Abstract
Stromal cell-derived factor-1α (SDF-1α ) is a member of the chemokine superfamily and functions as a growth factor and chemoattractant through activation of CXCR4/LESTR/Fusin, a G protein-coupled receptor. This receptor also functions as a coreceptor for T-tropic syncytium-inducing strains of HIV-1. SDF-1α antagonizes infectivity of these strains by competing with gp120 for binding to the receptor. The crystal structure of a variant SDF-1α ([N33A]SDF-1α ) prepared by total chemical synthesis has been refined to 2.2-Å resolution. Although SDF-1α adopts a typical chemokine β-β-β-α topology, the packing of the α-helix against the β-sheet is strikingly different. Comparison of SDF-1α with other chemokine structures confirms the hypothesis that SDF-1α may be either an ancestral protein from which all other chemokines evolved or the chemokine that is the least divergent from a primordial chemokine. The structure of SDF-1α reveals a positively charged surface ideal for binding to the negatively charged extracellular loops of the CXCR4 HIV-1 coreceptor. This ionic complementarity is likely to promote the interaction of the mobile N-terminal segment of SDF-1α with interhelical sites of the receptor, resulting in a biological response.
Chemokines are the largest superfamily of cytokines and are characterized by four conserved cysteines that form two disulfide bonds. Two major families have been defined based on the presence (CXC, or α-chemokines) or absence (CC, or β-chemokines) of an intervening amino acid between the first two cysteines. Proteins within the same family share a high degree of sequence homology and can compete for binding to the same receptor. Stromal cell-derived factor 1α (SDF-1α) is unusual in this respect. On the basis of sequence analysis, SDF-1α is equally divergent from both chemokine families, although it shares the α-chemokine CXC motif (1). However, the SDF-1α sequence is conserved remarkably across species. A single Ile → Val difference at position 18 results in 99% sequence identity between human and murine SDF-1α; other chemokines have interspecies identities that range between 60 and 85%. Furthermore, the gene for SDF-1α is located on chromosome 10 whereas all other human α- and β-chemokine genes localize to chromosomes 4 and 17, respectively (2). Deletion of this gene in mice is lethal; the mice die perinatally and have a severe defect in the cardiac ventricular septum (3). No other chemokine (or chemokine receptor) has been reported to be of such vital developmental importance. Although chemokines hitherto have been observed to be induced by inflammatory stimuli, SDF-1α mRNA is expressed constitutively in almost all organs (4, 5), and its levels are unaltered during inflammation (6). Moreover, no other mammalian chemokine has been known to compete with SDF-1α for binding to the CXCR4 receptor. These findings suggest that SDF-1α is unique among α- or β-chemokines.
Chemokines and their receptors are involved in HIV pathophysiology (7). The M-tropic, non-syncytium-inducing strains of HIV-1 appear to be involved in the primary infection. These strains use CCR5 as the coreceptor for viral entry and are inhibited by the physiological agonists of CCR5: macrophage inflammatory protein 1α (MIP-1α), MIP-1β, and regulated on activation, normal T cells expressed and secreted (RANTES) (7). The conversion of M-tropic to T-tropic HIV syncytium-inducing strains precedes the precipitous drop in CD4+ T-cells. The T-tropic HIV strains that infect T-cells use CXCR4, and infection of these T-cells is inhibited by SDF-1α (8).
In light of the unusually low sequence homology to other chemokines, a role in essential developmental processes, and inhibition of syncytium-inducing HIV-1 strains (8, 9), we have subjected SDF-1α to crystallographic analysis. Here we describe the 2.2-Å crystal structure of [N33A]SDF-1α, a variant of human SDF-1α .
MATERIALS AND METHODS
Preparation of [N33A]SDF-1α.
The N33A variant of SDF-1α was prepared by total chemical synthesis by using native chemical ligation of the N-terminal peptide (1–33) and C-terminal peptide (34–67) (refs. 10 and 11). The thioacid peptide (SDF-1α (1–33)-α-COSH) could not be generated readily on the standard α-thiocarboxylate resin because of the presence of an asparagine at the ligation site. Alanine, therefore, was substituted for asparagine. The two peptides were ligated as described, resulting in the full-length [N33A]SDF-1α variant (11). This variant is identical to wild-type SDF-1α in receptor-binding properties and biological activity (11).
Crystallization and Data Collection.
