Abstract
The study was to explore the power and feasibility of using programmable graphics processing units (GPUs) for real-time rendering and displaying large 3-D medical datasets for stereoscopic display workstation. Lung cancer screening CT images were used for developing GPU-based stereo rendering and displaying. The study was run on a personal computer with a 128 MB NVIDIA Quadro FX 1100 graphics card. The performance of rendering and displaying was measured and compared between GPU-based and CPU (central processing unit)-based programming. The results indicate that GPU-based programming was capable of rendering large 3-D datasets at real-time interactive rates with stereographic displays.
Keywords: programmable graphics processing units, stereoscopic, display, real time, image rendering
1. Introduction
Inherently 3-D medical imaging modalities, such as Computerized Tomography (CT) and Magnetic Resonance (MR) imaging systems, are generating an ever increasing volume of image data that must be reviewed by radiologists. This trend will almost certainly continue into the future as radiologists, in an effort to increase spatial resolution, depict 3-D volumes by using thinner, but more numerous, slices.
1.1. Current Display Paradigms
By far, the most common method used for viewing inherently 3-D data has been reading 2-D slices sequentially, from the 3-D dataset, in a slice-by-slice mode, a laborious and error prone process, or viewing the data as projections of thicker sections comprised of multiple adjacent slices.
It is known that the visibility of certain kinds of subtle features can be increased by presenting the data as a thicker 3-D volume [1,2], rendered with an appropriate projection algorithm. This is normally achieved by combining the thin slices directly acquired during volumetric imaging to form a thicker slab, and then projecting this slab onto a 2-D display, but increasing the thickness of projected volumes can cause ambiguities due to the superposition of tissues. Also, use of an averaging process to combine slices can reduce the contrast of features that are small relative to the thickness of the resulting slab. As slabs become thicker by adding more of the originally acquired thin slices, the contrast of smaller features, which often are visible on only one or two thin slices, may be reduced by averaging with the remaining thin slices [1]. As slices become thinner, the signal-to-noise ratio in individual slices is reduced making it more difficult to detect certain kinds of features, and at the same time, the number of slices that must be read increases. Furthermore, the process of reading individual slices sequentially forces viewers to reconstruct mentally the 3-dimensional structure, and does not permit the reader’s visual system to take full advantage of correlations between adjacent slices to improve apparent signal-to-noise ratios.
Various methods for 3-D display of volumetric radiographic datasets have been devised to make the reading process more efficient, but they have not been widely adopted because of certain performance limitations. Specifically, the task of rendering 3-D datasets in a form that is suitable for radiological applications is computationally intensive and it has not been possible to perform these calculations sufficiently fast to be able to provide radiologists with real-time interactive displays, except on superpremium computers. There is a consensus that, without real-time interactivity, volumetric display (monoscopic or stereoscopic) is often not justified by the added complexity.
1.2. Potential Role of Stereographic Displays
Stereographic display of 3-D radiographic datasets, which takes full advantage of readers’ binocular vision, may provide benefits beyond those attributed to monoscopic 3-D display [3]. Certain kinds of objects can be detected in a stereo 3-D display of data, which cannot be detected when the data is viewed in a slice-by-slice manner. Stereo projection can improve the visibility of objects by enhancing features that are correlated between slices, while reducing noise in a manner analogous to the signal-to-noise improvements obtained by averaging slices or MIPs - but stereo projection does not introduce tissue superposition ambiguities that would be caused by these methods [4]. Nevertheless, stereographic presentation has received even less attention than monoscopic 3-D because it further increases the computational burden.
1.3. Application of GPUs
With the evolution of commodity graphics processing units (GPUs) for accelerating games on personal computers, over the past couple of years, the amount of computing power that is available for rendering complex scenes has been rapidly increasing. GPUs may be capable of performing a wide range of reconstruction, volume reformatting and stereo projection in real-time under user control. In particular, the most recent GPUs are approaching a performance level where real-time interactivity with stereographic displays is feasible.
GPUs are organized as pipelined parallel processors. They differ from general purpose processors, that basically perform one instruction at a time and need to have the result returned immediately, in that they process parallel streams of independent data and can wait for an individual result as long as the entire dataset is processed quickly [5]. In this sense, they are ideal for tasks that are computationally intensive in volumetric rendering of 3-D datasets. Dietrich, et al, report that they were able to achieve real-time rendering of a 512 × 512 × 100 liver CT dataset on a 2 GHz Pentium 4, with a ATI 9800 GPU, though they were primarily concerned with only the volume clipping component of the rendering algorithm [6].
