Abstract
Steroid hormones exert profound effects on cell growth, development, differentiation, and homeostasis. Their effects are mediated through specific intracellular steroid receptors that act via multiple mechanisms. Among others, the action mechanism starting upon 17β-estradiol (E2) binds to its receptors (ER) is considered a paradigmatic example of how steroid hormones function. Ligand-activated ER dimerizes and translocates in the nucleus where it recognizes specific hormone response elements located in or near promoter DNA regions of target genes. Behind the classical genomic mechanism shared with other steroid hormones, E2 also modulates gene expression by a second indirect mechanism that involves the interaction of ER with other transcription factors which, in turn, bind their cognate DNA elements. In this case, ER modulates the activities of transcription factors such as the activator protein (AP)-1, nuclear factor-κB (NF-κB) and stimulating protein-1 (Sp-1), by stabilizing DNA-protein complexes and/or recruiting co-activators. In addition, E2 binding to ER may also exert rapid actions that start with the activation of a variety of signal transduction pathways (e.g. ERK/MAPK, p38/MAPK, PI3K/AKT, PLC/PKC). The debate about the contribution of different ER-mediated signaling pathways to coordinate the expression of specific sets of genes is still open. This review will focus on the recent knowledge about the mechanism by which ERs regulate the expression of target genes and the emerging field of integration of membrane and nuclear receptor signaling, giving examples of the ways by which the genomic and non-genomic actions of ERs on target genes converge.
Key Words: Estrogen, estrogen receptors, genomic and non-genomic action mechanism, gene transcription
1. INTRODUCTION
The principle estrogenic hormone, 17β-estradiol (E2), synthesized by testosterone aromatization in the ovary and in other tissues, plays a central role in the control of sexual behavior and reproductive functions. At present it is well recognized that the impact of E2 in human physiology is wider than previously thought impact including the differentiation of several tissues and organs, the modulation of inflammation, and brain and cardiovascular functions as well [see 1–3].
E2 regulates human physiology via diffusion through the plasma membrane of target cells and signaling through intra-cellular hormone-specific estrogen receptors (ERs). Two distinct types of signaling can be mediated, often referred to as genomic and non-genomic or non-genotropic pathways. In the genomic pathway, estrogens bind to ERs in the nucleus, inducing a conformational change in the receptors that cause dissociation from chaperones, dimerization, and activation of the receptor transcriptional domain [4–6].
The canonical model for ER-mediated regulation of gene expression involves the direct binding of dimeric ER to DNA sequences known as estrogen response elements (EREs), which are specific, inverted palindromic sequences [7]. In addition, ER can indirectly associate with promoters through protein-protein interactions with other DNA-binding transcription factors [8–10]. In either case, interaction of ERs with E2 leads to transcriptional activation of the associated genes via recruitment of coactivators and components of the basal transcriptional machinery [11–14]. In addition to the nuclear ERs, plasma membrane-associated ERs mediate the non-genomic signaling pathway [see 15–19], which can lead both to cytoplasmic alterations and to regulation of gene expression [16, 20, 1].
Regulation of transcription by nuclear ER is more complicated than the classical paradigm would predict [5, 18]. The two nuclear ERs, ERα and ERβ, exhibit distinct transcriptional properties and can form both homodimers and heterodimers [22–24]. Recent studies point to the fact that signaling pathways modulate both ERs and some co-regula- tory molecules activities [13, 25].
To understand the connection between physiological and molecular functions of ERs, the field requires an in-depth understanding of the spectrum of genes regulated in each tissue and cell type. This review will focus on the current state of knowledge about the mechanism by which ERs regulate the expression of target genes and the emerging field of integration of membrane and nuclear receptor signaling, giving examples of the ways by which the genomic and non-genomic actions of ERs on target genes converge.
2. THE STRUCTURE OF ESTROGEN RECEPTORS
Human ERα and ERβ are encoded by different genes located on different chromosomes (locus 6q25.1 and locus 14q23-24.1, respectively) [26–29]. ERα and ERβ, like all the members of the nuclear receptor super-family, are modular proteins sharing common regions, named A/B, C, D, and E/F, as well as a high sequence homology (Fig. (1A)). These regions participate in the formation of independent but interacting functional domains. The N-terminal domain (A/B region) is involved in both inter-molecular and intra-molecular interactions as well as in the activation of gene transcription. The DNA binding domain (DBD, C region) allows ER to dimerize and to bind to the specific ERE sequence on DNA through its two “zinc finger” structures (Fig. (1B)). The hinge domain (D region) has a role in receptor dimerization and in binding to chaperone heat-shock proteins (Hsp). The ligand binding domain (LBD, E/F region, C-terminal) comprises the E2-binding domain and works, synergistically with the N-terminal domain in the regulation of gene transcription [5, 30–32].
Fig. (1).
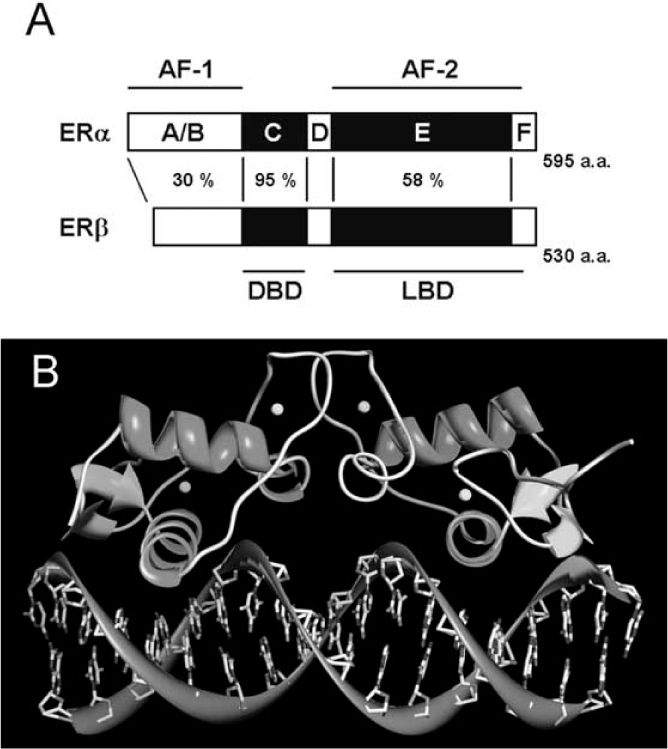
Domain organization of human ERαand ERβ (A). ERs consist of the N- terminal region involved in transactivation (A/B domains, AF-1), the DNA binding domain (DBD, C domain), the hinge region involved in dimerization (D domain), the C-terminal region containing ligand binding domain (LBD, E/F domain, AF-2) and transactivation function-2 (AF-2). The percentage indicates the homology between ERα and ERβ. (B) Binding mode of ERE to dimeric ERα (PDB ID:1HCQ) [163]. Spheres indicate the zinc atoms. For details, see text.
ERs contain two regions called activation functions (AFs) important for ligand-dependent transcriptional activity (Fig.(1A)) [5, 30–32]. AF-1 and AF-2 regions of ERs, interacting with a number of transcription co-activators, can activate with one another in a promoter- and cell-context specific manner [33].
AF-1 could be activated even in a ligand-independent manner, depending on the phosphorylation status of ER. In particular, the Ser118 residue in the AF-1 region of ERα, as well as residues Ser106 and Ser124 in the AF-1 region of ERβ, are the phosphorylation sites essential for the ligandindependent activation of ERs through the Ras-mitogen activated protein kinase (MAPK) signaling cascade [see 34,35].
Recent progress in studies on genomic and cDNA sequences has accelerated the identification of gene splice variants in the NR super-family. Numerous mRNA splice variants exist for both ERs and the best-characterized splice variants are ERα46 and ERβcx, which are frequently coexpressed with their wild-type counterparts. The exact function and potential role of these and other ERs splice variants in physiology and human disease remain to be elucidated [see 36].
3. ESTROGEN RECEPTOR GENOMIC ACTIVITY
3.1. Direct Association to DNA
The pioneering work by O’Malley and colleagues demonstrated that ERs function as ligand-activated transcription factors [37]. The trans-activation activity of ERs initiate through the ligand-bound receptor to its cognate, cis-acting enhancers, ERE [38]. The consensus palindromic element ERE was initially described based on the estrogen-respon- sive sequence in the Xenopus laevis vitellogenin A2 promoter: 5’-GGTCACAGTGACC-3’ [39–41]. This “perfect” ERE sequence was shown to function in an orientation- and distance-independent manner, both of which are properties of an enhancer [7, 42]. When ER directly interacts with the promoter/enhancer, binding to a full ERE is apparently the dominant mode of interaction. The human full EREs have a 3-bp spacer between the two half-sites, the exceptions being response elements in the human transforming growth factor (TGF)-α promoter, with a 4-bp spacer, and in the promoter of the rat luteinizing hormone β gene, with a 5-bp spacer [42]. Controversy still exists concerning ER DNA binding via ERE half sites, although a number of examples exist [43–46].
Since the identification of a canonical ERE, several computational approaches have been undertaken to identify target genes based on the presence of EREs within promoter proximal regions [47, 48]. For instance, for the 38 estrogen-responsive genes reviewed by Klinge [7], most of the functional EREs located within the promoters or 3’-untranslated regions are not the traditional consensus sequence. Thus, many target genes contain response elements that bear little similarity to consensus EREs. In one of the most comprehensive studies, Bourdeau and coworkers screened for all EREs in the human and mouse genomes and identified in excess of 70,000 EREs within the human genome, over 17,000 of which were within 15 kb of mRNA start sites [48]. Elimination of EREs that were not conserved between the human and mouse genomes reduced the number of gene proximal EREs to 660. A number of these sites were validated as genuine ER interaction sites, supporting the use of computational models to predict putative ER target genes to some degree [49].
The sequence of the response element affects the affinity that a given receptor has for binding DNA. ERα binds with the highest affinity the canonical ERE sequence found within the vitellogenin A2 gene, and less well the imperfect EREs found within the vitellogenin B1 (5’-AGTCACTGTGACC-3’) [39], pS2 (GGTCACGGTGGCC-3’) [50], and oxytocin (5’-GGTCAAGGTCACC-3’) [51] genes. This explains, at least in part, how the sequence of the response element can be one important determinant of the extent to which ERs can activate gene expression [52–55].
The conformation of transcription factors can be altered through binding to DNA [see 56]. The specific ERE sequences could exert distinct, allosteric effects on the conformation of ERα and ERβ [52, 57, 58]. Just as ligand-induced changes in ER conformation influence ER interactions with co-activators, consensus and imperfect EREs also influence the ability of ERs to bind co-activators. Note that the steroid receptor coactivator-2 (SRC-2) interacts better with ERα bound to EREs from the vitellogenin A2 than from the vitellogenin B1 gene [54].