[N33A]SDF-1α (12 mg/ml) was crystallized by vapor diffusion against 1.9 M ammonium sulfate and 0.1 M Tris (pH 8.5). Crystals belonged to space group P212121 with cell dimensions a = 38.84 Å, b = 50.47 Å, and c = 64.72 Å. Because crystals larger than 0.15 mm appeared twinned, we harvested the smaller crystals in solutions of 2.2 M ammonium sulfate and 0.1 M Tris (pH 8.5). For data collection, crystals were equilibrated with cryoprotectant (in 25% glycerol in mother liquor) for 10–15 min and were frozen rapidly in a stream of N2 at −160°C. Diffraction data were collected on a RAXIS-IIC image plate detector (Rigaku, Tokyo) with a Rigaku RU200 rotating anode x-ray generator and at beamline X12B of the National Synchrotron Light Source at the Brookhaven National Laboratories on a MAR-Research (Hamburg) 300-image plate-detection system. Data were processed by using denzo and scalepack (12).
Structure Determination.
Heavy atom derivatives were prepared by soaking crystals in 10-mM solutions of KAuCl4 (3 days) or K2PtCl4 (30 min) or KAuCl4 and K2PtCl4 (3 days in KAuCl4 and 30 min in K2PtCl4). Crystals for the platinum derivatives were preequilibriated in phosphate buffer (pH 7) before heavy atom soaking. Heavy atom positions were located by Patterson methods by using the CCP4 (13) and phases (W. Furey, Univ. of Pittsburgh) packages. mlphare (14) was used to refine heavy atom positions and to calculate initial phases followed by iterative solvent flattening and density modification protocols incorporated in dm (15).
Model Building and Refinement.
Two protein chains, one for each monomer in the crystallographic asymmetric unit, were built into density by using the modeling program O (16). The initial model was refined by using conjugate gradient and simulated annealing methods incorporated in xplor (17). The final refined model contained 989 protein atoms and 66 water molecules, with an Rfree of 28.5%.
Structural Comparisons.
Superposition of SDF-1α on the other known chemokine structures (from the first cysteine to the C-terminus) was performed by least squares minimization of the differences as incorporated in the program lsqman (16). An initial framework for the superposition consisted of the residues Cys-9, Cys-34, Cys-50, and Trp-57 in SDF-1α and the equivalent residues in the other chemokines.
RESULTS AND DISCUSSION
Structure of SDF-1α.
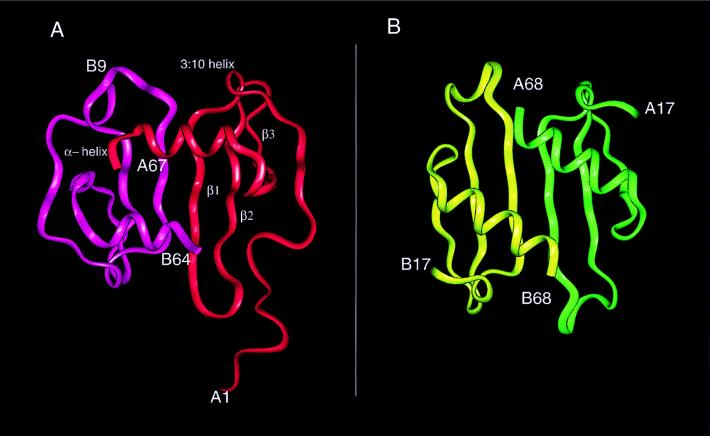
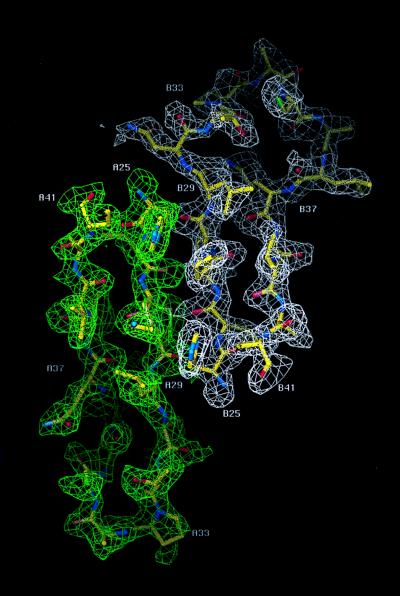
The structure of SDF-1α was determined by multiple isomorphous replacement and anomalous dispersion combined with solvent flattening and density modification (Table 1). The SIRAS phases calculated with the gold derivative did not permit an unambiguous interpretation of electron density maps. However, on inclusion of phases from the platinum derivative and the gold-platinum double derivative, secondary structural features and the loops connecting them were more clearly visible. A solvent content of 40% was assumed to improve the phases with several rounds of solvent flattening and density modification. The final model had a crystallographic R-factor of 22.5% (Rfree = 28.5%). There