Several researchers, including our own, have shown the potential benefit of GPUs for efficient image manipulation and visualization within medical applications [7-12]. For example, Briggs, et al, have demonstrated a display for volumetric electrical impedance tomography [7]. While their datasets are smaller than many that occur in radiology, they were able to achieve real-time performance. A Doppler-ultrasound display was implemented by Heid, et al, by exploiting the performance of a GPU [8]. GPU-based programming has been implemented for interactive 4-D motion segmentation and volume rendering of cardiac data and has resulted efficient data processing and visualizing with high quality and at real-time speeds [9]. GPUs were also demonstrated to be efficient in generating high quality reconstructed radiographs from portal images and CT volume data for radiation therapy [10]. Sorensen, et al, have also applied the technique to surgical simulation of the liver to achieve a real-time performance, where surface rendering involving dynamic geometric transformations and texture manipulations were implemented on a GPU [11]. These specialized applications can often achieve significant levels of performance by optimizing their systems for the application, but these systems do not necessarily retain that performance when used in a different context.
We have tested the feasibility and efficacy of performing renderings on GPUs for stereo display of medical 3-D dataset. Previously, we prestaged and prerendered stereo pair renderings of lung CT images for display. Because of different viewing positions and viewing volumes, rendering a complete set of image pairs for a case took a substantial amount of time and consumed vast storage space. Such a practice may work within certain research environments, but is not practical for the general clinical settings, where real-time rendering and manipulation are necessary for prompt and accurate diagnosis.
While GPUs have been applied to a number of radiological imaging tasks, their potential performance characteristics are not well understood. This study is an attempt to measure frame rates that can be achieved for stereographic rendering on a GPU and compares these to rates that can be achieved on central processing units (CPUs) alone.
2. Materials and Methods
2.1. Data set
Images used for developing GPU-based rendering and display were obtained from a 4-detector CT scanner (LightSpeed Plus, GE medical Systems, Milwaukee, WI) for lung cancer screening program. The CT images were acquired in the axial plane and reconstructed to a thickness of 2.5 mm/slice with lung kernel reconstruction algorithm provided by GE standard software. The pixel size on each slice ranges from 0.63-mm × 0.63-mm to 0.92-mm × 0.92-mm. There are approximately 512×512×100 data voxels for a typical lung CT case in our dataset.
2.2. Hardware
The study was run on an off-the-shelf personal computer with a 2.0 GHz AMD Athlon 64 3200+ processor and 512 MB of RAM. The computer is equipped with a 128 MB NVIDIA Quadro FX 1100 graphics card, which has build-in support for stereographic buffering system to hold left- and right-eye images in separate frame buffers and to swap frame buffers for a frame-swapped display. The stereo image pairs are viewed either on CRT monitors via shutterglasses controlled by frame-swapping signals or on superimposed cross-polarized displays via passive polarizing eyeglasses.
2.3. Volume rendering for stereo display
Two rendering methods, Maximum Intensity Projection (MIP) and averaging, have been implemented to generate stereo pairs of the lung CT images. Because lesions must be detected before they can be evaluated, high contrast MIP images were preferable for lesion detection while images rendered by averaging, which preserves local geometry, were preferable for lesion evaluation [12,13]. The rendering process for both MIP and averaging in this application involves perspective transformation [14], transparency modeling based on optical occlusion/distance characteristics (depth-based brightness weighting), and ray casting [12-13,15-18]. All the rendering processes that we have previously performed on CPU card can be now processed on a programmable GPU card.
2.4. Rendering on GPUs
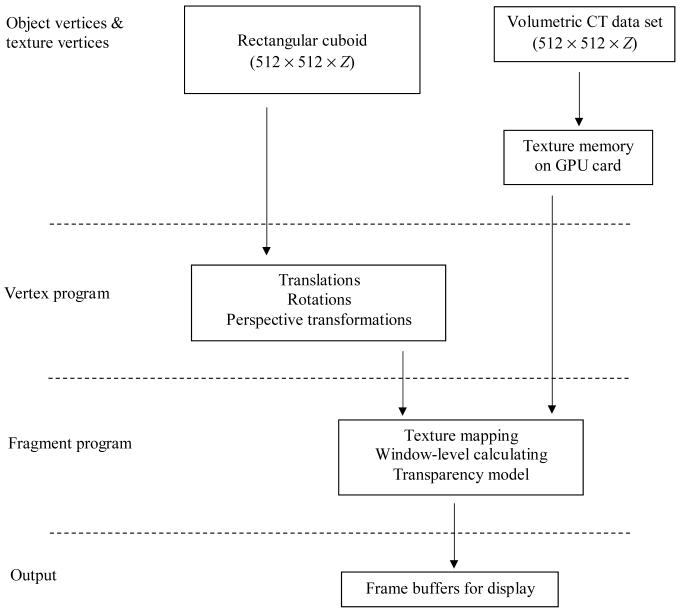
Stereographic compositing and display was implemented and compiled in the OpenGL and Cg languages on NVIDIA programmable GPUs. A flowchart, shown in Figure 1, illustrates the operations performed on GPU card.