3.2. Indirect Association to DNA
The ER signaling mechanisms discussed until now provide an explanation for the regulation of genes in which a functional ERE-like sequence can be documented within the promoter. Another category of gene promoters, lacking any ERE-like sequences, requires a second DNA-binding transcription factor to mediate ER association with the DNA [42]. This mechanism is generally referred to as “transcriptional cross-talk” [59–60]. Roughly 35% of the categorized human primary E2-responsive genes are transcripted via ER-indirect DNA association [42].
Stimulating protein-1 (Sp-1) is the predominant mediator of ER-DNA indirect binding [42] and increasing numbers of genes are found to be induced by E2 via this mechanism including the low-density lipoprotein (LDL) receptor [8], endothelial nitric oxide sinthase (eNOS) [61], c-fos [62], cyclin D1 [63], and the retinoic acid receptor-1α genes [64, 65]. In response to estrogenic stimulation, ER enhances the binding of Sp-1 to its site, containing GC-rich promoter sequences [46] and contributes to co-activator recruitment. The DNA-binding domain of ER is dispensable for such activation [42, 66, 67].
Another example is the interaction between ERα and the c-rel subunit of the nuclear factor-κ B (NF-κB) complex. This interaction prevents NF-κB from binding to and stimulating expression from the interleukin-6 (IL-6) promoter [68]. In this way, E2 inhibits expression of the cytokine IL-6 [68–70].
Other intermediary factors through which ER can associate with promoters/enhancers include: activating transcription factor (ATF)-2/c-jun or ATF-2/cAMP response element binding protein (CREB) for the cyclin D1 gene, ATF-1/CREB for the Bcl-2 gene, and nuclear transcription factorY for the mouse E2F1 gene [42].
ERs utilize protein-protein interactions also to enhance transcription of genes that contain activator protein-1 (AP-1) sites [71] related, but not identical, to those for the ATF/CREB transcription factors. The AP-1 complex, composed of Jun protein dimers and of Jun/Fos heterodimers, plays an important role in cell proliferation. Notably, ERα, activation of IGF-1 and collagenase expression is mediated through the interaction of receptor with Fos and Jun at AP-1 binding sites [42]. Collagenase, insulin-like growth factor (IGF)-1 receptor, ovalbumin, and cyclin D1 are examples of genes activated by the ERα-E2 complex via AP-1 [72, 73].
ERα and ERβ have been shown to signal in opposite ways at AP-1 sites. ER#x03B1; activates transcription in the presence of E2, whereas ERβ -E2 inhibits AP-1-dependent transcription [74, 60]. Studies show that ERβ -E2 activation of AP-1-responsive elements requires both AF-1 and AF-2 domains of the receptor, which bind and enhance the activity of the p160 components (e.g. SRC-1 and SRC-2) of the co-activator complex recruited to the site by Fos/Jun. Interestingly, human ERβ, which lacks a functional AF-1, is unable to activate transcription of AP-1-regulated genes when bound with ER agonists, indicating the possibility of distinct physiological actions of the two ERs via the regulation of unique subsets of genes [4]. Similar to AP-1, E2 binding to ERα induces transcriptional activation when associated with Sp-1 in GC-rich regions. However, E2 interaction with ERβ does not result in the formation of a transcriptionally active complex at a promoter containing Sp-1 elements. As an example ERα and ERβ, in the presence of E2, oppose each other’s function in the regulation of the cyclin D1 promoter [75]. There is considerable evidence that cyclin D1, important for progression of cells through the G1 phase of the cell cycle, is a well-defined target for ERβ-E2 action in mammary carcinoma cells [76–78], although no detectable “perfect” or ERE-like sequence in the cyclin D1 gene promoter has been reported [79]. Deletion of AP-1 and Sp-1 responsive element motifs in the cyclin D1 gene promoter resulted in attenuation of promoter responsiveness to E2 [72, 80]. Unlike ERα, E2-bound ERβ represses cyclin D1 expression [81] and blocks ERα-E2-mediated induction when both receptor isoforms are present [22]. Consequently, these differences in transcriptional activity between the ERα and ERβ may account for the major differences in their tissue specific biologic actions. This complexity is further enhanced by the presence of different ERβ splicing forms, by the ability of ERs to form homodimers and heterodimers, and by their capacity to interact with different co-regulators [82].
3.3. Transcriptional Co-Factors
Both in the direct and indirect action modes, the ligand-activated ERs are not the transcription controllers. In fact, ERs need to interact with co-regulatory proteins (co-activators or co-repressors) to form a platform upon which additional proteins are assembled [12, 13]. Cofactors interact with ERs through their Leu rich motif (i.e., Leu-Xxx-Xxx-Leu-Leu, where Xxx is any amino acid). Several classes of ER cofactors have been identified. The first identified and well-characterized co-activator family consists of three related members SRC-1, which is the founding member of the family, SRC-2, and SRC-3 [see 83]. A large co-activator complex, referred to as thyroid hormone receptor associated protein/vitamin D receptor-integrating protein (TRAP/DRIP) complex, could connect ERs directly to the basal transcription machinery via its intrinsic chromatin remodeling functions. In addition, histone acetyl transferase (e.g., CBP/p300), histone methyl transferase (e.g., CARM1 and PRMT1) and the nucleosome remodeling complexes (e.g., SWI/SNF) are necessary to release the chromatin-dependent inhibition of gene transcription [13].
Although there are far fewer nuclear receptor co-repre- ssors, these (macro)molecules serve important roles in negatively regulating ER-dependent gene expression. Two AF-2 interacting proteins, receptor-interacting protein-140 and short hetero-dimer partner, exhibit negative co-regulatory functions because they can antagonize SRC-1 co-activators in vivo and compete for AF-2 binding in vitro [84–86]. On the other hand, ERs could also associate with specific transcriptional repressors such as the nuclear receptor corepres-sor and specific histone deacetylase complexes[13].
The relative expression of co-activators and co-repre- ssors, within a cell, influences the ability of ER ligands (e.g., E2 and selective ER modulators (SERMs)) to regulate gene expression [2, 13, 87].
Because of the homology in their AF-2 domains (see Fig. (1A)), ERα and ERβ should be similar in co-activator recruitment, but certain differences have been reported. For E2-bound receptors, ERβ, but not ERα, binds well to the receptor-interacting component of the mammalian mediator complex, TRAP220. There are differences between the relative affinities of ERα and ERβ for members of the p160 co-activator family [13, 88]. More pronounced differences are observed in the case of SERM-bound ERs [see 2, 13]. For ERE-dependent gene expression, the SERM 4-hydroxy- tamoxifen is a partial agonist of ERα, but is generally unable to stimulate ERα transcriptional activity [89–91]. Conversely, when assessing ER activity on AP-1 containing reporter genes, 4-hydroxytamoxifen will stimulate ERα and ERβ transcriptional activity in a cell-dependent fashion [74].
A mechanism for shuttling off transcription involves the covalent post-translational modification of ERs and co-activators (e.g., lysine acetylation and arginine methylation), which can inhibit the binding of co-activators to nuclear receptors or other transcriptional activators by altering critical protein-protein interaction surface [see 13, 92]. Thus, the acetylation of SRC-3 by p300 has been shown to cause a disruption of receptor-co-activator complexes, leading to a decrease in receptor-mediated gene activation [see 92]. Using a variety of biochemical and cell-based assays, Krauss and co-workers have shown ERα, but not ERβ, is a target for acetylation by p300 and have identified acetylation as modulator of the ligand-dependent gene regulatory activity of ERα [93]. A number of cellular signaling pathways also influence the ER-dependent gene expression modulating ER conformational changes or co-regulators recruitment [5, 13]. It has been recognized only recently that both co-activators and co-repressors are also substrates for kinases, their phosphoryla-tion affects their ability to interact with steroid receptors [94, 95].
4. ESTROGEN RECEPTOR NON-GENOMIC ACTIVITY
The “genomic action” of steroid hormones occurs after a time-lag of at least 2 hours after E2 stimulation and explains some of hormone functions in physiological and pathological situations [see 96, 97]. This picture was challenged when a physiological dose of E2 was reported to increase the uterine cAMP level in ovariectomized rats within 15 seconds [98], an effect too rapid to be accounted for genomic action(s). This event was not abrogated by transcriptional inhibitors and was termed “rapid or non-genomic”. Actually the term “non-genomic” is not adequate when referring to rapid changes that may also initiate new gene transcription [see 96, 99].
Various signaling pathways are activated upon E2 binding to ERs. These rapid events may be classified into four main signaling cascade: phospholipase C (PLC)/protein kinase C (PKCs) [100–106], Ras/Raf/MAPK [72, 107–113], phosphatidyl inositol 3 kinase (PI3K)/AKT [15, 16, 80, 81, 97, 114–118], and cAMP/ protein kinase A (PKA) [104, 119–123].
These pathways present numerous interactions with several other pathways. The ERα-E2 complex interacts with the IGF-1 receptor, leading to IGF-1 receptor activation and hence to MAPK signaling pathway activation [124]. In addition, the ERα-E2 complex activates the EGF receptor by a mechanism that involves activation of guanine nucleotide exchange proteins (G-proteins), Src, and matrix metallopro-teinases, leading to an increase in extracellular regulated kinases (ERK) and PI3K/AKT activities [109, 125–129]. In endothelial cells the Src/PI3K/AKT pathway mediates rapid E2-dependent activation of eNOS and the release of nitric oxide. AKT and PKC could also modulate the MAPK pathway through Raf phosphorylation [97, 116, 130, 131].
It is important to note that activation of signaling pathways by E2 is cell type-specific. Indeed, the effect of E2 on PKC activity has been observed in the preoptic area of female rat brain slices, but not in the hypothalamus or cortex [132]. The activation of G-protein/Src/PI3K/MAPK pathway by E2 was evident in late, but not early, differentiated rat pre-adipocytes [109]. The differential requirement of Src/ PI3K or intracellular calcium for MAPK activation is also observed in diverse cell types [15, 109, 129]. Different PKC isoforms are rapidly activated by E2 in HepG2 and MCF7 cells [102]. As a whole, these studies indicate that the rapid actions of E2 depend on a number of conditions such as the set of signal transduction molecules and downstream targets present in the target cell, thus the responses are likely to be diverse.
All these results point to the concept that ERα is the primary endogenous mediator of rapid E2 actions. Less information is available on the role played by the ERβ-E2 complex to activate rapid non-genomic mechanisms. A subpopu-lation of ERβ transfected into Chinese Hamster ovary cells is capable of activating inositol tris-phosphate production, ERK and JNK phosphorylation [133]. Geraldes and coworkers reported that E2 reduces ERK activity through ERβ stimulation in porcine smooth muscle cells [134]. We have recently reported the ability of the ERβ-E2 complex to activate the p38 member of MAPK family, but not ERK or AKT, in human colon cancer cells [81, 135]. Although the scarce information does not allow a complete discussion on the contribution of ERβ in E2-induced rapid signals, these data indicate that also ERβ could originate cell-specific signal transduction cascade.