were two monomers in the asymmetric unit that associated to form a noncrystallographic dimer, similar to but not identical to interleukin (IL)-8 (18) (Fig. 1). Electron density for the entire polypeptide chain (residues 1–67) was observed for one of the two monomers with only the side chains of Val-3 and Arg-8 being undefined. For the second monomer, continuous density was observed only for residues 9 to 64. The monomers each had the conventional chemokine topology consisting of three antiparallel β-strands (β1, residues 24–31; β2, residues 35–42; and β3, residues 45–49) followed by a C-terminal α-helix (residues 56–64) that was packed against the β-sheet. In addition, there was a single turn of a 310 helix (residues 19–22) that preceded the first β-strand (Fig. 1). The overall rms difference between the two monomers was 1.2 Å, based on Cα atoms; the largest conformational difference was found in a loop composed of residues 29–37. An example of the electron density for both monomers in this region, displayed as an 2Fo-Fc omit map, is shown in Fig. 2. Chemokines are known to be predominantly in the monomeric form at physiological concentrations, and monomeric analogs are biologically active (19, 20). Moreover, sedimentation equilibrium and NMR studies of SDF-1α have indicated that this chemokine is a monomer even at high concentrations (21). The monomer of SDF-1α with electron density for the entire polypeptide was used for all subsequent analysis and discussion.
Table 1.
Data collection, phasing, and structure refinement statistics
| Native | KAuCl4 | K2PtCl4 | KAuCl4/K2PtCl4 | |
|---|---|---|---|---|
| Wavelength, Å | 0.95 | 1.04 | 1.54 | 1.54 |
| Resolution limit, Å | 2.0 | 2.0 | 2.5 | 2.5 |
| Measured reflections | 22,469 | 234,964 | 31,058 | 25,236 |
| Unique reflections | 7,600 | 9,129 | 4,846 | 4,753 |
| Completeness, % | 97.6 | 99.0 | 99.3 | 97.8 |
| Overall I/σ(I) | 9.3 | 10.1 | 5.4 | 7.0 |
| Rsym, % | 4.6 | 4.4 | 9.0 | 6.7 |
| Riso, % | — | 19.0 | 20.6 | 21.8 |
| Sites, no. | — | 1 | 1 | 2 |
| Rc,isom | — | 0.50 | 0.74 | 0.51 |
| Rc,anom | — | 0.65 | — | — |
| Phasing power | — | 2.71 | 1.52 | 2.74 |
| Refinement statistics | ||||
| Resolution range, Å | 5–2.2 | |||
| R-value/Rfree, % | 22.5/28.5 | |||
| rms deviation | ||||
| Bond length, Å | 0.010 | |||
| Bond angles, degrees | 2.0 |
Figure 1.
(A) Structure of the SDF-1α dimer with the secondary structure assignments for monomer 1 based on the program insightii (Biosym Technologies, San Diego) shows the extended N-terminal loop proceeding into the single turn of a 310 helix and strands β1, β2, and β3 followed by the C-terminal helix packing almost orthogonally to the three-stranded antiparallel β-sheet. (B) The IL-8 dimer is shown to highlight the differences between the IL-8 and SDF-1α dimers.
Figure 2.
2Fo-Fc omit map contoured at 1 σ between residues 25 to 41 that make up β strands 1 and 2 and the interconnecting loop. The electron density for monomer 1 (A25 to A41) is drawn in green and monomer 2 (B25 to B41) is drawn in white. This figure was created with the graphics program o (16).
Comparisons with Other Chemokine Structures.
