Abstract
The E2F transcription factors are critical regulators of genes required for appropriate progression through the cell cycle, and in special circumstances they can also promote the expression of another class of genes that function in the apoptotic program. Since E2Fs can initiate both cell proliferation and cell death, it is not surprising that the pro-apoptotic capacity of these proteins is subject to complex regulation. Recent study has expanded our knowledge both of the factors influencing E2F-induced apoptosis, as well as downstream targets of E2F in this process.
Introduction
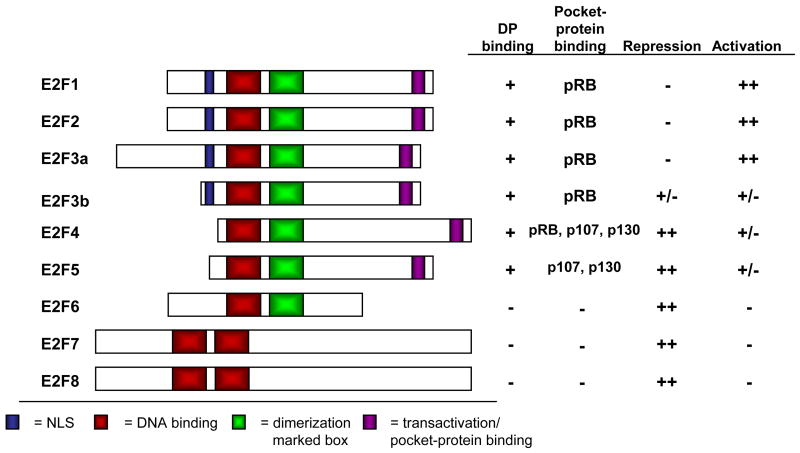
The E2F proteins regulate the cell cycle by controlling the coordinate transcription of a large number of targets, including genes involved in DNA replication and cell cycle progression. These genes are expressed at low levels during quiescence, and are induced as cells traverse the G1/S transition and enter the cell cycle. Eight E2F genes encode nine major protein species, which can be generally classified as either activators or repressors of transcription based on their functional properties as well as structural features [1] (Figure 1). E2F1, E2F2, and E2F3a are generally considered the “activating E2Fs”, based on their ability to potently activate transcription when overexpressed, and localization to the promoters of E2F target genes coincident with their transcriptional activation in G1/S phase. The “repressive E2Fs”, E2F4 and E2F5, bind their targets coincident with their repression in G0/G1, and only modestly activate transcription when overexpressed, despite containing a transcriptional activation domain homologous to the activating E2Fs. E2F3b is a second product of the E2f3 locus, which is identical in sequence to E2F3a through most of the protein, yet lacks the N-terminal domain characteristic of the activating E2Fs (Figure 1). Indeed, E2F3b protein is present throughout the cell cycle, similar to E2F4 and E2F5; it is this property that led to the hypothesis that E2F3b functions as a repressive E2F.
Figure 1. The E2F Family of Transcription Factors.
The E2F family is divided into three groups based on their structure and function. All E2Fs contain a homologous DNA-binding domain (red). The “activating E2Fs” E2F1, E2F2, and E2F3a each contain both a nuclear localization signal (NLS, blue) and a transactivation/pocket protein-binding domain (purple). The “repressive E2Fs” lack the N-terminal domain present in the activating E2Fs, and E2F4 and E2F5 accordingly lack NLS sequences. E2F6, E2F7, and E2F8 lack sequences required for transactivation and pocket protein-binding, and therefore are pocket protein-independent repressors of transcription.
The transcriptional activity of E2Fs1-5 is regulated primarily via their interaction with the “pocket protein” family, which includes the retinoblastoma protein (pRB), and its homologs p107 and p130. Pocket protein binding to E2F inhibits its transcriptional activity in two ways: by masking key residues required for transcriptional activation by E2F, and by recruiting co-repressor complexes to E2F-responsive promoters. E2F6, E2F7, and E2F8 lack sequences required for pocket protein-binding, yet are able to repress transcription via other mechanisms (Figure 1). E2F6 participates in the repression of E2F target genes whose expression decreases as cells exit S phase [2]. The silencing function of E2F6 appears to be due to its ability to associate with members of the Polycomb group of transcriptional repressors [1]. E2F7 and E2F8 also behave as repressors, and may function in a manner analogous to E2F6 [3], yet the biological functions of these proteins are just beginning to be understood. Interestingly, E2F7 and E2F8 are unique among E2Fs in their ability to bind DNA in the absence of interaction with a DP subunit, the normal dimerization partner of E2Fs. Rather, E2F7 and E2F8 contain a duplication of the DNA-binding domain in place of the dimerization domain present in other E2Fs (Figure 1); this second DNA-binding domain is likely to substitute for the function of the DP subunit in DNA binding.
Most classic E2F target genes are regulated by the cyclical repression or activation by different E2F species, according to the cell cycle stage (Figure 2). It is well established that in G0/G1 phase, the repressive E2Fs, E2F4 and E2F5, in complex with the pocket proteins p107, p130, and perhaps pRB, associate with the promoters of E2F-responsive genes and actively repress their transcription [1]. At the same time, the activating E2Fs are bound by pRB, inhibiting their potential to activate transcription. Whether complexes containing pRB and activating E2Fs are involved in the repression of E2F target genes is still subject to debate [4]. As cells are stimulated to enter the cell cycle, cyclin-CDK activity increases and targets the pocket proteins for phosphorylation, causing a disruption of pocket protein-E2F complexes. Because they lack nuclear localization signals, in the absence of pocket protein-binding E2F4 and E2F5 dissociate from DNA and are exported from the nucleus. Activating E2Fs, relieved of their pRB-mediated inhibition, bind the promoters of their targets and activate transcription [1]. The more limited analysis of E2F6, 7 and 8 suggests that they can also contribute to the regulation of these genes, but the precise timing of their action, versus E2Fs1-5, has yet to be fully established.