The rapidity by which E2 induces rapid signals as well as the localization of signaling complex raises the requirement of a plasma membrane ER. Debate continues over whether structural changes target nuclear ERs in separate pools localizing them to the membrane [61, 97, 99, 117, 136], or whether membrane ER represents a novel receptor [137–142]. Besides these data, much evidence favors the idea that the membrane-localized ER is the same protein as the nuclear-localized receptor [72, 80, 133, 143, 144]. Even if the definitive proof that membrane and nuclear ER are the same protein requires isolation and “sequencing” of the two receptor pools, ERα and ERβ must be considered a population of protein(s) which localization in the cell is able to dynamically change, shuttling from membrane to cytosol and to the nucleus, depending on ligand binding [87,97, 135,145].
Current evidence indicates that a small population of ERα and ERβ localize at the plasma membrane exists within caveolar rafts. It is at the plasma membrane that E2-liganded ER associates with the scaffolding protein caveolin-1 and a variety of signal transduction cascade activation occurs [e.g., PLC, PKC, ERK, PI3K, and nitric oxide synthase (NOS)]. ERs do not contain a trans-membrane domain [15, 18], thus the ability of ERα and ERβ to associate with the plasma membrane could be due to its association with membrane proteins and/or by post-translational addition of lipids to ERα [16, 146].
Fatty acids and isoprenoids are two of the most common lipid moieties found on post-translational modified proteins bound to membranes. No consensus sequences for N-acyla- tion (i.e., miristoylation) or S-prenylation have been found in ERα and ERβ [147]. On the contrary, S-acylation (i.e., palmitoylation) does not require any consensus sequence, but just reactive Cys residues [148].
Cys residues present in the ERα and ERβ LBD could undergo S-acylation. In particular, the amino acid sequence encompassing the Cys447 residue of ERα and Cys399 of ERβ is highly homologous to that surrounding the S-palmitoylated Cys132 residue of human caveolin-1 [147]. Based on this observation we demonstrated that ERα undergo S-palmitoylation which represents the major determinant for its residence at the plasma membrane and in its association with caveolin-1 [146, 147]. It is noteworthy that ERβ is also a palmitoylable protein [Marino M., unpublished results].
Because ERα has no intrinsic kinase domains the localization of ERs at the plasma membrane facilitate the association between ER and signaling proteins allowing the activation of rapid events. Src, Shc, proline-, glutamic acid-, leucine- rich protein /modulator of non-genomic activity of estrogen receptor (PELP1/MNAR), the p85α subunit of PI3K, receptor tyrosine kinases (i.e., EGF and IGF-1 receptors), as well as G-protein isoforms (i.e., Gαs and Gαq) have all been reported to serve as components of large complexes of interacting proteins. Through the mediation of these molecules, E2 activates the MAPK and PI3K/AKT pathways [16, 136,149–151].
Although the list of signaling and adapter proteins interacting with ER is growing, protein-protein complex formation occurs only 5 to 15 min after E2 stimulation [152]. Thus, the conformational changes of the ER LBD domain, which follows E2 entry into the cell, seems to be important in allowing the ER-E2 complex to detach from the membrane and allocate with growth factor receptors or adapter proteins to activate downstream signals.
4.1. Cell Functions Regulated by Non-Genomic Signals
The rapid activities of ERs are widely accepted and disagreement on the involvement of nuclear receptors is quite settled. However, other controversies in this field are still present and related to whether or not all of these rapid effects are of physiological relevance [153]. The main difficulties are linked to the experimental models used. In fact, the study of signaling pathways can be done mainly on isolated, often immortalized, cells and it is very complicated to obtain similar information on a whole organism in which the use of signaling inhibitors could have many side effects other than to inhibit just one kinase.
Nevertheless, the physiological significance of rapid membrane-starting pathways has been clarified at least for some E2 targets. In the nervous system, E2 affects neural functions (e.g., cognition, behavior, stress responses, and reproduction) in part by inducing such rapid responses [96]. In the skeleton, ERα, present in caveolae of bone-forming osteoblasts, transmits survival signals through activation of the Src/Shc/ERK pathway and prolongs the life span of os-teoblasts [21]. At the same time, E2 delivers a pro-apoptotic signal to bone-resorbing osteoclasts, shortening their life span [21]. Although these studies have been done mainly in cell-culture systems, their results suggest that ER rapid signaling actions have also a role in vivo. In the liver, rapid E2-induced signals (i.e., PLC/PKC) are deeply linked to the expression of the LDL receptor and to a decreased level of serum LDL-cholesterol [103]. Finally, vascular protection by E2 in ischemia/reperfusion injury in vivo requires E2-induced activation of endothelial NOS, as mediated by the PI3K/AKT pathway [117, 130].
The mechanism(s) by which E2 exerts proliferative effects is assumed to be exclusively mediated by rapid membrane-starting actions [72, 80, 101, 102, 114, 115]. E2 treatment of mammary-derived MCF-7 cells triggers the association of ERα with Src and p85α leading to DNA synthesis [115]. In HepG2 cells multiple and parallel membrane-starting pathways are rapidly activated by the ERα-E2 complex [72, 80, 101] and the blockade of PLC/PKC, ERK, and PI3K/AKT pathways completely prevents the E2-induced DNA synthesis [72, 80]. ERK/MAPK and PI3K/AKT pathways, rapidly activated by the ERα-E2 complex, also have a critical role in E2 action as a survival agent. In fact, these pathways enhance the expression of the anti-apoptotic protein Bcl-2, block the activation of the p38/MAPK, reduce the pro-apoptotic caspase-3 activation, and promote G1-to-S phase transition via the enhancement of the cyclin D1 expression [72, 80, 81].
What is the contribution of ERβ to E2-induced cell proliferation? ERβ appears to act as a dominant regulator in E2 signaling, and when co-expressed with ERα it causes a concentration-dependent reduction of ERα-mediated transcriptional activation [22] and the repression of ERα-mediated effects including cell proliferation. Consistent with this notion, E2 increases cell proliferation and causes tumor formation in MCF-7 cells expressing only ERα [22]. On the other hand, ERβ inhibits the E2-induced proliferation of trans-fected MCF-7 cells and prevents tumor formation in a mouse xenograft model in response to E2 [154]. This effect is linked to the ERβ repressive effect on ERα-induced gene transcription by binding to other transcription factors (e.g., AP-1, Sp-1) [22]. Recently, ERβ has been reported to rapidly induce a persistent membrane-initiated activation of p38/ MAPK without any interference on survival proliferative pathways, thus impairing the activation of cell cycle components (i.e., cyclin D1 expression) [81].
5. INTEGRATION OF NUCLEAR AND EXTRA-NUCLEAR ACTION OF E2
Even though the membrane ERs and associated non-genomic actions is an area of active research, the nuclear effects of membrane ERs has not received much attention [see 16, 19]. In human vascular smooth muscle cells transiently transfected with ERα an E2-dependent and an E2-independent translocation of ERα from the membrane to the nucleus was observed. The latter was blocked by MAPK inhibitors [155]. The ability of membrane ER and/or the growth factor receptor tyrosine kinases to signal via multiple kinases to the nucleus undoubtedly impacts all aspects of cellular function.
E2-induced ERK activation up regulates AP-1 mediated genes (e.g., c-fos) [156]. This results in part from serum response factor/elk-1 stimulation by E2, and in part by recruitment of nuclear ER and co-activators to AP-1 sites on gene promoters [16]. Other targets include several members of the signal transducer and activators of transcription (STAT) family such as STAT1, STAT3 and STAT5. In en-dothelial cells, activation of both STAT3 and STAT5 by E2 was mediated through signaling pathways involving MAPK, PI3K and Src and it functions to regulate β -casein expression [15].
Similarly, PI3K activation by E2-induced signaling from the membrane ER rapidly up regulates hundreds of genes in a target cell [157]. Microarray analysis of gene expression in vascular endothelial cells showed that about 250 genes were up-regulated 40 min after treatment. This effect could be prevented by the PI3K inhibitor, LY294,002 [157]. Interestingly, the transcriptional activity of the ERα-E2 complex is inhibited by a pre-treatment with the ERK inhibitors PD98,059 and U0126 [20, 114]. This suggests that stimulation of some gene expression (i.e., cyclin D1 and prolactin) by E2 occurs through ERK and PI3K activation.
CREB is the most studied of the several transcription factors rapidly activated by E2. In a hippocampal cell line [158], adipocyte cells [109], and colonic carcinoma cells [159], CREB transcriptional activity can be induced by E2 or E2-BSA through MAPK pathway, independently from the PKA pathway. Such activation of CREB induces expression of several genes (e.g., c-fos, uncoupling protein-2). In contrast, in neuroblastoma cells activation of CREB by ER-mediated rapid signals is dependent on the cAMP/PKA pathway, leading to neurotensin gene expression [160].
In addition, ERs are possible ER-mediated rapid signal targets. Indeed, it has been long known that E2 treatment can increase the phosphorylation state of ERs, via ERK and PI3K, the mutation of important phosphorylation sites reduces their transcription activity [19]. The rapid E2-evoked phosphorylation of ER contributes to the stimulation of ER dimerization and its nuclear translocation. As an example, the phosphorylation of ERα on Ser305 enhances cyclin D1 transcription in breast cancer. E2 also induces phosphoryla-tion of ERα in Ser118, Ser167, and Tyr537 residues through the non-genomic activation of the MAPK signaling pathway [19]. Furthermore, the Ser167 residue of ERα also can be phosphorylated in response to rapid E2-mediated PI3K/AKT activation, whereas E2-induced p38/MAPK phosphorylation of Thr311 promotes ERβ nuclear localization and interaction with specific receptor coactivators [19].
Besides these functions, the complexity of the mechanism of ER action suggests a more finely tuned control exerted by E2-induced rapid signals on cellular molecular events. In particular, the extra-nuclear signals induced by E2 occur before the appearance of nuclear effects and the cell context in which the genomic events occur will be different depending on which signal pathway is activated. Thus, the integration between these molecular events is required to obtain the complete cellular response.
The complex relationship between membrane and nuclear effects induced by E2 also involves membrane-initiating phosphorylation of co-activators recruiting these proteins to the nuclear transcriptosome [13, 16]. This augments the recruitment of co-activator proteins, such as SRC-1 by ER [13]. One can envisage a carefully controlled modulation of nuclear ER-induced transcription, depending upon which signaling pathway(s) are activated by E2 in a given cell context. It is likely that discrete signaling pathways regulate the access of co-repressors to target gene promoters, although this mechanism is not well studied. As a corollary to this, phosphorylation of co-activators at discrete motifs could be inhibitory as well.
The possible convergence of ER genomic and non-genomic activities at multiple response elements provides an extremely fine degree of control for the regulation of transcription by ERs. It has been estimated that more than 500 kinases are encoded within the human genome. The ability of ER-E2 membrane starting signals and/or growth factor receptor to signal through multiple cascades to the nucleus, undoubtedly has an impact on all aspects of cellular function, contributing to E2-induced cell proliferation and survival, all essential features of cell physiology as well as of tumor biology [16].