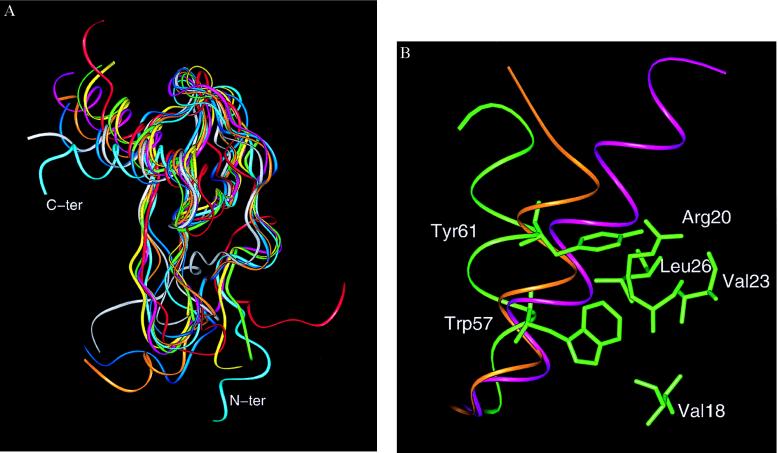
Although the topology of SDF-1α is similar to other chemokines, there are significant structural differences. Pairwise rms differences between SDF-1α and the α- and β-chemokines ranged between 2.0–2.2 Å and 1.9–2.2 Å, respectively (Table 2). These differences are substantially greater than those observed between all other known chemokine structures. The pairwise rms deviation among the β-chemokines MIP-1β (22), monocyte chemoattractant protein 1 (23), and RANTES (24) ranged between 1.2 and 1.4 Å; a similar comparison for the α-chemokines IL-8 (18), neutrophil-activating peptide 2 (25), platelet factor-4 (PF-4) (26), and gro-α/melanoma growth-stimulating activity (27) resulted in pairwise rms deviations that ranged from 1.0 to 1.9 Å. The source of the large rms difference of SDF-1α with respect to the other chemokines largely was caused by the orientation and packing of the α-helix in relation to the β-sheet. An overall superposition of the eight chemokine structures is given in Fig. 3A and shows that the largest differences are at the N-terminal region and the C-terminal helix. The angle between the helix and the axis defined by the β-strands was ≈90° for SDF-1α and was 70–85° for the other α- and β-chemokines. Although the N-terminal CXC motif in SDF-1α places this protein in the α-chemokine family, the angle between the helix and β-strands was more similar to the β-chemokines RANTES and MIP-1β. These two chemokines had angles of 80° and 83°, respectively. Because of its orientation, the helix in SDF-1α also adopts a more open conformation (Figs. 3 A and B). For instance, a region encompassing the α-helix (residues 55–65) of SDF-1α had a solvent accessible area of 741 Å2 as compared with 690 Å2 and 657 Å2 for RANTES and IL-8, respectively. To minimize the effects of the different side chains, the same ranking order was maintained when only the solvent accessible surface area of the main chain atoms for this region was calculated. The greater solvent exposure of the SDF-1α helix was caused by packing of the aromatic side chains of Trp-57 and Tyr-61. The indole nitrogen of Trp-57 formed a hydrogen bond with the carbonyl of Val-18, and Tyr-61 formed van der Waals interactions with Leu-26 in SDF-1α (Fig. 3B). The interactions of these bulky side chains forced the helix to move further away from the core of the protein and resulted in a helical conformation that was different from that of other chemokines. Tyr-61 appears to be particularly important for the movement of the helix away from the core. The two other chemokines with a helical orientation most similar to that of SDF-1α (MIP-1β and RANTES) also had a tyrosine at the equivalent position. In contrast, IL-8 and monocyte chemoattractant protein 1 had a valine and serine, respectively, at this position. It is, therefore, apparent from this analysis that SDF-1α has an N-terminal CXC motif most similar to α-chemokines and a C-terminal helix most similar to β-chemokines.
Table 2.