Figure 1.
A diagram of stereo image rendering process on GPU card. Z is the depth measure of a given rendering volume.
For a given slab thickness, a vertex block with dimensions of 512×512×thickness was generated to include all vertices for perspective transformation and texture-coordinates. The dimensions of each vertex were approximated so as to be isotropic in all three axes (x, y and z) based on acquired x and y dimensions. Vertex-coordinates and texture-coordinates were then specified and interpolated during rasterization before being input to the vertex and fragment programs.
A sufficient number of interpolated slices were generated to provide continuity of display in the axial direction. Typically, for a dataset such as the one employed in this project, 3 interpolated slices are generated for every real slice.
Perspective projection in ray casting was performed in vertex program for each input vertex. The matrices for perspective transformation were determined by a presetting of eye-offsets and viewing distance. In the case of stereo compositing, the projection centers for the left- and right-eye images are offset laterally relative to each other. The parallax value for each eye-offset is set close to 1° to achieve stereo depth perception while avoiding excessive eyestrain. The rotation transform was also performed in vertex program. Transformed vertices that were out of the clip volume were not used for display. An example of Cg vertex program for vertex transformations is shown in Code 1.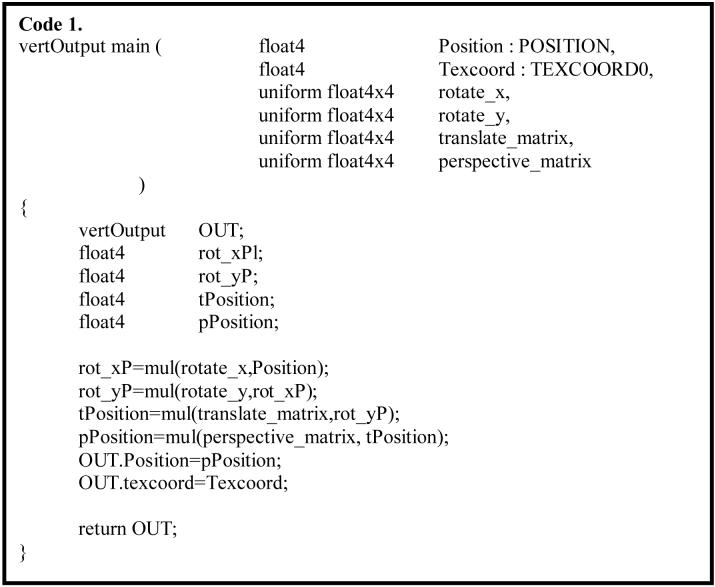
Once a vertex has been geometrically transformed to a proper position, texture mapping for the vertex takes place in a fragment program. The 16-bit lung CT volume data (approximately 512×512×100) was loaded into the graphics memory to serve as a 3-D texture map. Texture values were automatically interpolated in the texture map with the OpenGL linear filter function for a given texture-coordinate. Occlusion/distance based transparency and window-level settings were also implemented in the fragment program. A Cg code fragment implementation is shown in Code 2.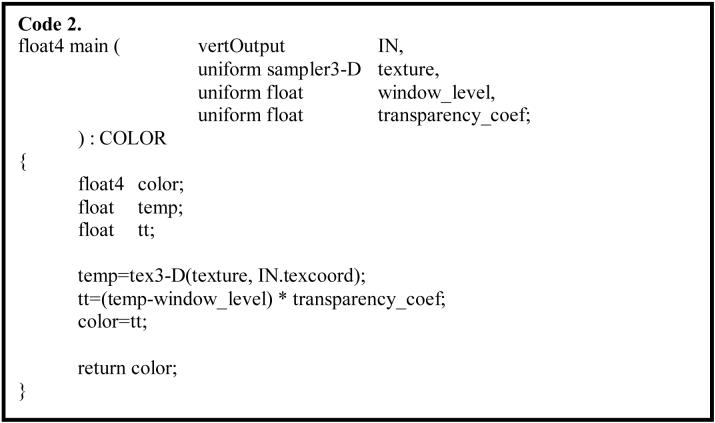
The final rendering process for displayed pixels was actualized by implementing the OpenGL blending functions. For MIP rendering the display value of each pixel was rendered by taking the maximum value among the points of a projection ray using MAX blending function, while for averaging rendering the display value was rendered by adding distance-weighted fractions of each fragment along a projection ray to the pixel using the ADD blending function.