Examples of such fine-tuned ER multiple control action are cyclin D1 and vascular endothelial growth factor (VEGF) genes. Cyclin D1, a well-defined target for E2 in mammary gland, is important for the progression of cells throughout the G1 phase of the cell cycle. The cyclin D1 promoter is complex and contains binding sites for several transcription factors, but no ERE-like sequences have been identified [79]. It has been suggested that activation of the cyclin D1 gene transcription by E2 results from different ER activities: direct ERα/Sp-1 or ERα/AP-1 interaction [161] as well as ER-dependent non-genomic mechanisms [72, 80]. The cyclin D1 promoter also contains binding sites for STAT5 and NF-κB, and these could be targets for ERs through both genomic and non-genomic actions [15]. The VEGF gene is another example of cross-talk between ERs non-genomic and genomic action. In fact, VEGF gene promoter contains both an ERE-like variant and GC-rich sequences that bind ER and ER-Sp-1 complex [42]. Both must be occupied for the E2 maximal activation [15].
As a whole, these data strongly suggest that E2-induced rapid signaling reaches to the nucleus through these and other, undiscovered, pathways and synergize each other to provide plasticity for cell response to sex steroids (see Fig. (2)).
Fig. (2).
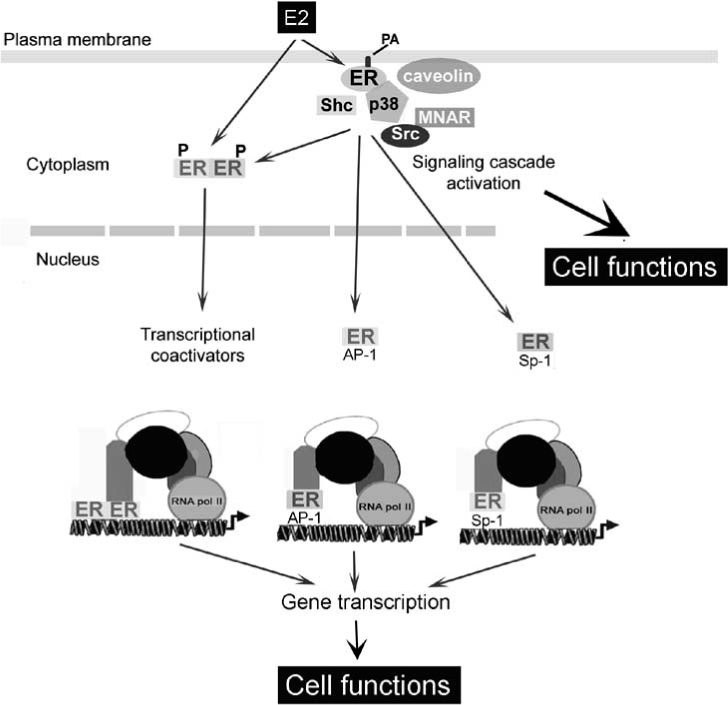
Schematic model illustrating the relationship between rapid, intermediate, and long term actions of E2 on target cells. Palmitoyla-tion (PA) allows the estrogen receptor (ER) localization at the plasma membrane. 17β -estradiol (E2) binding induces ER relocalization, association to signaling proteins, and triggers the activation of signaling cascades. The kinase activations phosphory-late ER, modulate transcriptional coactivators recruitment, and enhance AP-1 and Sp-1 activation. After dimerization ERs directly interact with ERE on DNA. ERs-DNA indirect association occurs through protein-protein interactions with the Sp-1 and AP-1 transcription factors. AP-1, activating protein-1; MNAR, modulator of non-genomic activity of ERα ; PA, palmitic acid; Sp-1, stimulating factor-1. For details, see text.
6. CONCLUSIONS
The regulation of gene expression by E2 is a multi-factorial process, involving both genomic and non-genomic actions that converge at certain response elements located in the promoters of target genes. The final gene responses, however, could depend on a number of conditions such as the combination of transcription factors bound to a specific gene promoter, the cellular localization of ERs, the levels of various co-regulator proteins and signal transduction components, as well as the nature of extra-cellular stimuli. These variables are highly specific for cell types. Thus, E2 could use different signaling pathways depending both on the cellular type and on the physiological status of the cell. In this way E2 evokes distinct gene responses in different types of target cells [15, 16, 97, 162].
The possibility that E2 could act on ER pools localized in different cell compartments (i.e., membrane versus cytoso-lic) gives rise to questioning the ability of these different ER pools to send parallel or synergic signals to the nucleus. For example, it has been observed that a naturally occurring variant of the metastatic tumor antigen 1 sequesters ER in the cytoplasm of breast cancer cells. The result of this cyto-solic retention is the reduction of E2-mediated transcription and the enhancement of E2-initiated ERK activation [136]. These data suggest that the same ER molecule is involved in genomic and in rapid signal transduction cascade. More data are needed to confirm this hypothesis and the use of dynamic imaging in the near future will help to clarify this issue.
Based upon findings highlighted in this review, one may envisage a dynamic integrated model of action for ERs inside the cell. In this model, ERs would shuttle from cell membrane to the cytoplasm and to the nucleus, in a dynamic equilibrium between different cell compartments. Each could play a different role in a multi step process of target gene activation by ER and co-activators from their upstream non-genomic to their downstream genomic responses would lead to activation of transcription (Fig. (2)).
The cell context specific environment (e.g., differentiation, ER level, and ER co-expression) has an impact on the integration of rapid signaling by E2 from the membrane and on subsequent nuclear transcription. This leads to different signal cascades, different gene expression in response to the same hormone, and different cell biological outcome.
The field is moving quickly. The challenges in the near future are to continue identifying the discrete actions of each ER intracellular pool, in order to clarify the role of ERβ, and to identify the potential cross-talk between ERs and other nuclear receptors. As we gain a deeper understanding of the complex controls exerted by ER and start identifying the critical players, it is likely that some of these putative molecules might emerge target candidates for therapeutic development in the treatment of hormone-responsive diseases, such as for different types of cancer.
ACKNOWLEDGEMENTS
Some experimental concepts described in the current paper are based on work conducted in the laboratories of the authors. These experimental studies were supported by grants from the Ministry of Education, University, and Research of Italy (PRIN-COFIN 2004 to M.M.). The Authors wish to thank past and present members of their laboratory who contributed with data and discussions to the ideas presented here.
ABBREVIATIONS
- AF
Activation function
- AP-1
Activator protein-1
- CREB
cAMP responsive element binding protein
- DBD
DNA-binding domain
- EGF
Epidermal growth factor
- ER
Estrogen receptor
- ERE
Estrogen responsive element
- ERK
Extracellular regulated kinase
- E2
17β-estradiol
- G-proteins
Guanine nucleotide exchange proteins
- Hsp
Heath shock protein
- IGF-1
Insulin-like growth factor-1
- IL-6
Interleukin-6
- LDL
Low dendity lipoprotein
- LBD
Ligand-binding domain
- MAPK
Mitogen-activated protein kinase
- MNAR
Modulator of non-genomic activity of estrogen receptor (also named Pro-, Glu-, and Leu-rich protein-1 PELP1)
- NF-κB
Nuclear factor κB
- PI3K
Phosphatidyl inositol 3-kinase
- NOS
Nitric oxide synthase
- PKA
Protein kinase A
- PKC
Protein kinase C
- PLC
Phospholipase C
- SERMs
Selective estrogen modulators
- Sp-1
Stimulating protein-1
- SRC
Steroid receptor co-activator family
- STAT
Signal transducers and activators of transcription
REFERENCES
- 1.Gruber CJ, Tschugguel W, Schneeberger C, Huber JC. Production and actions of estrogens. New Engl J Med. 2002;346:340–352. doi: 10.1056/NEJMra000471. [DOI] [PubMed] [Google Scholar]
- 2.Pearce ST, Jordan VC. The biological role of estrogen receptors α and β in cancer. Crit Rev Oncol Hematol. 2004;50:3–22. doi: 10.1016/j.critrevonc.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Deroo BJ, Korach KS. Estrogen receptors and human disease. J Clin Invest. 2006;116:561–570. doi: 10.1172/JCI27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall JM, Couse JF, Korach KS. The multifaceted mechanisms of estradiol and estrogen receptor signaling. J Biol Chem. 2001;276:36869–36872. doi: 10.1074/jbc.R100029200. [DOI] [PubMed] [Google Scholar]
- 5.Nilsson S, Makela S, Treuter E, Tujague M, Thomsen J, Andersson G, Enmark E, Pettersson K, Warner M, Gustafsson JÅ. Mechanisms of estrogen action. Physiol Rev. 2001;81:1535–1565. doi: 10.1152/physrev.2001.81.4.1535. [DOI] [PubMed] [Google Scholar]
- 6.Hall JM, McDonnell DP. Coregulators in nuclear estrogen receptor action: from concept to therapeutic targeting. Mol Interv. 2005;5:343–357. doi: 10.1124/mi.5.6.7. [DOI] [PubMed] [Google Scholar]
- 7.Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001;29:2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li C, Briggs MR, Ahlborn TE, Kraemer FB, Liu J. Requirement of Sp1 and estrogen receptor-interaction in 17β-estradiol-mediated transcriptional activation of the low density lipoprotein receptor gene expression. Endocrinology. 2001;142:1546–1553. doi: 10.1210/endo.142.4.8096. [DOI] [PubMed] [Google Scholar]
- 9.Safe S. Transcriptional activation of genes by 17β-estradiol through estrogen receptor-Sp1 interactions. Vitam Horm. 2001;62:231–252. doi: 10.1016/s0083-6729(01)62006-5. [DOI] [PubMed] [Google Scholar]
- 10.Stossi F, Likhite VS, Katzenellenbogen JA, Katzenellenbogen BS. Estrogen-occupied estrogen receptor represses cyclin G2 gene expression and recruits a repressor complex at the cyclin G2 promoter. J Biol Chem. 2006;281:16272–16278. doi: 10.1074/jbc.M513405200. [DOI] [PubMed] [Google Scholar]
- 11.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 12.McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 13.Smith CL, O'Malley BW. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- 14.Kato S, Sato T, Watanabe T, Takemasa S, Masuhiro Y, Ohtake F, Matsumoto T. Function of nuclear sex hormone receptors in gene regulation. Cancer Chemother Pharmacol. 2005;56(Suppl.1):4–9. doi: 10.1007/s00280-005-0102-8. [DOI] [PubMed] [Google Scholar]
- 15.Björnström L, Sjöberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol. 2005;19:833–842. doi: 10.1210/me.2004-0486. [DOI] [PubMed] [Google Scholar]