Pairwise analysis of known chemokine structures
| Molecule | SDF–1α | IL-8 | NAP-2 | PF-4 | MGSA | MIP-1β | MCP-1 | RANTES |
| SDF-1α | — | 2.0/48 | 2.0/51 | 2.0/51 | 2.0/45 | 2.0/51 | 1.9/45 | 2.2/51 |
| IL-8 | 27% | — | 1.0/56 | 1.5/56 | 1.7/56 | 1.6/51 | 1.2/50 | 1.5/48 |
| NAP-2 | 21% | 46% | — | 1.2/56 | 1.6/57 | 1.5/49 | 1.7/63 | 1.3/47 |
| PF-4 | 19.4% | 32% | 51.4% | — | 1.7/45 | 1.9/50 | 1.7/50 | 1.9/48 |
| MGSA | 22.3% | 42% | 57% | 31.4% | — | 1.6/41 | 1.6/47 | 1.6/48 |
| MIP-1β | 21% | 22.2% | 13.2% | 12% | 12% | — | 1.3/50 | 1.3/52 |
| MCP-1 | 16.4% | 21% | 17.1% | 14% | 11.4% | 37% | — | 1.4/54 |
| RANTES | 24% | 24% | 10% | 11.4% | 12.3% | 50% | 23.2% | — |
The least squares superposition was done by using the Cα atoms of the four CXC chemokines IL-8 (18), NAP-2 (25), PF-4 (26), and MGSA (27), and the three CC chemokines MIP-1β (22), MCP-1 (23), and RANTES (24). The initial framework of residues used for the alignment consisted of Cys-9, Cys-34, Cys-50, and Trp-57 in SDF-1α and their counterparts in the other chemokines. The upper quadrant of the table lists the rms difference followed by the total number of aligned residues in the superposition for each pair of chemokines. Percentage sequence identities are shown in the bottom quadrant in bold lettering. The rms difference was calculated with the lsqman option in o (16).
Figure 3.
(A) Diagram of the superimposed monomers of SDF-1α (cyan), gro-α/melanoma growth-stimulating activity (red), neutrophil-activating peptide 2 (yellow), platelet factor 4 (green), IL-8 (pink), RANTES (white), monocyte chemoattractant protein 1 (ochre), and MIP-1β (blue), highlighting differences at the N-terminal region and the C-terminal helix. (Figure generated with the graphics program insightii). The Cα superposition was based on lsqman (16) with an initial framework of Cys-9, Cys-34, Cys-50, and Trp-57 in SDF-1α and their equivalent counterparts in the other chemokines. The angle of the helix with respect to the β-sheet was measured by using the Cα from the three residues 49 (from β3), 54 (from the beginning of the helix), and 66 (from the end of the helix) in SDF-1α. The equivalent residues were used for measuring the angles in the other chemokine structures. (B) Packing of the C-terminal helix and relevant side chains in SDF-1α (green). The ribbon diagram of the α-helix of IL-8 (purple) and RANTES (orange) is shown for reference. The van der Waals interactions involving Tyr-61 are believed to be pivotal in the “open” orientation of the helix of SDF-1α.
Presumed Receptor Binding Site.
The three-dimensional structure of SDF-1α also is essential for understanding the receptor interactions that result in growth factor and chemoattractant activities (1, 28). The interactions with the receptor are particularly important in light of the role of CXCR4 in HIV-1 infection (29) and the ability of SDF-1α to inhibit viral entry via CXCR4 (8, 9). Mutational analysis has indicated that the N terminus is involved in receptor binding and biological activity (8, 21), which is consistent with the critical role of the N terminus for all other examined chemokines (7). However, the N terminus alone is insufficient for receptor binding and activation (30). These studies indicate that additional binding sites on the ligand are required for chemokine receptor binding. For SDF-1α, a second binding site consisting of an RFFESH motif (residues 12–17) has been identified (21).