2.5. Rendering on CPU card
The stereo image pairs for a lung CT case were prestaged and precalculated for all volume sizes between 1 up to 45 interpolated slices (see column 1 in table 1 and table 2) at all axial viewing positions. The detailed methods can be found in the references [13, 19]. In brief, we used trilinear interpolation to resample the data for a given volume of CT images to achieve final pixel dimension close to isotropic. Perspective transformation and ray casting based on compositing methods were performed for each pixel on stereo images. For MIP rendering the highest voxel value along a projection ray was used for projection value, while for averaging rendering each voxel value along a projection ray contributed a fraction to the final projection value.
Table 1.
Frame rates measured as stereo pairs per second for rendering on GPU card and CPU card at different number of interpolated slices
| Number of images for rendering | GPU (stereo pairs per second) | CPU (stereo pairs per second) |
|---|---|---|
| 1 | 103.3 | - |
| 9 | 20.1 | 1.3 |
| 15 | 13.2 | 0.8 |
| 21 | 10.1 | 0.5 |
| 33 | 6.6 | 0.3 |
| 45 | 5.0 | 0.2 |
Table 2.
Frame rates measured as stereo pairs per second for rendering on GPU card with and without rotation implementation
| Number of images for rendering | Rotation implemented (stereo pairs per second) | Without rotation (stereo pairs per second) |
|---|---|---|
| 1 | 103.3 | 103.3 |
| 3 | 44.4 | 44.4 |
| 5 | 33.7 | 33.7 |
| 9 | 20.1 | 20.1 |
| 15 | 13.2 | 13.2 |
| 21 | 10.1 | 10.1 |
| 33 | 6.6 | 6.6 |
| 45 | 5.0 | 5.0 |
2.6. Other functionality
An OpenGL based window display was built for displaying both GPU- and CPU-based stereo images. Specifically, window-level adjustment, viewing volume and viewing position selection and choice between MIP rendering and averaging rendering, were implemented. Image rotations were only performed by GPU-based rendering.
3. Results
The GPU-based program achieved real-time rendering and real-time display rates without any perceptible delay in the display of successive frames, following a user controlled frame switch command. We found no difference in frame rates between renderings by MIP and by averaging. A comparison of the rendering rates between GPU- and CPU-rendering, for our lung CT dataset, is shown in Tables 1 and 2. Table 1 lists the frame rate measurements of stereo compositing on GPU card as well as on CPU card at various volume sizes. The highest volume we rendered for lung CT images is 45 images, which is equivalent to a 37.5mm-thick slab of CT images or 15 CT slices at 2.5mm thickness per slice. When we reviewed various stereo images with several experienced radiologists, we found that the preferred viewing volume for detection and diagnosis ranged from 3 to 7 CT slices (i.e., 9 to 21 interpolated images), and 15 CT slices (i.e., 45 interpolated images) contained too much information to be useful for detection and diagnosis. Even with volume of 15 CT slices, which has more than 23 million vertex rendering processings (512×512×45×2 stereo images), we still achieved a rate at 5-frames per second. Rendering performed on the CPU card resulted in much slower frame rates and would not give the impression of real-time interactivity. If we precalculate all of these stereo image pairs for a case, it would take less than a minute on the GPU card versus more then 20 minutes on the CPU card. Implementing rotation on the GPU card did not measurably reduce frame rates for the data volumes used in this study, as shown in Table 2.
4. Discussion
Traditionally, 3-D medical image datasets are rendered predominantly on CPUs to generate precalculated images that can be prestaged for reading by radiologists. This preprocessing procedure puts many constraints on the review process and, at the same time, consumes a substantial amount of storage space and CPU time. These CPU-based processes most likely will be replaced in the near future by processes performed on the advanced graphics cards, due to the fact that these cards are becoming readily available and their real-time processing speed and improved arithmetic precision is making them suitable for the processing of many types of radiological images. The study presented in this paper shows that GPU-based rendering can achieve real-time interactive stereo display rates for lung CT images up to volumes larger than the optimal volume used for diagnosis. Monoscopic rendering rates, though not measured in this study, would likely be nearly double the stereoscopic rates for a given volume.