Figure 2. Regulation of cell cycle-dependent transcription by the pRB-E2F pathway.
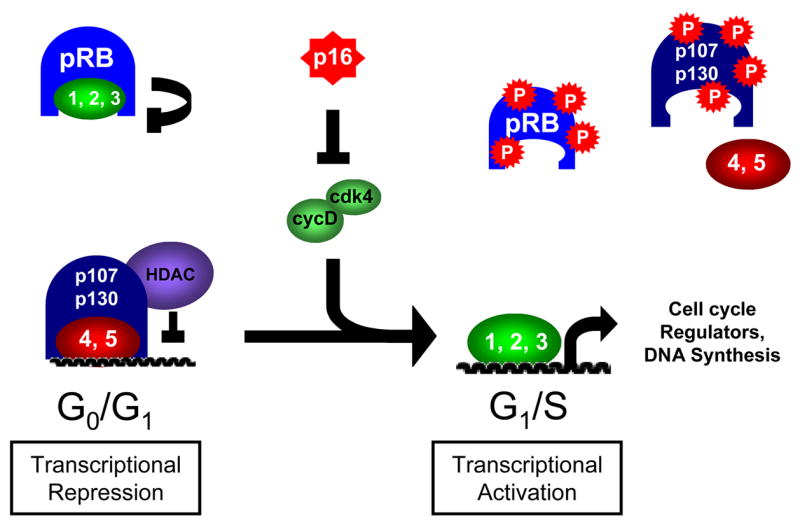
E2F target genes are regulated by repression of transcription in G0/G1 phase, followed by activation in G1/S phase. Repressive complexes containing primarily E2F4 and p107/p130 occupy promoters in G0/G1 phase, and association with chromatin remodeling enzymes such as histone deacetylases (HDAC) contributes to transcriptional repression. pRB binds to and inhibits the transcriptional activity of the activating E2Fs 1-3. Upon cell cycle entry, cyclin-CDK complexes such as cyclin D-CDK4 overcome inhibition by CDK inhibitors such as p16INK4a and phosphorylate the pocket proteins. This phosphorylation leads to disruption of the pocket protein-E2F complexes, causing E2F4 to be exported from the nucleus, and activating E2Fs to bind the promoters of cell cycle-regulated target genes and activate their transcription.
Thus, every cell cycle, there is essentially a coordinated switch from the repressive to the activating E2Fs that enables the simultaneous activation of genes involved in DNA replication and cell cycle progression. Notably, this wave of expression appears to be largely synonymous with the restriction point, defined as the point where cells become committed to divide even in the absence of mitogenic stimuli. Consistent with this model, the exogenous expression of any individual activating E2F in cell culture is sufficient to stimulate DNA synthesis in the absence of growth signals [5–7]. Remarkably, in addition to causing cell cycle entry, E2F overexpression induces extensive apoptosis [7–9]. In most cases, this effect is associated with activation of p53, a key regulator of the apoptotic response [7]; however, E2F can also induce apoptosis in a p53-independent manner [10,11]. More recently, much research has been devoted to elucidating the mechanisms by which E2F contributes to apoptotic signaling. The ability of E2F to activate p53-dependent apoptosis was partially explained by the finding that E2F1 activates transcription of the p53 regulator, p19Arf, which indirectly increases p53 protein levels by inhibiting its degradation [12,13]. Since this discovery, further gene expression analyses, including genome-wide microarray experiments, have found that E2F can activate transcription of a large number of pro-apoptotic genes, including both upstream signaling proteins and core members of the apoptotic machinery [14•], (reviewed in [15]).
Paradoxically, E2F activity can promote both cell proliferation and cell death. Interestingly, most tumors display excess levels of both cell division and apoptosis. Since E2F activity is implicated in regulation of both of these processes, it may be a key factor in the decision between these two fates. An important element of this choice is likely to be the distinction between pro-apoptotic and cell cycle-regulated gene activation by E2F. How does E2F avoid activation of apoptosis when its activity increases during cell cycle entry, yet faithfully contribute to cell death in response to cell stress? For the remainder of this article, we will review recent progress in the study of E2F-mediated cell death, focusing on the intricacies of E2F regulation in this pathway.
Specificity of E2F induction of apoptosis
Since the discovery that E2F1 could trigger apoptosis, there have been conflicting reports as to whether this is a specific property of E2F1, or if other E2Fs can contribute. Initial studies indicated that overexpression of either E2F2 or E2F3a, unlike E2F1, was unable to induce apoptosis [12,16]. However, subsequent studies performed in a similar manner found that activation of E2F2 or E2F3a could also lead to cell death, although possibly not to the same degree as E2F1 expression [8,9]. Since E2F2 and E2F3a can activate pro-apoptotic gene transcription, at least when overexpressed [9,12,17•], the unique tendency of E2F1 to potentiate apoptosis in certain scenarios may depend on other specific properties of E2F1. Consistent with this idea, many E2F1-interacting proteins that specifically regulate E2F1’s ability to induce apoptosis fail to bind E2F2 and E2F3a (see below).
Overexpression of E2F can also lead to apoptosis in vivo. Targeted expression of an E2F1 transgene to the squamous epithelium led to p53-dependent apoptosis in this tissue, and E2F1-dependent apoptosis also suppresses papilloma formation in a ras-driven skin carcinogenesis model [18, Pierce, 1999 #779]. Similar to E2F1, overexpression of E2F3a causes hyperproliferation in vivo, and leads to p53-independent apoptosis [19,20]. Intriguingly, E2F3a was unable to cause apoptosis, either in vitro or in vivo, in the absence of E2f1 [21••]. This result can be interpreted in several ways. At one extreme, it is possible that E2F1 is the sole member of the E2F family that can directly stimulate apoptosis, and that E2F2 and E2F3a act in an indirect manner by promoting the expression of E2F1, itself an E2F-responsive gene. At the other extreme, the ability to initiate the apoptotic response may depend on the overall level of activating E2F activity, rather than the level of any individual E2F. Lack of E2f1 would cause an overall reduction in E2F levels, such that the expression of E2F3a was no longer sufficient to overcome the threshold level of E2F activity necessary to initiate the apoptotic cascade. Likely, the answer lies somewhere in between; E2F1 may play a particularly important role in the apoptotic response but E2F2 and E2F3a also contribute. This also appears to be context dependent and, as we will describe below, differs considerably in response to oncogenic stress versus DNA damage.