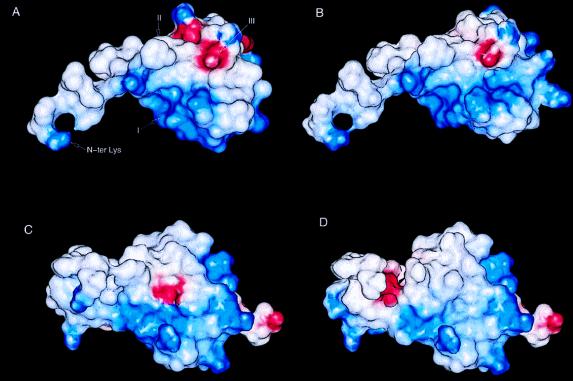
The recognition and affinity between chemokines and their receptors is thought to be initiated by charge complementarity (31, 32). In this regard, SDF-1α is one of the most basic chemokines, with an overall charge of +8. The corresponding net charge of the extracellular regions of CXCR4 is −9. Analysis of the electrostatic potential of SDF-1α revealed a surface of high positive potential that arises from the clustering of the basic residues Lys-24, His-25, Lys-27, and Arg-41 (Fig. 4A). Although the side chain of Arg-8 was not observed and, therefore, was not included in the calculation, the orientation of the Cβ for its side chain indicates that the guanidinium group could extend into this region and further increase the positive potential. Adjacent to this positively charged region is an extended hydrophobic crevice consisting of residues Pro-10, Phe-13, Val-18, Val-27, Leu-29, Val-39, Leu-42, and Val-49, which is flanked by Arg-12, Glu-15, His-17, and Arg-47. The pattern of a hydrophobic surface surrounded by a constellation of ionic residues appears to be a common characteristic of the chemokines. In IL-8 and murine MIP-2, such a combination of hydrophobic and charged residues has been proposed to be involved in receptor binding (31, 32). Unlike the prototypical G protein-coupled receptors, such as the β-adrenergic and rhodopsin receptors that have extracellular regions that do not appear to participate in ligand or cofactor binding (33), the extracellular loops of chemokine receptors play a crucial role in ligand binding and recognition (7, 34). It is, therefore, likely that the extracellular loops of CXCR4 form extensive interactions with the charged residues and hydrophobic surface, thereby promoting interactions of the highly mobile N-terminal region of SDF-1α with the interhelical transmembrane sites of the receptor.
Figure 4.
Electrostatic potential map of (A) the SDF-1α monomer, (B) the gro-α/SDF-1α chimera based on the SDF-1α structure, (C) the gro-α/SDF-1α chimera based on the gro-α structure (27), and (D) gro-α. The chimera for B and C is based on results from Crump et al. (21) and has the sequence KPSVSLSYRC PCRFFESHIH PKNIQHLKIL NTPNCAQTEV IATLKNGRKA CLNPASPIVK KIIEKMLNSD KSN, where the underlined residues are from SDF-1α and the remaining residues are gro-α. The electrostatic potential was calculated by using delphi (Biosym Technologies) and displayed with insightii. For the calculation, lysine and arginine side chains were assigned an overall charge of +1.0, histidine was assigned an overall charge of +0.5, and aspartic acid and glutamic acid were assigned an overall charge of −1.0. The regions of positive potential are shown in blue, and the regions of negative potential in red. In A, region I consists of residues Lys-24, His-25, Lys-27, and Arg-41; region II consists of Pro-10, Phe-13, Val-18, Val-27, Leu-29, Val-39, Leu-42, Val-49; and region III consists of Arg-12, Glu-15, His-17, and Arg-47.
A recently reported chimera of gro-α and SDF-1α was observed to have equivalent activity in a Ca2+ mobilization assay as wild-type SDF-1α (21). To assess the role played by the positively charged groups of SDF-1α in receptor binding, electrostatic potential maps were calculated for SDF-1α , the gro-α/SDF-1α chimera and gro-α (Fig. 4). Because there is no structure of the gro-α/SDF-1α chimera, it was modeled in the context of both the SDF-1α (Fig. 4B) and gro-α (Fig. 4C) structures. Analysis of the electrostatic potential maps revealed that the region of high positive potential (defined as region I in Fig. 4A) is retained in the chimera and that this region is of lower potential and covers a smaller area in gro-α. These results suggest that the positive electrostatic potential may contribute to receptor binding and activation. Furthermore, it is interesting to note that three other molecules that interact with CXCR4 are positively charged (35, 36). The V3 loop of gp120 from T-tropic strains of HIV-1 have a high net positive charge relative to V3 sequences from M-tropic strains (37). Two small molecule antagonists belonging to distinct chemical families share properties that also suggest that these molecules mimic the surface of SDF-1α that exhibits a positive electrostatic potential. One antagonist is a bicyclam derivative containing a cluster of eight amines that inhibits SDF-1α responses at submicromolar concentrations (35). The other antagonist is n-α-acetyl-nona-d-arginine amide, a polypeptide consisting of nine d-arginine residues with protected N and C termini that is active at micromolar concentrations (36). Both of these compounds are able to inhibit CXCR4-mediated entry of HIV-1. It is tempting to speculate that the compounds interact with the negatively charged extracellular loops of CXCR4.
Conclusions.
SDF-1α displays relatively large structural differences with respect to other chemokines. These differences are consistent with the low sequence homology between SDF-1α and other chemokines and indicate that SDF-1α is equally divergent from both α- and β-chemokines. The N-terminal CXC sequence of SDF-1α places this protein in the α-chemokine family whereas the conformation and packing of the C-terminal helix is most similar to β-chemokines. These observations support the proposition that SDF-1α may define a new subfamily and may be most closely related to the evolutionary ancestor of the chemokine superfamily (1).
