangement with conveniently fabricated continuous wavy features on poly(dimethylsiloxane). Langmuir. 2002;18:3273–3280. [Google Scholar]
- 22.Bowden N, Brittain S, Evans AG, Hutchinson JW, Whitesides GM. Spontaneous formation of ordered structures in thin films of metals supported on an elastomeric polymer. Nature. 1998;393:146–149. [Google Scholar]
- 23.Cerda E, Mahadevan L. Geometry and physics of wrinkling. Phys Rev Lett. 2003;90:074302. doi: 10.1103/PhysRevLett.90.074302. [DOI] [PubMed] [Google Scholar]
- 24.Noria S, Xu F, McCue S, Jones M, Gotlieb AI, Langille BL. Assembly and reorientation of stress fibers drives morphological changes to endothelial cells exposed to shear stress. Am J Pathol. 2004;164:1211–1223. doi: 10.1016/S0002-9440(10)63209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang JH, Goldschmidt-Clermont P, Wille J, Yin FC. Specificity of endothelial cell reorientation in response to cyclic mechanical stretching. J Biomech. 2001;34:1563–1572. doi: 10.1016/s0021-9290(01)00150-6. [DOI] [PubMed] [Google Scholar]
- 26.Curtze S, Dembo M, Miron M, Jones DB. Dynamic changes in traction forces with DC electric field in osteoblast-like cells. J Cell Sci. 2004;117:2721–2729. doi: 10.1242/jcs.01119. [DOI] [PubMed] [Google Scholar]
- 27.Zamir E, Geiger B. Components of cell-matrix adhesions. J Cell Sci. 2001;114:3577–3579. doi: 10.1242/jcs.114.20.3577. [DOI] [PubMed] [Google Scholar]
- 28.Zamir E, Geiger B. Molecular complexity and dynamics of cell-matrix adhesions. J Cell Sci. 2001;114:3583–3590. doi: 10.1242/jcs.114.20.3583. [DOI] [PubMed] [Google Scholar]
- 29.Geiger B, Bershadsky A, Pankov R, Yamada KM. Transmembrane crosstalk between the extracellular matrix--cytoskeleton crosstalk. Nat Rev Mol Cell Biol. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 30.Kong HJ, Mooney DJ. Microenvironmental regulation of biomacromolecular therapies. Nat Rev Drug Discov. 2007;6:455–463. doi: 10.1038/nrd2309. [DOI] [PubMed] [Google Scholar]
- 31.Dias MS, Li V, Landi M, Schwend R, Grabb P. The embryogenesis of congenital vertebral dislocation: early embryonic buckling? Pediatr Neurosurg. 1998;29:281–289. doi: 10.1159/000028738. [DOI] [PubMed] [Google Scholar]
- 32.Freyman TM, Yannas IV, Pek YS, Yokoo R, Gibson LJ. Micromechanics of fibroblast contraction of a collagen-GAG matrix. Exp Cell Res. 2001;269:140–153. doi: 10.1006/excr.2001.5302. [DOI] [PubMed] [Google Scholar]
- 33.Volokh KY. Tissue morphogenesis: a surface buckling mechanism. Int J Dev Biol. 2006;50:359–365. doi: 10.1387/ijdb.052039kv. [DOI] [PubMed] [Google Scholar]
- 34.Toepke MW, Beebe DJ. PDMS absorption of small molecules and consequences in microfluidic applications. Lab Chip. 2006;6:1484–1486. doi: 10.1039/b612140c. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen KT, Shukla KP, Moctezuma M, Tang L. Cellular and molecular responses of smooth muscle cells to surface nanotopography. J Nanosci Nanotechnol. 2007;7:2823–2832. doi: 10.1166/jnn.2007.610. [DOI] [PubMed] [Google Scholar]
- 36.Au HT, Cheng I, Chowdhury MF, Radisic M. Interactive effects of surface topography and pulsatile electrical field stimulation on orientation and elongation of fibroblasts and cardiomyocytes. Biomaterials. 2007;28:4277–4293. doi: 10.1016/j.biomaterials.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamilton DW, Brunette DM. The effect of substratum topography on osteoblast adhesion mediated signal transduction and phosphorylation. Biomaterials. 2007;28:1806–1819. doi: 10.1016/j.biomaterials.2006.11.041. [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto S, Tanaka M, Sunami H, Ito E, Yamashita S, Morita Y, et al. Effect of honeycomb-patterned surface topography on the adhesion and signal transduction of porcine aortic endothelial cells. Langmuir. 2007;23:8114–8120. doi: 10.1021/la7003326. [DOI] [PubMed] [Google Scholar]
- 39.Kidambi S, Udpa N, Schroeder SA, Findlan R, Lee I, Chan C. Cell adhesion on polyelectrolyte multilayer coated polydimethylsiloxane surfaces with varying topographies. Tissue Eng. 2007;13:2105–2117. doi: 10.1089/ten.2006.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J, Shi R. Fabrication of patterned multi-walled poly-l-lactic acid conduits for nerve regeneration. J Neurosci Methods. 2007;165:257–264. doi: 10.1016/j.jneumeth.2007.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Diagram of the process of molding substrate wells. (a) PDMS was poured into a square Petri dish, allowed to cure, and material was cut out (b). (c) Epoxy was poured into the cut-out regions, UV cured, and the surrounding PDMS removed, leaving an epoxy mold. (d) PDMS was poured over the epoxy, cured, and regions cut out around the epoxy, resulting in customized-sized PDMS wells.