- 16.Levin ER. Integration of the extra-nuclear and nuclear actions of estrogen. Mol Endocrinol. 2005;19:1951–1959. doi: 10.1210/me.2004-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marino M, Ascenzi P, Acconcia F. S-palmitoylation modulates estrogen receptor alpha localization and functions. Steroids. 2006;71:298–303. doi: 10.1016/j.steroids.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 18.Ascenzi P, Bocedi A, Marino M. Structure-function relationship of estrogen receptor α and β: impact on human health. Mol Aspects Med. 2006;27:299–402. doi: 10.1016/j.mam.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Zhang D, Trudeau VL. Integration of membrane and nuclear estrogen receptor signaling. Comp Biochem Physiol A Mol Integr Physiol. 2006;144:306–315. doi: 10.1016/j.cbpa.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 20.Acconcia F, Marino M. Synergism between genomic and non-genomic estrogen action mechanisms. IUBMB Life. 2003;55:145–150. doi: 10.1080/1521654031000110172. [DOI] [PubMed] [Google Scholar]
- 21.Kousteni S, Han L, Chen JR, Almeida M, Plotkin LI, Bellido T, Manolagas SC. Kinase-mediated regulation of common transcription factors accounts for the bone-protective effects of sex steroids. J Clin Invest. 2003;111:1651–1664. doi: 10.1172/JCI17261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matthews J, Gustafsson JÅ. Estrogen signalling: A subtle balance between ERα and ERβ. Mol Interventions. 2003;3:281–292. doi: 10.1124/mi.3.5.281. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Huang J, Yi P, Bambara RA, Hilf R, Muyan M. Single-chain estrogen receptors (ERs) reveal that the ERα/β heterodimer emulates functions of the ERα dimer in genomic estrogen signaling pathways. Mol Cell Biol. 2004;24:7681–7694. doi: 10.1128/MCB.24.17.7681-7694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monroe DG, Secreto FJ, Subramaniam M, Getz BJ, Khosla S, Spelsberg TC. Estrogen receptor α and β heterodimers exert unique effects on estrogen-and tamoxifen-dependent gene expression in human U2OS osteosarcoma cells. Mol Endocrinol. 2005;19:1555–1568. doi: 10.1210/me.2004-0381. [DOI] [PubMed] [Google Scholar]
- 25.Mendelsohn ME. Genomic and nongenomic effects of estrogen in the vasculature. Am J Cardiol. 2002;90:3F–6F. doi: 10.1016/s0002-9149(02)02418-9. [DOI] [PubMed] [Google Scholar]
- 26.Gosden JR, Middleton PG, Rout D. Localization of the human oestrogen receptor gene to chromosome 6q24-q27 by in situ hybridization. Cytogenet Cell Genet. 1986;43:218–220. doi: 10.1159/000132325. [DOI] [PubMed] [Google Scholar]
- 27.Enmark E, Pelto-Huikko M, Grandien K, Lagercrantz S, Lagercrantz J, Fried G, Nordenskjold M, Gustafsson JÅ. Human estrogen receptor β-gene structure, chromosomal localization, and expression pattern. J Clin Endocrinol Metab. 1997;82:4258–4265. doi: 10.1210/jcem.82.12.4470. [DOI] [PubMed] [Google Scholar]
- 28.Luisi S, Galleri L, Marini F, Ambrosiani G, Brandi ML, Petraglia F. Estrogen receptor gene polymorphisms are associated with recurrence of endometriosis. Fertil Steril. 2006;85:764–766. doi: 10.1016/j.fertnstert.2005.08.038. [DOI] [PubMed] [Google Scholar]
- 29.Zhou W, Liu Z, Wu J, Liu JH, Hyder SM, Antoniou E, Lubahn DB. Identification and characterization of two novel splicing isoforms of human estrogen-related receptor β. J Clin Endocrinol Metab. 2006;91:569–579. doi: 10.1210/jc.2004-1957. [DOI] [PubMed] [Google Scholar]
- 30.Mosselman S, Polman J, Dijkema R. ERβ: identification and characterization of a novel human estrogen receptor. FEBS Lett. 1996;392:49–53. doi: 10.1016/0014-5793(96)00782-x. [DOI] [PubMed] [Google Scholar]
- 31.Claessens F, Gewirth DT. DNA recognition by nuclear receptors. In: McEwan IJ, editor. Essay in Biochemistry. London: The Nuclear Receptor Superfamily Portland Press; 2004. pp. 59–72. [DOI] [PubMed] [Google Scholar]
- 32.Kumar R, Johnson BH, Thompson EB. Overview of the structural basis for transcription regulation by nuclear hormone receptors. In: McEwan IJ, editor. Essay in Biochemistry. London: The Nuclear Receptor Superfamily Portland Press; 2004. pp. 27–39. [DOI] [PubMed] [Google Scholar]
- 33.McEwan IJ. Sex, drugs and gene expression: signalling by members of the nuclear receptor superfamily. In: McEwan IJ, editor. Essays in Biochemistry: the Nuclear Receptor Superfamily. London: Portland Press; 2004. pp. 1–10. [DOI] [PubMed] [Google Scholar]
- 34.Ortì E, Bodwell JE, Munck A. Phosphorylation of steroid hormone receptors. Endocr Rev. 1992;13:105–128. doi: 10.1210/edrv-13-1-105. [DOI] [PubMed] [Google Scholar]
- 35.Lannigan DA. Estrogen receptor phosphorylation. Steroids. 2003;68:1–9. doi: 10.1016/s0039-128x(02)00110-1. [DOI] [PubMed] [Google Scholar]
- 36.Herynk MH, Fuqua SA. Estrogen receptor mutations in human disease. Endocr Rev. 2004;25:869–898. doi: 10.1210/er.2003-0010. [DOI] [PubMed] [Google Scholar]
- 37.O'Malley BW. A life-long search for the molecular pathways of steroid hormone action. Mol Endocrinol. 2005;19:1402–1411. doi: 10.1210/me.2004-0480. [DOI] [PubMed] [Google Scholar]
- 38.Beato M. Gene regulation by steroid hormones. Cell. 1989;56:335–344. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- 39.Walker P, Germond JE, Brown-Luedi M, Givel F, Wahli W. Sequence homologies in the region preceding the transcription initiation site of the liver estrogen-responsive vitellogenin and apo-VLDLII genes. Nucleic Acids Res. 1984;12:8611–8626. doi: 10.1093/nar/12.22.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klein-Hitpass L, Schorpp M, Wagner U, Ryffel GU. An estrogen-responsive element derived from the 5′ flanking region of the Xenopus vitellogenin A2 gene functions in transfected human cells. Cell. 1986;46:1053–1061. doi: 10.1016/0092-8674(86)90705-1. [DOI] [PubMed] [Google Scholar]
- 41.Ponglikitmongkol M, Green S, Chambon P. Genomic organization of the human oestrogen receptor gene. EMBO J. 1988;7:3385–3388. doi: 10.1002/j.1460-2075.1988.tb03211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Lone R, Frith MC, Karlsson EK, Hansen U. Genomic targets of nuclear estrogen receptors. Mol Endocrinol. 2004;18:1859–1875. doi: 10.1210/me.2003-0044. [DOI] [PubMed] [Google Scholar]
- 43.Kato S, Tora L, Yamauchi J, Masushige S, Bellard M, Chambon P. A far upstream estrogen response element of the ovalbumin gene contains several half palindromic 5′-TGACC-3′ motifs acting synergistically. Cell. 1992;68:731–742. doi: 10.1016/0092-8674(92)90148-6. [DOI] [PubMed] [Google Scholar]
- 44.Klinge CM, Bodenner DL, Desai D, Niles RM, Traish AM. Binding of type II nuclear receptors and estrogen receptor to full and half-site estrogen response elements in vitro. Nucleic Acids Res. 1997;25:1903–1912. doi: 10.1093/nar/25.10.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Porter W, Wang F, Wang W, Duan R, Safe S. Role of estrogen receptor/Sp1 complexes in estrogen-induced heat shock protein 27 gene expression. Mol Endocrinol. 1996;10:1371–1378. doi: 10.1210/mend.10.11.8923463. [DOI] [PubMed] [Google Scholar]
- 46.Porter W, Saville B, Hoivik D, Safe S. Functional synergy between the transcription factor Sp1 and the estrogen receptor. Mol Endocrinol. 1997;11:1569–1580. doi: 10.1210/mend.11.11.9916. [DOI] [PubMed] [Google Scholar]
- 47.Bajic VB, Tan SL, Chong A, Tang S, Strom A, Gustafsson JÅ, Lin CY, Liu ET. Dragon ERE Finder version 2: A tool for accurate detection and analysis of estrogen response elements in vertebrate genomes. Nucleic Acids Res. 2003;31:3605–3607. doi: 10.1093/nar/gkg517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bourdeau V, Deschenes J, Métivier R, Nagai Y, Nguyen D, Bretschneider N, Gannon F, White JH, Mader S. Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol Endocrinol. 2004;18:1411–1427. doi: 10.1210/me.2003-0441. [DOI] [PubMed] [Google Scholar]
- 49.Carroll JS, Brown M. Estrogen receptor target gene: an evolving concept in molecular endocrinology. Mol Endocrinol. 2006;20:1707–1714. doi: 10.1210/me.2005-0334. [DOI] [PubMed] [Google Scholar]
- 50.Nunez AM, Jakowlev S, Briand JP, Gaire M, Krust A, Rio MC, Chambon P. Characterization of the estrogen-induced pS2 protein secreted by the human breast cancer cell line MCF-7. Endocrinology. 1987;121:1759–1765. doi: 10.1210/endo-121-5-1759. [DOI] [PubMed] [Google Scholar]
- 51.Sausville E, Carney D, Battey J. The human vasopressin gene is linked to the oxytocin gene and is selectively expressed in a cultured lung cancer cell line. J Biol Chem. 1985;260:10236–10241. [PubMed] [Google Scholar]
- 52.Loven MA, Wood JR, Nardulli AM. Interaction of estrogen receptors α and β with estrogen response elements. Mol Cell Endocrinol. 2001;181:151–163. doi: 10.1016/s0303-7207(01)00491-9. [DOI] [PubMed] [Google Scholar]
- 53.Loven MA, Likhite VS, Choi I, Nardulli AM. Estrogen response elements alter coactivator recruitment through allosteric modulation of estrogen receptor β conformation. J Biol Chem. 2001;276:45282–45288. doi: 10.1074/jbc.M106211200. [DOI] [PubMed] [Google Scholar]
- 54.Wood JR, Likhite VS, Loven MA, Nardulli AM. Allosteric modulation of estrogen receptor conformation by different estrogen response elements. Mol Endocrinol. 2001;15:1114–1126. doi: 10.1210/mend.15.7.0671. [DOI] [PubMed] [Google Scholar]