In most cases of HIV-1 infection, the decrease in CD4+ T cells and the development of clinical symptoms of AIDS follows the conversion of HIV-1 from M-tropic to T-tropic strains. The arrangement of hydrophobic and ionic groups in the SDF-1α structure along with further mutational analysis will provide the basis for the rational design of new classes of molecules that exhibit receptor antagonism and inhibit CXCR4-mediated HIV-1 viral entry. These CXCR4 antagonists may prove, therefore, to be useful in delaying the onset of disease.
Acknowledgments
We acknowledge Bill Furey (University of Pittsburgh), Timothy Springer (Harvard University), and Ian Tickle (Birkbeck College) for helpful discussions, Malcolm Capel (Brookhaven National Laboratory), Pat Fleming and Paul Pepin for technical help, and Stephen B. H. Kent (Gryphon Sciences), Steven C. Almo (Albert Einstein School of Medicine) and Robert F. Tilton (ArQule, Inc.) for critical reading of the manuscript.
ABBREVIATION
- SDF-1α
stromal cell-derived factor 1α
- MIP
macrophage inflammatory protein
- RANTES
regulated on activation, normal T cells expressed and secreted
- IL
interleukin
Footnotes
C.D. and E.J.F. contributed equally to this work and should each be considered first authors.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Chemistry Department, Brookhaven National Laboratory, Upton, NY 11973 (reference 1A15).
References
- 1.Bleul C C, Fuhlbrigge R C, Casasnovas J M, Aiuti A, Springer T A. J Exp Med. 1996;184:1101–1109. doi: 10.1084/jem.184.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shirozu M, Nakano T, Inazawa J, Tashiro K, Tada H, Shinohara T, Honjo T. Genomics. 1995;28:495–500. doi: 10.1006/geno.1995.1180. [DOI] [PubMed] [Google Scholar]
- 3.Nagasawa T, Hirota S, Tachibana K, Takakura N, Nishikawa S, Kitamura Y, Yoshida N, Kikutani H, Kishimoto T. Nature (London) 1996;382:635–638. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
- 4.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 5.Tashiro K, Tada H, Heilker M, Shirozu M, Nakano T, Honjo T. Science. 1993;261:600–603. doi: 10.1126/science.8342023. [DOI] [PubMed] [Google Scholar]
- 6.Godiska R, Chandry D, Dietsch G N, Gray P W. J Neuroimmunol. 1995;58:167–176. doi: 10.1016/0165-5728(95)00008-p. [DOI] [PubMed] [Google Scholar]
- 7.Baggiolini M, Dewald B, Moser B. Annu Rev Immunol. 1997;15:675–705. doi: 10.1146/annurev.immunol.15.1.675. [DOI] [PubMed] [Google Scholar]
- 8.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. Nature (London) 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 9.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J-L, Arenzana-Seisdedos F, Schwartz O, Heard J-M, Clark-Lewis I, Legler D, et al. Nature (London) 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 10.Dawson P E, Muir T W, Clark-Lewis I, Kent S B H. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 11.Ueda H, Siani M, Wanghua G, Thompson D A, Brown G B, Wang J M. J Biol Chem. 1997;272:24966–24970. doi: 10.1074/jbc.272.40.24966. [DOI] [PubMed] [Google Scholar]
- 12.Otwinowski Z. In: Proceedings of the CCP4 Study Weekend. Sawyer L, Isaac N, Borley S, editors. Daresbury Laboratory, Daresbury, U.K.: Science and Engineering Research Council; 1993. pp. 56–62. [Google Scholar]
- 13.Collaborative Computing Project No. 4. Acta Crystallogr. 1994;D50:760–763. [Google Scholar]
- 14.Otwinowski Z. Daresbury Study Weekend Proceedings. Daresbury Laboratory, Daresbury, U.K.: Science and Engineering Research Council; 1991. pp. 80–89. [Google Scholar]