Results from several different mutant mouse models also support a physiological role of E2F in regulation of apoptosis. E2f1−/− mice display hypercellularity of the thymus, due to a defect in the ability of E2f1−/− thymocytes to undergo apoptosis during negative selection [22–24]. This may be due to the inability of the mutant cells to induce Arf and p53 during this process [23]. Also, the appearance of diverse tumor types in aging E2f1−/− mice hints at a requirement of E2F1-induced apoptosis in the suppression of endogenous tumorigenesis in vivo [25].
E2F is also required for apoptosis caused by loss of Rb in mice. Mutation of either E2f1 or E2f3 suppresses both the increased proliferation and apoptosis in the lens and nervous system of Rb−/− embryos, suggesting that both E2F1 and E2F3 are required for induction of apoptosis due to loss of pRb [26,27]. Intriguingly, a portion of Rb−/−;E2f3+/− embryos exhibited suppression of the increased apoptosis, but not the ectopic proliferation in the peripheral nervous system (PNS) [27]. This suggests that E2F3 has a primary role in induction of apoptosis due to pRb-loss, rather than simply contributing to abnormal proliferation and resultant apoptosis of Rb-mutant cells. More recently, it has been determined that many of the defects in the Rb−/− embryos are indirectly due to an essential role of Rb in placental development; however, the defects in the lens and PNS appear to be cell-autonomous, suggesting that E2F may indeed play a direct role in regulation of proliferation and apoptosis in these tissues [28].
The requirement for any individual E2F in induction of apoptosis could also depend on the specific stress involved. It has been reported that E2f1-loss has no effect on apoptosis due to overexpression of c-Myc, either in vivo or in vitro [29]. This result could indicate either that E2F1 is not involved in apoptotic signaling downstream of Myc activation, or that other E2Fs can compensate for this function in the absence of E2F1. Surprisingly, a similar study did find a specific requirement of E2F1, but not E2F2 or E2F3, in mediating Myc-induced apoptosis in vitro [30]. This discrepancy is difficult to reconcile, but an important difference in the experiments may be illustrative. Two different targeted E2f1 alleles were used in these studies, which have also been shown to differentially modify the effects of a p53 mutation in the mouse [31,32]. Therefore, it is possible that one or both of these E2f1 alleles produces a protein product that may have a biologically relevant role in modulating the apoptotic response.
In general, genetic experiments attempting to define a requirement for any individual E2F in apoptosis are complicated by the extensive compensation occurring in E2F-mutant cells. Similarly, the overexpression of these proteins likely leads to neomorphic functions unrepresentative of their normal biological role. At the very least, since E2F1, 2 and 3a are all themselves E2F-responsive targets, it is difficult to unequivocally establish the separate biological properties of the individual E2Fs using an over-expression approach. In vivo, the crosstalk between the activating E2Fs likely provides an amplification mechanism that is critical in yielding a coordinated and robust biological response, both in normal cell cycle progression and also programmed cell death. At the experimental level, it makes the puzzle hard to unravel. More sensitive examination of the functions of the endogenous E2Fs in the response to apoptotic signaling may be crucial to understanding the normal role of these proteins.
Pro-apoptotic E2F target genes
Since the discovery that E2F could induce the transcription of Arf and p73, many additional pro-apoptotic targets have been identified (reviewed in [15]). Of these targets, most striking is the large number of Bcl-2 homology region 3 (BH3)-only proteins induced by E2F. In fact, at this time nearly all BH3-only proteins have been shown to be transcriptionally up-regulated by ectopically expressed E2F [15,33]. In addition to pro-apoptotic signaling pathways, much of the core apoptotic machinery is regulated by E2F, including both initiator (caspase-8 and -9) and effector caspases (caspase-3 and -7) [17•]. Notably, the genes encoding these caspases and several BH3-only proteins are regulated by endogenous E2F proteins, and accordingly display growth-regulated expression as cells enter the cell cycle [17•,33].
In addition to causing elevated p53 levels due to its induction of p19Arf, E2F can contribute to p53-dependent apoptosis in the absence of Arf. First, E2F is able to induce the expression of ASPP1 and ASPP2 [34,35]; ASPPs (apoptosis-stimulating proteins of p53) are co-factors that enhance p53 transcriptional activity, along with increasing its selectivity for pro-apoptotic targets [36]. E2F also cooperates with p53 in the transcriptional induction of many pro-apoptotic target genes, such as PERP [37], SIVA [38], Apaf1 [39], and the BH3-only proteins PUMA and NOXA [33]. Finally, E2F activates transcription of the p53-homolog p73, leading to an induction of p53 target genes in a p53-independent manner [40,41]. This concerted action of the E2F1 and p53 apoptotic response may provide a mechanism to ensure full activation of programmed cell death in the presence of apoptotic stimuli.
The induction of apoptosis involves sensing of cellular or environmental cues, leading to the activation of intracellular signaling pathways. Importantly, while E2F causes an elevation of the level of many proteins involved in the apoptotic response, it does not necessarily cause their activation. Caspases, especially, require post-translational activation to become catalytically active and mediate cell death. Therefore, it is likely that transcriptional activation of the apoptotic machinery by E2F merely sensitizes the cell to other pro-apoptotic signals.