- 55.Yi P, Driscoll MD, Huang J, Bhagat S, Hill R, Bambara RA, Muyan M. The effects of estrogen-responsive element- and ligand-induced structural changes on the recruitment of cofactors and transcriptional responses by ERα and ERβ. Mol Endocrinol. 2002;16:674–693. doi: 10.1210/mend.16.4.0810. [DOI] [PubMed] [Google Scholar]
- 56.Lefstin JA, Yamamoto KR. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- 57.Hall JM, McDonnell DP, Korach KS. Allosteric regulation of estrogen receptor structure, function, and coactivator recruitment by different estrogen response elements. Mol Endocrinol. 2002;16:469–486. doi: 10.1210/mend.16.3.0814. [DOI] [PubMed] [Google Scholar]
- 58.Ikeda M, Wilcox EC, Chin WW. Different DNA elements can modulate the conformation of thyroid hormone receptor heterodimer and its transcriptional activity. J Biol Chem. 1996;271:23096–23104. doi: 10.1074/jbc.271.38.23096. [DOI] [PubMed] [Google Scholar]
- 59.Gottlicher M, Heck S, Herrlich P. Transcriptional cross-talk, the second mode of steroid hormone receptor action. J Mol Med. 1998;76:480–489. doi: 10.1007/s001090050242. [DOI] [PubMed] [Google Scholar]
- 60.Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev. 2001;81:1269–1304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- 61.Chambliss KL, Shaul PW. Rapid activation of endothelial NO synthase by estrogen: evidence for a steroid receptor fast-action complex (SRFC) in caveolae. Steroids. 2002;67:413–419. doi: 10.1016/s0039-128x(01)00177-5. [DOI] [PubMed] [Google Scholar]
- 62.Duan R, Porter W, Safe S. Estrogen-induced c-fos protooncogene expression in MCF-7 human breast cancer cells: role of estrogen receptor Sp1 complex formation. Endocrinology. 1998;139:1981–1990. doi: 10.1210/endo.139.4.5870. [DOI] [PubMed] [Google Scholar]
- 63.Castro-Rivera E, Samudio I, Safe S. Estrogen regulation of cyclin D1 gene expression in ZR-75 breast cancer cells involves multiple enhancer elements. J Biol Chem. 2001;276:30853–30861. doi: 10.1074/jbc.M103339200. [DOI] [PubMed] [Google Scholar]
- 64.Sun G, Porter W, Safe S. Estrogen-induced retinoic acid receptor α1 gene expression: role of estrogen receptor-Sp1 complex. Mol Endocrinol. 1998;12:882–890. doi: 10.1210/mend.12.6.0125. [DOI] [PubMed] [Google Scholar]
- 65.Zhang J, Hu X, Lazar MA. A novel role for helix 12 of retinoid X receptor in regulating repression. Mol Cell Biol. 1999;19:6448–6457. doi: 10.1128/mcb.19.9.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Batistuzzo de Medeiros SR, Krey G, Hihi AK, Wahli W. Functional interaction between the estrogen receptor and the transcription activator Sp1 regulate the estrogen-dependent transcriptional activity of the vitellogenin A1 promoter. J Biol Chem. 1997;272:18250–18260. doi: 10.1074/jbc.272.29.18250. [DOI] [PubMed] [Google Scholar]
- 67.Qin C, Singh P, Safe S. Transcriptional activation of insulin-like growth factor-binding protein-4 by 17β-estradiol in MCF-7 cells: role of estrogen receptor-SP1 complexes. Endocrinology. 1999;140:2501–2508. doi: 10.1210/endo.140.6.6751. [DOI] [PubMed] [Google Scholar]
- 68.Galien R, Garcia T. Estrogen receptor impairs interleukin-6 expression by preventing protein binding on the NF-κB site. Nucleic Acids Res. 1997;25:2424–2429. doi: 10.1093/nar/25.12.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ray A, Prefontaine KE, Ray P. Down-modulation of interleukin-6 gene expression by estradiol 17β in the absence of high affinity DNA binding by the estrogen receptor. J Biol Chem. 1994;269:12940–12946. [PubMed] [Google Scholar]
- 70.Kalaitzidis D, Gilmore TD. Transcription factor cross-talk: the estrogen receptor and NF-κB. Trends Endocrinol Metab. 2005;16:46–52. doi: 10.1016/j.tem.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 71.Gaub MP, Bellard M, Scheuer I, Chambon P, Sassone-Corsi P. Activation of the ovalbumin gene by the estrogen receptor involves the fos-jun complex. Cell. 1990;3:1267–1276. doi: 10.1016/0092-8674(90)90422-b. [DOI] [PubMed] [Google Scholar]
- 72.Marino M, Acconcia F, Bresciani F, Weisz A, Trentalance A. Distinct nongenomic signal transduction pathways controlled by 17β-estradiol regulate DNA synthesis and cyclin D1 gene transcription in HepG2 cells. Mol Biol Cell. 2002;13:3720–3729. doi: 10.1091/mbc.E02-03-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fujimoto N, Honda H, Kitamura S. Effects of environmental estrogenic chemicals on AP-1 mediated transcription with estrogen receptors α and β. J Steroid Biochem Mol Biol. 2004;88:53–59. doi: 10.1016/j.jsbmb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 74.Paech K, Webb P, Kuiper GG, Nilsson S, Gustafsson J, Kushner PJ, Scanlan TS. Differential ligand activation of estrogen receptors ERα and ERβ at AP-1 sites. Science. 1997;277:1508–1510. doi: 10.1126/science.277.5331.1508. [DOI] [PubMed] [Google Scholar]
- 75.Liu MM, Albanese C, Anderson CM, Hilty K, Webb P, Uht RM, Price RH, Jr, Pestell RG, Kushner PJ. Opposing action of estrogen receptors α and β on cyclin D1 gene expression. J Biol Chem. 2002;277:24353–24360. doi: 10.1074/jbc.M201829200. [DOI] [PubMed] [Google Scholar]
- 76.Altucci L, Addeo R, Cicatiello L, Dauvois S, Parker MG, Truss M, Beato M, Sica V, Bresciani F, Weisz A. 17β-Estradiol induces cyclin D1 gene transcription, p36D1-p34cdk4 complex activation and p105Rb phosphorylation during mitogenic stimulation of G1-arrested human breast cancer cells. Oncogene. 1996;12:2315–2324. [PubMed] [Google Scholar]
- 77.Foster JS, Wimalasena J. Estrogen regulates activity of cyclin-dependent kinases and retinoblastoma protein phosphorylation in breast cancer cells. Mol Endocrinol. 1996;10:488–498. doi: 10.1210/mend.10.5.8732680. [DOI] [PubMed] [Google Scholar]
- 78.Prall OW, Sarcevic B, Musgrove EA, Watts CK, Sutherland RL. Estrogen-induced activation of Cdk4 and Cdk2 during G1-S phase progression is accompanied by increased cyclin D1 expression and decreased cyclin-dependent kinase inhibitor association with cyclin E-Cdk2. J Biol Chem. 1997;272:10882–10894. doi: 10.1074/jbc.272.16.10882. [DOI] [PubMed] [Google Scholar]
- 79.Herbert B, Truss M, Beato M, Müller R. Inducibile regulatory elements in the human cyclin D1 promoter. Oncogene. 1994;9:1295–1304. [PubMed] [Google Scholar]
- 80.Marino M, Acconcia F, Trentalance A. Biphasic estradiol induced AKT-phosphorylation is modulated by PTEN via MAP kinase in HepG2 cells. Mol Biol Cell. 2003;14:2583–2591. doi: 10.1091/mbc.E02-09-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Acconcia F, Totta P, Ogawa S, Cardillo I, Inoue S, Leone S, Trentalance A, Muramatsu M, Marino M. Survival versus apoptotic 17β-estradiol effect: role of ERα and ERβ activated non-genomic signalling. J Cell Physiol. 2005;203:193–201. doi: 10.1002/jcp.20219. [DOI] [PubMed] [Google Scholar]
- 82.Bardin A, Boulle N, Lazennec G, Vignon F, Pujol P. Loss of ERβ expression as a common step in estrogen-dependent tumor progression. Endocr Relat Cancer. 2004;11:537–551. doi: 10.1677/erc.1.00800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McKenna NJ, Lanz RB, O'Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 84.Cavailles V, Dauvois S, L'Horset F, Lopez G, Hoare S, Kushner PJ, Parker MG. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Treuter E, Johansson L, Thomsen JS, Wärnmark A, Leers J, Pelto-Huikko M, Sjöberg M, Wright AP, Spyrou G, Gustafsson JÅ. Competition between thyroid hormone receptor-associated protein (TRAP) 220 and transcriptional intermediary factor (TIF) 2 for binding to nuclear receptors. Implications for the recruitment of TRAP and p160 coactivator complexes. J Biol Chem. 1999;274:6667–6677. doi: 10.1074/jbc.274.10.6667. [DOI] [PubMed] [Google Scholar]
- 86.Johansson L, Bavner A, Thomsen JS, Farnegardh M, Gustafsson JÅ, Treuter E. The orphan nuclear receptor SHP utilizes conserved LXXLL-related motifs for interactions with ligand-activated estrogen receptors. Mol Cell Biol. 2000;20:1124–1133. doi: 10.1128/mcb.20.4.1124-1133.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Leclercq G, Lacroix M, Laios I, Laurent G. Estrogen receptor α: impact of ligands on intracellular shuttling and turnover rate in breast cancer cells. Curr Cancer Drug Targets. 2006;6:39–64. doi: 10.2174/156800906775471716. [DOI] [PubMed] [Google Scholar]
- 88.Wärnmark A, Almlöf T, Leers J, Gustafsson JÅ, Treuter E. Differential recruitment of the mammalian mediator subunit TRAP220 by estrogen receptors ERα and ERβ. J Biol Chem. 2001;276:23397–23404. doi: 10.1074/jbc.M011651200. [DOI] [PubMed] [Google Scholar]
- 89.Barkhem T, Carlsson B, Nilsson Y, Enmark E, Gustafsson JÅ, Nilsson S. Differential response of estrogen receptor α and estrogen receptor β to partial estrogen agonists/antagonists. Mol Pharmacol. 1998;54:105–112. doi: 10.1124/mol.54.1.105. [DOI] [PubMed] [Google Scholar]
- 90.McInerney EM, Weis KE, Sun J, Mosselman S, Katzenellenbogen BS. Transcription activation by the human estrogen receptor subtype β (ER β) studied with ERα and ERβ receptor chimeras. Endocrinology. 1998;139:4513–4522. doi: 10.1210/endo.139.11.6298. [DOI] [PubMed] [Google Scholar]
- 91.Hall JM, McDonnell DP. The estrogen receptor β-isoform (ERβ) of the human estrogen receptor modulates ERα transcriptional activity and is a key regulator of the cellular response to estrogens and antiestrogens. Endocrinology. 1999;140:5566–5578. doi: 10.1210/endo.140.12.7179. [DOI] [PubMed] [Google Scholar]