The benefit of using GPUs processing power can be widely appreciated in medical image detection and diagnosis. As show in Table 1, GPU-based programming renders stereo pairs in real-time for as many as 45 images interpolated from 15 CT slices at 2.5mm/slice (more than 23 million vertices) and gives no perceptional delay between frame changes. The capability of real-time process eliminates the constraints from prestaged paradigms.
Viewing angles, for example, can be important for detection and differentiation of an object. It is, however, impractical and impossible to prestage and precalculate all viewing angles for a set of images, or to perform smooth rotations. Whereas programmable GPUs perform real-time renderings, rotation functionality can be seamlessly and smoothly implemented during rendering process and consumes negligible GPU processing time compared to the overall processing time, as shown in Table 2.
From research conducted by others and our previous studies, we have observed that no single algorithm can meet all the requirements of clinical tasks. We have demonstrated that for stereo display, MIP rendering is the best for detection owing to the high contrast of rendered images, but not optimal for classification because of lack of local geometric fidelity in the rendered images. On the other hand, rendering by averaging will preserves local geometry despite providing low contrast of the rendered images. The two renderings can be used for different tasks during medical image interpretations. We have implemented this mechanism in CPU-based prestaged calculations and display, and the results were satisfactory at the expense of longer processing time and much more storage space. The GPU-based programming not only naturally solved this problem of dynamically switching between MIP and averaging renderings, but can also, in general, implement any algorithms, whichever needed, specific to the task in real time. This will dramatically improve efficacy of image presentation and diagnostic performance.
5. Summary
The amount of volumetric data being acquired in radiology is rapidly increasing. To maintain performance and efficiency in reading this data, it is desirable to be able to display the data as 3-D monoscopic- or stereoscopic-renderings, with real-time interactive control by radiologists. This paradigm has not been widely adopted because of the difficulty and expense of providing the required computational resources. With the availability of newer commodity graphics processing units for personal computers, it may be possible to overcome the computational impediments to interactive 3-D displays. This study compared the frame rates that can be achieved on CPUs to those that can be achieved by exploiting GPUs, and finds that GPUs are capable of rendering large 3-D datasets at real-time interactive rates.
Acknowledgement
This work is sponsored in part by the US Army Medical Research Acquisition Center, 820 Chandler Street, Fort Detrick, MD 21702-5014 under Contract PR043488, and also by grant CA109101-01A1 from the National Cancer Institute, National Institutes of Health. The content of the contained information does not necessarily reflect the position or the policy of the government, and no official endorsement should be inferred.
Biographies
Xiao Hui Wang
Xiao Hui Wang received her M.D. from Shanghai Second Medical College in 1982, after which she practiced as a pediatrician for five years in Shanghai Reijing Hospital. In 1994, she received her Ph.D. in neuroscience from Medical College of Pennsylvania, and in 2001, she received her MS degree in Information Sciences from the University of Pittsburgh. She is currently a Research Assistant Professor with the Department of Radiology at the University of Pittsburgh. Her research interests include medical image processing, visualization, and computer-assisted detection on medical images. Her current projects include interactive stereoscopic display for medical 3D images, volumetric dataset analysis and differential geometry based volumetric image segmentation for Radiology.
Walter F. Good
Walter Good received his B.S. Degree in mathematics from Case Western Reserve University in 1973 and his M.S. and Ph.D. from the University of Pittsburgh in 1984 and 1986, where he is currently a Research Professor in the Department of Radiology. Previously he held the position of Director of the Division of Imaging Technology at Allegheny University of the Health Sciences and subsequently the position of Director of Imaging Research in the Department of Radiology at the University of Pittsburgh. His current research interests include: development of display systems for Radiology, with a major emphasis on the use of stereographic displays for the efficient display of large 3D datasets; application of high performance computing on microprocessor based; analysis and display of temporal sequences of screening mammograms; computer-aided diagnosis in volumetric datasets; and, the application of methods based on differential geometry and human perception to the problem of image segmentation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Xiao Hui Wang, University of Pittsburgh, 300 Halket Street, Suite 4200, Pittsburgh, PA 15231, Fax: 412 641 2582, Phone: 412 641 2561, Email: xwang@mail.magee.edu.