E2F1 and the DNA damage response
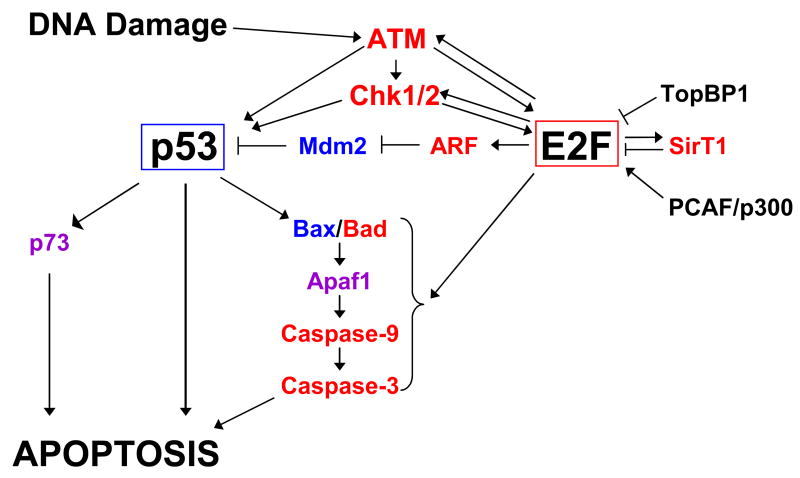
The first clue that E2F1 could contribute to the cellular response to DNA damage came from the observation that E2F1, and not other E2Fs, accumulates at the protein level in response to multiple DNA damaging insults [42–44]. E2F1 is also phosphorylated after DNA damage, and this modification has been attributed to the DNA damage-response kinases ATM/ATR and Chk1/Chk2 [24,40,45] (Figure 3). ATM/ATR phosphorylation appears to occur in the N-terminal domain of E2F1, which is involved in targeting it for degradation by the ubiquitin-ligase complex SCFSkp2, and therefore this phosphorylation may abrogate E2F1 degradation. Chk1 and Chk2, on the other hand, phosphorylate serine 364 in the C-terminus [45], so it is unclear whether this modification directly contributes to E2F1 stabilization, or affects E2F1 activity in another manner. Phosphorylation of E2F1 is presumed to modulate its transcriptional activity, although exactly how this occurs is still unknown.
Figure 3. Complex regulation of E2F and p53 in the apoptotic response pathway.
Many proteins regulate the activity of E2F and p53 in the apoptotic response. In turn, E2F and p53 transcriptionally activate a large number of pro-apoptotic genes. Transcriptional targets of E2F are shown in red; targets of p53 are shown in blue; targets of both E2F and p53 are indicated in purple.
In addition to phosphorylation, DNA damage-induced acetylation of E2F1 dramatically affects its transcriptional activity. E2F1 is targeted by either p300/CBP or P/CAF acetylases, depending on the DNA-damaging agent [46–48]. Acetylation leads to stabilization of E2F1, as well as stimulation of its transcriptional activity. Intriguingly, DNA damage-induced acetylation of E2F1 by P/CAF also affects the target gene specificity of E2F1, increasing E2F1’s selectivity for pro-apoptotic targets such as p73 and APAF1 [49••]. The observation that E2F1 is acetylated during normal cell cycle progression supports the idea that DNA damage-response signals are present even during proper DNA replication [47].
E2F1 contributes to the apoptotic response to DNA damage by indirectly inducing the activation of the p53. E2F1 overexpression leads to phosphorylation and stabilization of p53; importantly, this effect is independent of the ability of E2F1 to induce Arf [50]. Rather, it may be due to the ability of E2F1 to transcriptionally activate the genes encoding the p53-kinases ATM and/or Chk2 [51] (Figure 3). E2F1-induced p53 phosphorylation is sensitive to inhibition of ATM and related kinases by caffeine, suggesting a primary role for these kinases in p53 activation by E2F1 [50]. Again, although E2F activity is clearly able to induce the protein levels of p53-kinases, how these enzymes become catalytically active is unclear. It is possible that E2F1 overexpression itself contributes to DNA damage by inducing unscheduled DNA replication. This is consistent with the hypothesis that oncogene-induced proliferation causes DNA damage and can lead to cellular senescence (reviewed in [52]).
Whether regulation of the DNA damage-response pathway by endogenous E2F proteins is important during the normal cell cycle has not been thoroughly investigated. However, genome-wide location analysis has confirmed that E2F1 and/or E2F4 bind the promoters of many genes with checkpoint or DNA repair function during the normal cell cycle [53]. This may reflect a role of E2F in preparing the cell for endogenous DNA damage, such as the replication stress encountered during normal proliferation.
Regulation of pro-apoptotic functions of E2F by interacting proteins
While some pro-apoptotic target genes appear to be induced by E2F during each cell cycle, other target genes, such as Arf, are activated only in the presence of additional apoptotic stimuli. Since there are no obvious differences in the E2F-binding sites or other cis-acting sequences in the promoters of pro-apoptotic versus cell cycle-regulated E2F target genes, it is likely that E2F-interacting proteins provide additional regulation of the pro-apoptotic function of E2F. Driven by this hypothesis, Hallstrom and Nevins [54•] sought to identify E2F-interacting proteins that bound the “marked box” domain of E2F, a region postulated to confer target gene specificity [55]. They identified a factor, Jab1, which interacts predominantly with E2F1, and whose expression augments the ability of E2F1 to induce cell death and Arf/p53 activation [54•]. Depletion of Jab1 impairs E2F1-induced activation of p53 and apoptosis. Since Jab1 does not enhance E2F1-mediated cell cycle entry or transcription of the cell cycle-associated transcript PCNA, it appears that only pro-apoptotic functions of E2F1 are affected by Jab1. It remains unclear whether Jab1 directly affects E2F1 target gene specificity as a transcriptional cofactor, or merely favors E2F1 modification or the formation of a complex affecting E2F1 activity.