- 15.Cowtan K. Joint CCP4 ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34. [Google Scholar]
- 16.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 17.Brunger A T. xplorVersion 3.1 Manual. Yale University, New Haven, CT: The Howard Hughes Medical Institute and Department of Biophysics and Biochemistry; 1992. [Google Scholar]
- 18.Clore M G, Appella E, Yamada M, Matsushima K, Gronenborn A. Biochemistry. 1990;24:1689–1696. doi: 10.1021/bi00459a004. [DOI] [PubMed] [Google Scholar]
- 19.Rajarathnam K, Sykes B D, Kay C M, Dewald B, Geiser T, Baggiolini M, Clark-Lewis I. Science. 1994;264:90–92. doi: 10.1126/science.8140420. [DOI] [PubMed] [Google Scholar]
- 20.Paolini J F, Willard D, Consler T, Luther M, Krangel M S. J Immunol. 1994;153:2704–2717. [PubMed] [Google Scholar]
- 21.Crump M P, Gong J H, Loetscher P, Rajarathnam K, Amara A, Arenzana-Seisdedos F, Virelizier J-L, Baggiolini M, Sykes B D, Clark-Lewis I. EMBO J. 1997;16:6996–7007. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lodi P, Garrett D, Kuszewski J, Tsang M, Weatherbeen J, Leonard W, Gronenborn A, Clore G. Science. 1994;263:1762–1767. doi: 10.1126/science.8134838. [DOI] [PubMed] [Google Scholar]
- 23.Handel T M, Domaille P J. Biochemistry. 1996;35:6569–6584. doi: 10.1021/bi9602270. [DOI] [PubMed] [Google Scholar]
- 24.Chung C, Cooke R M, Proudfoot A E I, Wells T N C. Biochemistry. 1995;34:9307–9314. doi: 10.1021/bi00029a005. [DOI] [PubMed] [Google Scholar]
- 25.Malkowski M G, Wu J Y, Lazar J B, Johnson P H, Edwards B F P. J Biol Chem. 1995;270:7088–7087. doi: 10.1074/jbc.270.13.7077. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Chen L, Bancroft D P, Lai C K, Maione T E. Biochemistry. 1994;33:8361–8366. doi: 10.1021/bi00193a025. [DOI] [PubMed] [Google Scholar]
- 27.Kim K-S, Clark-Lewis I, Sykes B D. J Biol Chem. 1994;269:32909–32915. [PubMed] [Google Scholar]
- 28.Nagasawa T, Kikutani H, Kishimoto T. Proc Natl Acad Sci USA. 1994;91:2305–2309. doi: 10.1073/pnas.91.6.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng Y, Broder C, Kennedy P, Berger E. Science. 1996;272:872–877. [Google Scholar]
- 30.Clark-Lewis I, Kim K S, Rajarathnam K, Gong J H, Dewald B, Moser B, Baggiolini M, Sykes B D. J Leukocyte Biol. 1995;57:703–711. doi: 10.1002/jlb.57.5.703. [DOI] [PubMed] [Google Scholar]
- 31.Jerva L, Sullivan G, Lolis E. Protein Sci. 1997;6:1643–1652. doi: 10.1002/pro.5560060805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammond M E W, Shymala V, Siani M A, Gallegos C A, Feucht P H, Abbott J, Lapointe G R, Moghadam M, Khoja J, Zakel J, et al. J Biol Chem. 1996;271:8228–8235. doi: 10.1074/jbc.271.14.8228. [DOI] [PubMed] [Google Scholar]
- 33.Dohlman H G, Thorner J, Caron M G, Lefkowitz R J. Annu Rev Biochem. 1991;60:653–688. doi: 10.1146/annurev.bi.60.070191.003253. [DOI] [PubMed] [Google Scholar]
- 34.Atchison R E, Gosling J, Monteclaro F S, Franci C, Digilio L, Charo I F, Goldsmith M A. Science. 1996;274:1924–1926. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- 35.Schols D, Struyf S, Van Damme J, Este J A, Henson G, De Clercq E. J Exp Med. 1997;186:1383–1388. doi: 10.1084/jem.186.8.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doranz B J, Grovit-Ferbas K, Sharron M P, Mao S-H, Goetz M B, Daar E S, Doms R W, O’Brien W A. J Exp Med. 1997;186:1395–1400. doi: 10.1084/jem.186.8.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Obrien W A, Sumner-Smith M, Mao S, Sadeghi S, Zhao J, Chen S Y. J Virol. 1996;70:2825–2831. doi: 10.1128/jvi.70.5.2825-2831.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]