- 92.Acevedo ML, Kraus WL. Transcriptional activation by nuclear receptors. In: McEwan IJ, editor. Essays in Biochemistry: the Nuclear Receptor Superfamily. London: Portland Press; 2004. pp. 73–88. [DOI] [PubMed] [Google Scholar]
- 93.Kim MY, Woo EM, Chong YT, Homenko DR, Kraus WL. Acetylation of estrogen receptor alpha by p300 at lysines 266 and 268 enhances the DNA binding and transactivation activities of the receptor. Mol Endocrinol. 2006;20:1479–1493. doi: 10.1210/me.2005-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rowan BG, Weigel NL, O'Malley BW. Phosphorylation of steroid receptor coactivator-1. Identification of the phosphorylation sites and phosphorylation through the mitogen-activated protein kinase pathway. J Biol Chem. 2000;275:4475–4483. doi: 10.1074/jbc.275.6.4475. [DOI] [PubMed] [Google Scholar]
- 95.Ko L, Cardona GR, Iwasaki T, Bramlett KS, Burris TP, Chin WW. Ser-884 adjacent to the LXXLL motif of coactivator TRBP defines selectivity for ERs and TRs. Mol Endocrinol. 2002;16:128–140. doi: 10.1210/mend.16.1.0755. [DOI] [PubMed] [Google Scholar]
- 96.Farach-Carson MC, Davis PJ. Steroid hormone interactions with target cells: cross talk between membrane and nuclear pathways. J Pharmacol Exper Therap. 2003;30:839–845. doi: 10.1124/jpet.103.055038. [DOI] [PubMed] [Google Scholar]
- 97.Marino M, Acconcia F, Ascenzi P. Estrogen receptor signalling: Bases for drug actions. Curr Drug Targets - Immune, Endocrine & Metabolic Disorders. 2005;5:305–314. doi: 10.2174/1568008054863763. [DOI] [PubMed] [Google Scholar]
- 98.Szego CM, Davis JS. Adenosine 3′,5′-monophosphate in rat uterus: acute elevation by estrogen. Proc Natl Acad Sci USA. 1967;58:1711–1718. doi: 10.1073/pnas.58.4.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kampa M, Castanas E. Membrane steroid receptor signaling in normal and neoplastic cells. Mol Cell Endocrinol. 2006;246:76–82. doi: 10.1016/j.mce.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 100.Morley P, Whitfield JF, Vanderhyden BC, Tsang BK, Schwartz JL. A new, nongenomic estrogen action: the rapid release of intracellular calcium. Endocrinology. 1992;131:1305–1312. doi: 10.1210/endo.131.3.1505465. [DOI] [PubMed] [Google Scholar]
- 101.Marino M, Pallottini V, Trentalance A. Estrogens cause rapid activation of IP3-PKC-α signal transduction pathway in HEPG2 cells. Biochem Biophys Res Commun. 1998;245:254–258. doi: 10.1006/bbrc.1998.8413. [DOI] [PubMed] [Google Scholar]
- 102.Marino M, Ficca R, Ascenzi P, Trentalance A. Nitric oxide inhibits selectively the 17β-estradiol-induced gene expression without affecting nongenomic events in HeLa cells. Biochem Biophys Res Commun. 2001;286:529–533. doi: 10.1006/bbrc.2001.5433. [DOI] [PubMed] [Google Scholar]
- 103.Marino M, Distefano E, Trentalance A, Smith CL. Estradiol induced IP3 mediate the estrogen receptor activity expressed in human cells. Mol Cell Endocrinol. 2001;182:19–26. doi: 10.1016/s0303-7207(01)00556-1. [DOI] [PubMed] [Google Scholar]
- 104.Picotto G, Vazquez G, Boland R. 17β-oestradiol increases intra-cellular Ca2+ concentration in rat enterocytes. Potential role of phospholipase C-dependent store-operated Ca2+ influx. Biochem J. 1999;339:71–77. [PMC free article] [PubMed] [Google Scholar]
- 105.Perret S, Dockery P, Harvey BJ. 17β-oestradiol stimulates capacitative Ca2+ entry in human endometrial cells. Mol Cell Endocrinol. 2001;176:77–84. doi: 10.1016/s0303-7207(01)00464-6. [DOI] [PubMed] [Google Scholar]
- 106.Incerpi S, D'Arezzo S, Marino M, Musanti R, Pallottini V, Pascolini A, Trentalance A. Short-term activation by low 17β-estradiol concentrations of the Na+/H+ exchanger in rat aortic smooth muscle cells: physiopathological implications. Endocrinology. 2003;144:4315–4324. doi: 10.1210/en.2003-0495. [DOI] [PubMed] [Google Scholar]
- 107.Watters JJ, Campbell JS, Cunningham MJ, Krebs EG, Dorsa DM. Rapid membrane effects of steroids in neuroblastoma cells: effects of estrogen on mitogen activated protein kinase signalling cascade and c-fos immediate early gene transcription. Endocrinology. 1997;138:4030–4033. doi: 10.1210/endo.138.9.5489. [DOI] [PubMed] [Google Scholar]
- 108.Russell KS, Haynes MP, Sinha D, Clerisme E, Bender JR. Human vascular endothelial cells contain membrane binding sites for estradiol, which mediate rapid intracellular signaling. Proc Natl Acad Sci USA. 2000;97:5930–5935. doi: 10.1073/pnas.97.11.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dos Santos EG, Dieudonne MN, Pecquery R, Le Moal V, Giudicelli Y, Lacasa D. Rapid nongenomic E2 effects on p42/p44 MAPK, activator protein-1, and cAMP response element binding protein in rat white adipocytes. Endocrinology. 2002;143:930–940. doi: 10.1210/endo.143.3.8678. [DOI] [PubMed] [Google Scholar]
- 110.Migliaccio A, Castoria G, Di Domenico M, de Falco A, Bilancio A, Auricchio F. Src is an initial target of sex steroid hormone action. AnnNY Acad Sci. 2002;963:185–190. doi: 10.1111/j.1749-6632.2002.tb04109.x. [DOI] [PubMed] [Google Scholar]
- 111.Tanaka Y, Gavrielides MV, Mitsuuchi Y, Fujii T, Kazanietz MG. Protein kinase C promotes apoptosis in LNCaP prostate cancer cells through activation of p38 MAPK and inhibition of the Akt survival pathway. J Biol Chem. 2003;278:33753–33762. doi: 10.1074/jbc.M303313200. [DOI] [PubMed] [Google Scholar]
- 112.Klinge CM, Blankenship KA, Risinger KE, Bhatnagar S, Noisin EL, Sumanasekera WK, Zhao L, Brey DM, Keynton RS. Resveratrol and estradiol rapidly activate MAPK signaling through estrogen receptors α and β in endothelial cells. J Biol Chem. 2005;280:7460–7468. doi: 10.1074/jbc.M411565200. [DOI] [PubMed] [Google Scholar]
- 113.Woo CH, Lim JH, Kim JH. VCAM-1 upregulation via PKCδ-p38 kinase-linked cascade mediates the TNF-α-induced leukocyte adhesion and emigration in the lung airway epithelium. Am J Physiol Lung Cell Mol Physiol. 2005;288:L307–L316. doi: 10.1152/ajplung.00105.2004. [DOI] [PubMed] [Google Scholar]
- 114.Castoria G, Barone MV, Di Domenico M, Bilancio A, Ametrano D, Migliaccio A, Auricchio F. Non-trascriptional action of oestradiol and progestin triggers DNA synthesis. EMBO J. 1999;18:2500–2510. doi: 10.1093/emboj/18.9.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Castoria G, Migliaccio A, Bilancio A, Di Domenico M, de Falco A, Lombardi M, Fiorentino R, Varricchio L, Barone MV, Auricchio F. PI3-kinase in concert with Src promotes the S-phase entry of oestradiol-stimulated MCF-7 cells. EMBO J. 2001;20:6050–6059. doi: 10.1093/emboj/20.21.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chambliss KL, Simon L, Yuhanna IS, Mineo C, Shaul PW. Dissecting the basis of nongenomic activation of eNOS by estradiol: role of ERα domains with known nuclear functions. Mol Endocrinol. 2005;19:277–289. doi: 10.1210/me.2004-0008. [DOI] [PubMed] [Google Scholar]
- 117.Simoncini T, Hafezi-Moghadam A, Brazil DP, Ley K, Chin WW, Liao JK. Interaction of oestrogen receptor with the regulatory subunit of phosphatidylinositol-3-OH kinase. Nature. 2000;407:538–541. doi: 10.1038/35035131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Alexaki VI, Charalampopoulos I, Kampa M, Nifli AP, Hatzoglou A, Gravanis A, Castanas E. Activation of membrane estrogen receptors induce pro-survival kinases. J Steroid Biochem Mol Biol. 2006;98:97–110. doi: 10.1016/j.jsbmb.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 119.Farhat MY, Abi-Younes S, Dingaan B, Vargas R, Ramwell PW. Estradiol increases cyclic adenosine monophosphate in rat pulmonary vascular smooth muscle cells by a nongenomic mechanism. J Pharmacol Exp Ther. 1996;276:652–657. [PubMed] [Google Scholar]
- 120.Gu Q, Moss RL. 17β-estradiol potentiates kainate-induced currents via activation of the cAMP cascade. J Neurosci. 1996;16:3620–3629. doi: 10.1523/JNEUROSCI.16-11-03620.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Picotto G, Massheimer V, Boland R. Acute stimulation of intestinal cell calcium influx induced by 17β-estradiol via the cAMP messenger system. Mol Cell Endocrinol. 1996;119:129–134. doi: 10.1016/0303-7207(96)03799-9. [DOI] [PubMed] [Google Scholar]
- 122.Chen ZJ, Yu L, Chang CH. Stimulation of membrane-bound guanylate cyclase activity by 17-β estradiol. Biochem Biophys Res Commun. 1998;252:639–642. doi: 10.1006/bbrc.1998.9716. [DOI] [PubMed] [Google Scholar]
- 123.Malyala A, Kelly MJ, Ronnekleiv OK. Estrogen modulation of hypothalamic neurons, activation of multiple signaling pathways and gene expression changes. Steroids. 2005;70:397–406. doi: 10.1016/j.steroids.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 124.Kahlert S, Nuedling S, van Eickels M, Vetter H, Meyer R, Grohe C. Estrogen receptor α rapidly activates the IGF-1 receptor pathway. J Biol Chem. 2000;275:18447–18453. doi: 10.1074/jbc.M910345199. [DOI] [PubMed] [Google Scholar]