The pro-apoptotic functions of E2F1 are also affected by its primary regulator during the normal cell cycle, pRB. E2F1 is regulated by a C-terminal domain of pRB that does not bind to other E2F proteins, and binding of pRB via this interaction domain is able to suppress the pro-apoptotic activity of E2F1 [56••]. This pRB-E2F1 interaction may be partially dependent on post-translational modification of pRB, which is acetylated within this domain in response to DNA damaging agents [57]. These two reports suggest that DNA damage causes a disruption of the pRB-E2F1 interaction, perhaps due to acetylation of pRB, allowing transcriptionally active E2F1 to drive the apoptotic response. In contrast, another study found that phosphorylation of pRB by Chk2 increased its affinity for E2F1 after DNA damage, leading to inhibition of its pro-apoptotic activity [58]. In fact, all these signals may converge to enable sensitive control over the pro-apoptotic activity of E2F1. Low doses, or certain types of DNA damaging agents may favor inhibition of E2F1 activity, suppressing both cell cycle progression and apoptosis, and permitting DNA repair. Greater levels of DNA damage could promote apoptosis by inducing pRB acetylation and subsequent activation of E2F1 and thereby pro-apoptotic targets.
Other protein-protein interactions may contribute to regulation of E2F1 in response to DNA damage (Figure 3). The N-terminus of E2F1, when phosphorylated by ATM, provides a binding site for the BRCT domains of TopBP1 [59]. TopBP1-mediated inhibition of E2F1 transcriptional activity, including its ability to induce cell death, occurs both by recruitment of a repressive complex containing E2F1, TopBP1 and Brg/Brm to E2F-responsive promoters, as well as sequestration of E2F1 in BRCA1-containing foci in the nucleus [59,60]. DNA damage also induces E2F1-mediated activation of the NAD-dependent deacetylase SirT1, which in turn binds to and inhibits the activity of E2F1, in an apparent negative-feedback mechanism [61•]. This effect may be dependent on the ability of SirT1 to cause deacetylation of E2F1. Finally, the ETS-related transcription factor GABP 1 also binds E2F1, specifically inhibiting its ability to activate pro-apoptotic gene transcription without affecting the general transactivation ability of E2F1 [62]. Interaction of E2F1 with these factors may be important for inactivating the pro-apoptotic activity of E2F1 after initiation of the apoptotic response.
Other regulators of E2F-induced apoptosis may act downstream of E2F-activated transcription. Dyson and colleagues identified the anti-apoptotic protein Api5 in a screen for suppressors of E2F-mediated apoptosis in Drosophila [63]. Api5 specifically affects E2F-driven apoptosis both in flies and human cells, having no effect on other inducers of cell death. Unlike the E2F1-binding proteins mentioned above, Api5 does not affect E2F1-mediated transcription, either of cell cycle-related or pro-apoptotic targets. The mechanism of Api5-mediated suppression of apoptosis remains elusive, yet the ability of this protein to inhibit E2F-driven apoptosis in multiple situations, and both in flies and in mammalian cells, suggests that the downstream apoptotic mechanisms activated by E2F must be remarkably well-conserved.
Conclusion and perspective
Major regulators of cell proliferation, the E2F transcription factors now have an increasingly-appreciated role as components of various cell death pathways. Although much has been learned about the function of E2F in programmed cell death—including upstream regulators of E2F’s pro-apoptotic role, as well as downstream mediators of its effects—some basic questions remain unanswered. Most notably, it is unclear what allows the distinction between E2F-mediated regulation of proliferation- versus apoptosis-inducing genes. The analysis of E2F1 regulation in response to DNA damage suggests that the convergence of many different post-translational modifications and interacting proteins can greatly affect the functions of E2F1, including its target gene specificity. However, many targets of E2F in the cell death and DNA damage response pathways are regulated by E2F even during the normal cell cycle [17•,33,53], suggesting that induction of apoptosis by overexpressed E2F1 may be merely over-stimulation of an already-present mechanism. In this case, elevated E2F activity, such as that present during oncogene-induced proliferation, could lead to an E2F-dependent increase in the levels of pro-apoptotic proteins, resulting in sensitization to programmed cell death.
Despite the fact that many pro-apoptotic E2F targets are also regulated in a cell cycle-dependent manner by E2F proteins, some E2F target genes, notably Arf and p73, are induced by E2F only in response to cellular stresses. It is currently unclear what distinguishes these pro-apoptotic genes from cell cycle-regulated E2F targets. Unlike most E2F target genes, Arf is regulated only by E2F3, and not other E2F proteins, in unstressed cells [64]. It is possible that this binding specificity may contribute to defining Arf as a pro-apoptotic gene. Indeed, we have found that several pro-apoptotic targets of E2F are regulated similarly to Arf; these genes are repressed specifically by E2F3 in normal cells, and induced by activating E2F proteins only in response to oncogenic stress (PJ Iaquinta and JA Lees, unpublished).
Still, it is not clear what signals allow E2F to mediate the activation of these genes in response to stress. A simple model would predict that a general increase in total E2F levels causes hyper-activation of pro-apoptotic genes that are normally targeted by E2F, and/or induction of genes not normally activated by E2F, such as Arf. Alternatively, particular post-translational modifications or E2F-binding proteins may influence its target gene specificity in response to DNA damage or oncogenic stress. The difference in target gene regulation is likely at the heart of the distinction between the pro-apoptotic and pro-proliferation affects of E2F activity. Indeed, understanding the dichotomy between these two functions of E2F may be the key to explaining the apparent paradox of the roles of E2Fs as both tumor suppressor genes and oncogenes, and lead to a greater appreciation of the affect of E2F activity on tumorigenesis.
Acknowledgments
We thank members of the Lees lab, especially Allison Landman, for their comments on this manuscript. We apologize to our colleagues whose work we were unable to cite due to space limitations. J.A.L. is a Daniel K. Ludwig Scholar and P.I. was supported by a grant from the NIH (PO1-CA42063) to J.A.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nature Reviews Mol Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- 2.Giangrande PH, Zhu W, Schlisio S, Sun X, Mori S, Gaubatz S, Nevins JR. A role for E2F6 in distinguishing G1/S- and G2/M-specific transcription. Genes Dev. 2004;18:2941–2951. doi: 10.1101/gad.1239304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Stefano L, Jensen MR, Helin K. E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. Embo J. 2003;22:6289–6298. doi: 10.1093/emboj/cdg613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stevaux O, Dyson NJ. A revised picture of the E2F transcriptional network and RB function. Curr Opin Cell Biol. 2002;14:684–691. doi: 10.1016/s0955-0674(02)00388-5. [DOI] [PubMed] [Google Scholar]
- 5.Qin XQ, Livingston DM, Kaelin WG, Jr, Adams PD. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci U S A. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shan B, Lee WH. Deregulated expression of E2F-1 induces S-phase entry and leads to apoptosis. Mol Cell Biol. 1994;14:8166–8173. doi: 10.1128/mcb.14.12.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kowalik TF, DeGregori J, Schwarz JK, Nevins JR. E2F1 overexpression in quiescent fibroblasts leads to induction of cellular DNA synthesis and apoptosis. J Virol. 1995;69:2491–2500. doi: 10.1128/jvi.69.4.2491-2500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vigo E, Muller H, Prosperini E, Hateboer G, Cartwright P, Moroni MC, Helin K. CDC25A phosphatase is a target of E2F and is required for efficient E2F-induced S phase. Mol Cell Biol. 1999;19:6379–6395. doi: 10.1128/mcb.19.9.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moroni MC, Hickman ES, Denchi EL, Caprara G, Colli E, Cecconi F, Muller H, Helin K. Apaf-1 is a transcriptional target for E2F and p53. Nat Cell Biol. 2001;3:552–558. doi: 10.1038/35078527. [DOI] [PubMed] [Google Scholar]
- 10.Hsieh JK, Fredersdorf S, Kouzarides T, Martin K, Lu X. E2F1-induced apoptosis requires DNA binding but not transactivation and is inhibited by the retinoblastoma protein through direct interaction. Genes Dev. 1997;11:1840–1852. doi: 10.1101/gad.11.14.1840. [DOI] [PubMed] [Google Scholar]
- 11.Phillips AC, Bates S, Ryan KM, Helin K, Vousden KH. Induction of DNA synthesis and apoptosis are separable functions of E2F- 1. Genes Dev. 1997;11:1853–1863. doi: 10.1101/gad.11.14.1853. [DOI] [PubMed] [Google Scholar]
- 12.DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci U S A. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, Vousden KH. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 14•.Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD, Helin K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15:267–285. doi: 10.1101/gad.864201. This study was one of the first to attempt to isolate a large number of E2F targets in an unbiased manner, using microarray analysis. Their identification of many genes involved in apoptosis, including Apaf1, Caspase-3 and Caspase-7, suggested that the induction of apoptosis due to E2F expression involved the concerted activation of many pro-apoptotic genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci. 2004;29:409–417. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Kowalik TF, DeGregori J, Leone G, Jakoi L, Nevins JR. E2F1-specific induction of apoptosis and p53 accumulation, which is blocked by Mdm2. Cell Growth Differ. 1998;9:113–118. [PubMed] [Google Scholar]
- 17•.Nahle Z, Polakoff J, Davuluri RV, McCurrach ME, Jacobson MD, Narita M, Zhang MQ, Lazebnik Y, Bar-Sagi D, Lowe SW. Direct coupling of the cell cycle and cell death machinery by E2F. Nat Cell Biol. 2002;4:859–864. doi: 10.1038/ncb868. This elegant study confirmed that many Caspase genes are targeted for activation by endogenous E2F proteins in response to oncogenic stress. Additionally, they found that the mRNA levels of caspases are induced during cell cycle entry, suggesting that they are regulated by E2F in a manner similar to most classic E2F target genes. [DOI] [PubMed] [Google Scholar]
- 18.Pierce AM, Gimenez-Conti IB, Schneider-Broussard R, Martinez LA, Conti CJ, Johnson DG. Increased E2F1 activity induces skin tumors in mice heterozygous and nullizygous for p53. Proc Natl Acad Sci U S A. 1998;95:8858–8863. doi: 10.1073/pnas.95.15.8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazzerini Denchi E, Attwooll C, Pasini D, Helin K. Deregulated E2F activity induces hyperplasia and senescence-like features in the mouse pituitary gland. Mol Cell Biol. 2005;25:2660–2672. doi: 10.1128/MCB.25.7.2660-2672.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paulson QX, McArthur MJ, Johnson DG. E2F3a stimulates proliferation, p53-independent apoptosis and carcinogenesis in a transgenic mouse model. Cell Cycle. 2006;5:184–190. doi: 10.4161/cc.5.2.2307. [DOI] [PubMed] [Google Scholar]
- 21••.Lazzerini Denchi E, Helin K. E2F1 is crucial for E2F-dependent apoptosis. EMBO Rep. 2005;6:661–668. doi: 10.1038/sj.embor.7400452. This study, a companion study to [19], showed found that overexpression of E2F3a specifically in the mouse pituitary gland led to hyperproliferation, hyperplasia, and apoptosis in this tissue. These effects were associated with increased expression of E2F1, and inactivation of E2f1 abrogated the ability of E2F3a to induce apoptosis. Expression of E2F3a in E2f1−/− mice led to increased hyperplasia, consistent with the idea that E2F-induced apoptosis is tumor-suppressive, at least in this tissue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Field SJ, Tsai FY, Kuo F, Zubiaga AM, Kaelin WG, Jr, Livingston DM, Orkin SH, Greenberg ME. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 23.Zhu JW, DeRyckere D, Li FX, Wan YY, DeGregori J. A role for E2F1 in the induction of ARF, p53, and apoptosis during thymic negative selection. Cell Growth Differ. 1999;10:829–838. [PubMed] [Google Scholar]