- 125.Improta-Brears T, Whorton AR, Codazzi F, York JD, Meyer T, McDonnell DP. Estrogen-induced activation of mitogen-activated protein kinase requires mobilization of intracellular calcium. Proc Natl Acad Sci USA. 1999;96:4686–4691. doi: 10.1073/pnas.96.8.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Driggers PH, Segars JH. Estrogen action and cytoplasmic signaling pathways: Part II. The role of growth factors and phosphorylation in estrogen signaling. Trends Endocrinol Metab. 2002;13:422–427. doi: 10.1016/s1043-2760(02)00634-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Razandi M, Alton G, Pedram A, Ghonshani S, Webb P, Levin ER. Identification of a structural determinant necessary for the localization and function of estrogen receptor α at the plasma membrane. Mol Cell Biol. 2003;23:1633–1646. doi: 10.1128/MCB.23.5.1633-1646.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang Z, Kumar R, Santen RJ, Song RXD. The role of adapter protein Shc in estrogen non-genomic action. Steroids. 2004;69:523–529. doi: 10.1016/j.steroids.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 129.Kupzig S, Walker SA, Cullen PJ. The frequencies of calcium oscillations are optimized for efficient calcium-mediated activation of Ras and the ERK/MAPK cascade. Proc Natl Acad Sci USA. 2005;102:7577–7582. doi: 10.1073/pnas.0409611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chambliss KL, Yuhanna IS, Anderson RG, Mendelsohn ME, Shaul PW. ERβ has nongenomic action in caveolae. Mol Endocrinol. 2002;16:938–946. doi: 10.1210/mend.16.5.0827. [DOI] [PubMed] [Google Scholar]
- 131.Kim KH, Bender JR. Rapid, estrogen receptor-mediated signaling: why is the endothelium so special? Sci STKE. 2005;14:pe28. doi: 10.1126/stke.2882005pe28. [DOI] [PubMed] [Google Scholar]
- 132.Ansonoff MA, Etgen AM. Estradiol elevates protein kinase C catalytic activity in the preoptic area of female rats. Endocrinology. 1998;139:3050–3056. doi: 10.1210/endo.139.7.6088. [DOI] [PubMed] [Google Scholar]
- 133.Razandi M, Pedram A, Greene GL, Levin ER. Cell membrane and nuclear estrogen receptors (ERs) originate from a single transcript: studies of ERα and ERβ expressed in Chinese hamster ovary cells. Mol Endocrinol. 1999;13:307–319. doi: 10.1210/mend.13.2.0239. [DOI] [PubMed] [Google Scholar]
- 134.Geraldes P, Sirois MG, Tanguay JF. Specific contribution of estrogen receptors on mitogen-activated protein kinase pathways and vascular cell activation. Circ Res. 2003;93:399–405. doi: 10.1161/01.RES.0000088640.18462.42. [DOI] [PubMed] [Google Scholar]
- 135.Marino M, Galluzzo P, Leone S, Acconcia F, Ascenzi P. Nitric oxide impairs the 17β-estradiol-induced apoptosis in human colon adenocarcinoma cells. Endocr Relat Cancer. 2006;13:559–569. doi: 10.1677/erc.1.01106. [DOI] [PubMed] [Google Scholar]
- 136.Acconcia F, Kumar R. Signaling regulation of genomic and non-genomic functions of estrogen receptors. Cancer Lett. 2005;238:1–14. doi: 10.1016/j.canlet.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 137.Ahola TM, Manninen T, Alkio N, Ylikomi T. G protein-coupled receptor 30 is critical for a progestin-induced growth inhibition in MCF-7 breast cancer cells. Endocrinology. 2002;143:3376–3384. doi: 10.1210/en.2001-211445. [DOI] [PubMed] [Google Scholar]
- 138.Filardo EJ, Quinn JA, Frackelton AR, Jr, Bland KI. Estrogen action via the G protein-coupled receptor, GPR30: stimulation of adenylyl cyclase and cAMP-mediated attenuation of the epidermal growth factor receptor-to-MAPK signaling axis. Mol Endocrinol. 2002;16:70–84. doi: 10.1210/mend.16.1.0758. [DOI] [PubMed] [Google Scholar]
- 139.Ropero AB, Soria B, Nadal A. A nonclassical estrogen membrane receptor triggers rapid differential actions in the endocrine pancreas. Mol Endocrinol. 2002;16:497–505. doi: 10.1210/mend.16.3.0794. [DOI] [PubMed] [Google Scholar]
- 140.Toran-Allerand CD, Guan X, MacLusky NJ, Horvath TL, Diano S, Singh M, Connolly ES, Jr, Nethrapalli IS, Tinnikov AA. ER-X: a novel, plasma membrane-associated, putative estrogen receptor that is regulated during development and after ischemic brain injury. J Neurosci. 2002;22:8391–8401. doi: 10.1523/JNEUROSCI.22-19-08391.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Thomas P, Pang Y, Filardo EJ, Dong J. Identity of an estrogen membrane receptor coupled to a G protein in human breast cancer cells. Endocrinology. 2005;146:624–632. doi: 10.1210/en.2004-1064. [DOI] [PubMed] [Google Scholar]
- 142.Vivacqua A, Bonofiglio D, Recchia AG, Musti AM, Picard D, Andò S, Maggiolini M. The G protein-coupled receptor GPR30 mediates the proliferative effects induced by 17β-estradiol and hydroxytamoxifen in endometrial cancer cells. Mol Endocrinol. 2006;20:631–646. doi: 10.1210/me.2005-0280. [DOI] [PubMed] [Google Scholar]
- 143.Pappas TC, Gametchu B, Watson CS. Membrane estrogen receptors identified by multiple antibody labeling and impeded-ligand binding. FASEB J. 1995;9:404–410. doi: 10.1096/fasebj.9.5.7896011. [DOI] [PubMed] [Google Scholar]
- 144.Norfleet AM, Thomas ML, Gametchu B, Watson CS. Estrogen receptor-α detected on the plasma membrane of aldehyde-fixed GH3/B6/F10 rat pituitary tumor cells by enzyme-linked immuno-cytochemistry. Endocrinology. 1999;140:3805–3814. doi: 10.1210/endo.140.8.6936. [DOI] [PubMed] [Google Scholar]
- 145.Dan P, Cheung JC, Scriven DR, Moore ED. Epitope dependent localization of estrogen receptor-α, but not -β, in en face arterial endothelium. Am J Physiol. 2003;284:H1295–H1306. doi: 10.1152/ajpheart.00781.2002. [DOI] [PubMed] [Google Scholar]
- 146.Acconcia F, Ascenzi P, Bocedi A, Spisni E, Tomasi V, Trentalance A, Visca P, Marino M. Palmitoylation-dependent estrogen receptor β membrane localization: regulation by 17β-estradiol. Mol Biol Cell. 2005;16:231–237. doi: 10.1091/mbc.E04-07-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Acconcia F, Bocedi A, Ascenzi P, Marino M. Does palmitoylation target estrogen receptors to plasma membrane caveolae? IUBMB Life. 2003;55:33–35. doi: 10.1080/1521654031000081256. [DOI] [PubMed] [Google Scholar]
- 148.Bijlmakers MJ, Marsh M. The on-off story of protein palmitoylation. Trends Cell Biol. 2003;13:32–42. doi: 10.1016/s0962-8924(02)00008-9. [DOI] [PubMed] [Google Scholar]
- 149.Kennedy AM, Shogren KL, Zhang M, Turner RT, Spelsberg TC, Maran A. 17β-estradiol-dependent activation of signal transducer and activator of transcription-1 in human fetal osteoblasts is dependent on Src kinase activity. Endocrinology. 2005;146:201–207. doi: 10.1210/en.2004-0486. [DOI] [PubMed] [Google Scholar]
- 150.Song RXD, Zhang Z, Santen RJ. Estrogen rapid action via protein complex formation involving ERα and Src. Trends Endocrinol Metab. 2005;16:347–353. doi: 10.1016/j.tem.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 151.Greger JG, Guo Y, Henderson R, Ross JF, Cheskis BJ. Characterization of MNAR expression. Steroids. 2006;71:317–322. doi: 10.1016/j.steroids.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 152.Song RXD, Barnes CJ, Zhang Z, Bao Y, Kumar R, Santen RJ. The role of Shc and insulin-like growth factor 1 receptor in mediating the translocation of estrogen receptor α to the plasma membrane. Proc Natl Acad Sci USA. 2004;101:2076–2081. doi: 10.1073/pnas.0308334100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Warner M, Gustafsson JÅ. Nongenomic effects of estrogen: why all the uncertainty? Steroids. 2006;71:91–95. doi: 10.1016/j.steroids.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 154.Paruthiyil S, Parmar H, Kerekatte V, Cunha GR, Firestone GL, Leitman DC. Estrogen receptor β inhibits human breast cancer cell proliferation and tumor formation by causing a G2 cell cycle arrest. Cancer Res. 2004;64:423–428. doi: 10.1158/0008-5472.can-03-2446. [DOI] [PubMed] [Google Scholar]
- 155.Lu Q, Ebling H, Mittler J, Baur WE, Karas RH. MAP kinase mediates growth factor-induced nuclear translocation of estrogen receptor α. FEBS Lett. 2002;516:1–8. doi: 10.1016/s0014-5793(02)02432-8. [DOI] [PubMed] [Google Scholar]
- 156.Yamakawa K, Arita J. Cross-talk between the estrogen receptor-, protein kinase A-, and mitogen-activated protein kinase-mediated signaling pathways in the regulation of lactotroph proliferation in primary culture. J Steroid Biochem Mol Biol. 2004;88:123–130. doi: 10.1016/j.jsbmb.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 157.Pedram A, Razandi M, Aitkenhead M, Hughes CC, Levin ER. Integration of the non-genomic and genomic actions of estrogen. Membrane-initiated signaling by steroid to transcription and cell biology. J Biol Chem. 2002;277:50768–50775. doi: 10.1074/jbc.M210106200. [DOI] [PubMed] [Google Scholar]
- 158.Wade CB, Dorsa DM. Estrogen activation of cyclic adenosine 5′- monophosphate response element-mediated transcription requires the extracellularly regulated kinase/mitogen-activated protein kinase pathway. Endocrinology. 2003;144:832–838. doi: 10.1210/en.2002-220899. [DOI] [PubMed] [Google Scholar]
- 159.Hennessy BA, Harvey BJ, Healy V. 17β-Estradiol rapidly stimulates c-fos expression via the MAPK pathway in T84 cells. Mol Cell Endocrinol. 2005;229:39–47. doi: 10.1016/j.mce.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 160.Watters JJ, Dorsa DM. Transcriptional effects of estrogen on neuronal neurotensin gene expression involve cAMP/protein kinase A-dependent signaling mechanisms. J Neurosci. 1998;18:6672–6680. doi: 10.1523/JNEUROSCI.18-17-06672.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Foster JS, Henley DC, Ahamed S. Estrogens and cell-cycle regulation in breast cancer. Trends Endocrinol Metabol. 2001;12:320–327. doi: 10.1016/s1043-2760(01)00436-2. [DOI] [PubMed] [Google Scholar]
- 162.Marino M, Ascenzi P. Estrogen receptor-α: Plasma membrane localization and functions. Immun Endoc & Metab Agents in Med Chem. 2006;6:281–286. [Google Scholar]
- 163.Schwabe JWR, Chapman L, Finch JT, Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]