- 24.Lin WC, Lin FT, Nevins JR. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 2001;15:1833–1844. [PMC free article] [PubMed] [Google Scholar]
- 25.Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson NJ. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 26.Tsai KY, Hu Y, Macleod KF, Crowley D, Yamasaki L, Jacks T. Mutation of E2f-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol Cell. 1998;2:293–304. doi: 10.1016/s1097-2765(00)80274-9. [DOI] [PubMed] [Google Scholar]
- 27.Ziebold U, Reza T, Caron A, Lees JA. E2F3 contributes both to the inappropriate proliferation and to the apoptosis arising in Rb mutant embryos. Genes Dev. 2001;15:386–391. doi: 10.1101/gad.858801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Bruin A, Wu L, Saavedra HI, Wilson P, Yang Y, Rosol TJ, Weinstein M, Robinson ML, Leone G. Rb function in extraembryonic lineages suppresses apoptosis in the CNS of Rb-deficient mice. Proc Natl Acad Sci U S A. 2003;100:6546–6551. doi: 10.1073/pnas.1031853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baudino TA, Maclean KH, Brennan J, Parganas E, Yang C, Aslanian A, Lees JA, Sherr CJ, Roussel MF, Cleveland JL. Myc-mediated proliferation and lymphomagenesis, but not apoptosis, are compromised by E2f1 loss. Mol Cell. 2003;11:905–914. doi: 10.1016/s1097-2765(03)00102-3. [DOI] [PubMed] [Google Scholar]
- 30.Leone G, Sears R, Huang E, Rempel R, Nuckolls F, Park C, Giangrande P, Wu L, Saavedra HI, Field SJ, et al. Myc requires distinct E2F activities to induce S phase and apoptosis. Mol Cell. 2001;8:105–113. doi: 10.1016/s1097-2765(01)00275-1. [DOI] [PubMed] [Google Scholar]
- 31.Wikonkal NM, Remenyik E, Knezevic D, Zhang W, Liu M, Zhao H, Berton TR, Johnson DG, Brash DE. Inactivating E2f1 reverts apoptosis resistance and cancer sensitivity in Trp53-deficient mice. Nat Cell Biol. 2003;5:655–660. doi: 10.1038/ncb1001. [DOI] [PubMed] [Google Scholar]
- 32.Wloga EH, Criniti V, Yamasaki L, Bronson RT. Lymphomagenesis and female-specific lethality in p53-deficient mice occur independently of E2f1. Nat Cell Biol. 2004;6:565–567. doi: 10.1038/ncb0704-565. author reply 567–568. [DOI] [PubMed] [Google Scholar]
- 33.Hershko T, Ginsberg D. Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J Biol Chem. 2004;279:8627–8634. doi: 10.1074/jbc.M312866200. [DOI] [PubMed] [Google Scholar]
- 34.Fogal V, Kartasheva NN, Trigiante G, Llanos S, Yap D, Vousden KH, Lu X. ASPP1 and ASPP2 are new transcriptional targets of E2F. Cell Death Differ. 2005;12:369–376. doi: 10.1038/sj.cdd.4401562. [DOI] [PubMed] [Google Scholar]
- 35.Hershko T, Chaussepied M, Oren M, Ginsberg D. Novel link between E2F and p53: proapoptotic cofactors of p53 are transcriptionally upregulated by E2F. Cell Death Differ. 2005;12:377–383. doi: 10.1038/sj.cdd.4401575. [DOI] [PubMed] [Google Scholar]
- 36.Samuels-Lev Y, O’Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S, Campargue I, Naumovski L, Crook T, Lu X. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell. 2001;8:781–794. doi: 10.1016/s1097-2765(01)00367-7. [DOI] [PubMed] [Google Scholar]
- 37.Attardi LD, Reczek EE, Cosmas C, Demicco EG, McCurrach ME, Lowe SW, Jacks T. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev. 2000;14:704–718. [PMC free article] [PubMed] [Google Scholar]
- 38.Fortin A, MacLaurin JG, Arbour N, Cregan SP, Kushwaha N, Callaghan SM, Park DS, Albert PR, Slack RS. The proapoptotic gene SIVA is a direct transcriptional target for the tumor suppressors p53 and E2F1. J Biol Chem. 2004;279:28706–28714. doi: 10.1074/jbc.M400376200. [DOI] [PubMed] [Google Scholar]
- 39.Fortin A, Cregan SP, MacLaurin JG, Kushwaha N, Hickman ES, Thompson CS, Hakim A, Albert PR, Cecconi F, Helin K, et al. APAF1 is a key transcriptional target for p53 in the regulation of neuronal cell death. J Cell Biol. 2001;155:207–216. doi: 10.1083/jcb.200105137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urist M, Tanaka T, Poyurovsky MV, Prives C. p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev. 2004;18:3041–3054. doi: 10.1101/gad.1221004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stiewe T, Putzer BM. Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat Genet. 2000;26:464–469. doi: 10.1038/82617. [DOI] [PubMed] [Google Scholar]
- 42.Huang Y, Ishiko T, Nakada S, Utsugisawa T, Kato T, Yuan ZM. Role for E2F in DNA damage-induced entry of cells into S phase. Cancer Res. 1997;57:3640–3643. [PubMed] [Google Scholar]
- 43.Blattner C, Sparks A, Lane D. Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Mol Cell Biol. 1999;19:3704–3713. doi: 10.1128/mcb.19.5.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meng RD, Phillips P, El-Deiry WS. p53-independent increase in E2F-1 expression enhances the cytotoxic effects of etoposide and of adriamycin. Int J Oncol. 1999;14:5–14. [PubMed] [Google Scholar]
- 45.Stevens C, Smith L, La Thangue NB. Chk2 activates E2F-1 in response to DNA damage. Nat Cell Biol. 2003;5:401–409. doi: 10.1038/ncb974. [DOI] [PubMed] [Google Scholar]
- 46.Martinez-Balbas MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. Embo J. 2000;19:662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galbiati L, Mendoza-Maldonado R, Gutierrez MI, Giacca M. Regulation of E2F-1 after DNA damage by p300-mediated acetylation and ubiquitination. Cell Cycle. 2005;4:930–939. doi: 10.4161/cc.4.7.1784. [DOI] [PubMed] [Google Scholar]
- 48.Ianari A, Gallo R, Palma M, Alesse E, Gulino A. Specific role for p300/CREB-binding protein-associated factor activity in E2F1 stabilization in response to DNA damage. J Biol Chem. 2004;279:30830–30835. doi: 10.1074/jbc.M402403200. [DOI] [PubMed] [Google Scholar]
- 49••.Pediconi N, Ianari A, Costanzo A, Belloni L, Gallo R, Cimino L, Porcellini A, Screpanti I, Balsano C, Alesse E, et al. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat Cell Biol. 2003;5:552–558. doi: 10.1038/ncb998. This study investigated the mechanism of E2F-regulation of pro-apoptotic versus cell cycle-regulated E2F target genes in response to DNA damage. E2F1 was required for the activation of the apoptotic targets p73 and Apaf1, while E2F4 enforced repression of classic E2F targets. Importantly, acetylation of E2F1 by PCAF was required to induce p73 activation by E2F1 after genotoxic stress. [DOI] [PubMed] [Google Scholar]
- 50.Rogoff HA, Pickering MT, Debatis ME, Jones S, Kowalik TF. E2F1 induces phosphorylation of p53 that is coincident with p53 accumulation and apoptosis. Mol Cell Biol. 2002;22:5308–5318. doi: 10.1128/MCB.22.15.5308-5318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rogoff HA, Pickering MT, Frame FM, Debatis ME, Sanchez Y, Jones S, Kowalik TF. Apoptosis associated with deregulated E2F activity is dependent on E2F1 and Atm/Nbs1/Chk2. Mol Cell Biol. 2004;24:2968–2977. doi: 10.1128/MCB.24.7.2968-2977.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hemann MT, Narita M. Oncogenes and senescence: breaking down in the fast lane. Genes Dev. 2007;21:1–5. doi: 10.1101/gad.1514207. [DOI] [PubMed] [Google Scholar]
- 53.Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54•.Hallstrom TC, Nevins JR. Jab1 is a specificity factor for E2F1-induced apoptosis. Genes Dev. 2006;20:613–623. doi: 10.1101/gad.1345006. This study characterized Jab1 as a specific binding partner for E2F1 that modulates its pro-apoptotic activities. Jab1 represents the first trans-acting factor able to alter E2F target gene specificity towards pro-apoptotic genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hallstrom TC, Nevins JR. Specificity in the activation and control of transcription factor E2F-dependent apoptosis. Proc Natl Acad Sci U S A. 2003;100:10848–10853. doi: 10.1073/pnas.1831408100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56••.Dick FA, Dyson N. pRB contains an E2F1-specific binding domain that allows E2F1-induced apoptosis to be regulated separately from other E2F activities. Mol Cell. 2003;12:639–649. doi: 10.1016/s1097-2765(03)00344-7. By utilizing extensive site-directed mutagenesis with the intent of disrupting the pRB-E2F interaction, the authors identified a second interaction domain on each protein, which is specific for the pRB-E2F1 interaction. Strikingly, interaction of pRB with E2F1 via this domain specifically regulated E2F1 regulation of pro-apoptotic targets, having little effect on its general transcription activity. This finding confirms that pRB is a major regulator of the pro-apoptotic activity of E2F. [DOI] [PubMed] [Google Scholar]
- 57.Markham D, Munro S, Soloway J, O’Connor DP, La Thangue NB. DNA-damage-responsive acetylation of pRb regulates binding to E2F-1. EMBO Rep. 2006;7:192–198. doi: 10.1038/sj.embor.7400591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Inoue Y, Kitagawa M, Taya Y. Phosphorylation of pRB at Ser612 by Chk1/2 leads to a complex between pRB and E2F-1 after DNA damage. Embo J. 2007;26:2083–2093. doi: 10.1038/sj.emboj.7601652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu K, Lin FT, Ruppert JM, Lin WC. Regulation of E2F1 by BRCT domain-containing protein TopBP1. Mol Cell Biol. 2003;23:3287–3304. doi: 10.1128/MCB.23.9.3287-3304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu K, Luo Y, Lin FT, Lin WC. TopBP1 recruits Brg1/Brm to repress E2F1-induced apoptosis, a novel pRb-independent and E2F1-specific control for cell survival. Genes Dev. 2004;18:673–686. doi: 10.1101/gad.1180204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61•.Wang C, Chen L, Hou X, Li Z, Kabra N, Ma Y, Nemoto S, Finkel T, Gu W, Cress WD, et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;8:1025–1031. doi: 10.1038/ncb1468. This study characterized the regulation of the SirT1 gene by E2F1. DNA damage activated E2F1 to induce SirT1 transcription. Subsequently, SirT1 binding to E2F1 suppresses its ability to activate transcription. Since SirT1-E2F1 complexes could be detected at E2F-responsive promoters, this inhibition may occur via direct repression of E2F1 transcriptional activation. Also, deacetylation of E2F1 by SirT1 could affect its activity, particularly its ability to activate pro-apoptotic gene transcription. [DOI] [PubMed] [Google Scholar]
- 62.Hauck L, Kaba RG, Lipp M, Dietz R, von Harsdorf R. Regulation of E2F1-dependent gene transcription and apoptosis by the ETS-related transcription factor GABPgamma1. Mol Cell Biol. 2002;22:2147–2158. doi: 10.1128/MCB.22.7.2147-2158.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morris EJ, Michaud WA, Ji JY, Moon NS, Rocco JW, Dyson NJ. Functional identification of Api5 as a suppressor of E2F-dependent apoptosis in vivo. PLoS Genet. 2006;2:e196. doi: 10.1371/journal.pgen.0020196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aslanian A, Iaquinta PJ, Verona R, Lees JA. Repression of the Arf tumor suppressor by E2F3 is required for normal cell cycle kinetics. Genes Dev. 2004;18:1413–1422. doi: 10.1101/gad.1196704. [DOI] [PMC free article] [PubMed] [Google Scholar]