Walter F. Good, University of Pittsburgh, 300 Halket Street, Suite 4200, Pittsburgh, PA 15231, Fax: 412 641 2582, Phone: 412 641 2560, Email: goodwf@upmc.edu.
References
- 1.Brown DG, Riederer SJ. Contrast-to-noise ratios in maximum intensity projection images. Magn Reson Med. 1992;23:130–137. doi: 10.1002/mrm.1910230114. [DOI] [PubMed] [Google Scholar]
- 2.Keller PJ, Drayer BP, Fram EK, et al. Angiography with two-dimensional acquisition and three-dimensional display. Work in progress. Radiology. 1989;173:527–32. doi: 10.1148/radiology.173.2.2798885. [DOI] [PubMed] [Google Scholar]
- 3.Roberts JW, Slattery OT. Display characteristics and the impact on usability for stereo. Proc. SPIE, Stereoscopic Displays and Virtual Reality Systems VIII. 2000;3957:128–138. [Google Scholar]
- 4.Maidment ADA, Bakic PR, Albert M. Effects of quantum noise and binocular summation on dose requirements in stereomammogry. Med Phys. 2003;30:3061–3071. doi: 10.1118/1.1621869. [DOI] [PubMed] [Google Scholar]
- 5.Ujval J, Kapasi UJ, Rixner S, et al. Programmable Stream Processors. Computer. 2003;36:54–62. [Google Scholar]
- 6.Dietrich CA, Nedel LP, Olabarriaga SD, et al. Real-time interactive visualization and manipulation of the volumetric data using GPU-based methods. SPIE Medical Imag. 2004;5367:181–191. [Google Scholar]
- 7.Briggs NM, Avis NJ, Kleinermann F. A real-time volumetric visualization system for electrical impedance tomography. Physiol. Meas. 2000;21:27–33. doi: 10.1088/0967-3334/21/1/304. [DOI] [PubMed] [Google Scholar]
- 8.Heid V, Evers H, Henn C, et al. Interactive real-time Doppler-ultrasound visualization of the heart. Stud Health Technol Inform. 2000;70:119–25. [PubMed] [Google Scholar]
- 9.Levin D, Aladl U, Germano G, et al. Techniques for efficient, real-time, 3-D visualization of multi-modality cardiac data using consumer graphics hardware. Computerized Medical Imaging and Graphics. 2005;29:463–475. doi: 10.1016/j.compmedimag.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 10.Khamene A, Bloch P, Wein W, et al. Automatic registration of portal images and volumetric CT for patient positioning in radiation therapy. Medical Image Analysis. 2006;10:96–112. doi: 10.1016/j.media.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Sørensen TS, Mosegaard J. Haptic feedback for the GPU-based surgical simulator. Medicine Meets Virtual Reality. 2006;14:523–528. [PubMed] [Google Scholar]
- 12.Wang XH, Good WF, Fuhrman CR, et al. Stereo Display for Chest CT. Proc SPIE EI2004. 2004;5291:17–24. [Google Scholar]
- 13.Wang XH, Good WF, Fuhrman CR, Sumkin JH, Britton CA, Warfel TE, Gur D. Projection Models for Stereo Display of Chest CT. Proc SPIE MI2004. 2004 In Press. [Google Scholar]
- 14.Mortenson ME. Computer Graphics Handbook: Geometry and Mathematics. Industrial Press; New York, NY: 1990. [Google Scholar]
- 15.Drebin RA, Carpenter L, Hanrahan P. Volume rendering. Comput Graphics. 1988;22:65–74. [Google Scholar]
- 16.Levoy M. Display of surfaces from volume data. IEEE Comput Graph Appl. 1988;8:29–37. [Google Scholar]
- 17.Ney DR, Drebin RA, Fishman EK, et al. Volumetric rendering of computed tomographic data: principles and techniques. IEEE Comput Graph Appl. 1990;10:24–32. [Google Scholar]
- 18.Ney DR, Fishman EK, Magid D, et al. Three-dimensional volumetric display of CT data: effect of scan parameters upon image quality. J Comput Assist Tomogr. 1991;15:875–885. doi: 10.1097/00004728-199109000-00033. [DOI] [PubMed] [Google Scholar]
- 19.Wang XH, Walter FG, Fuhrman CR, et al. Stereo CT Image Compositing Methods for Lung Nodule Detection and Characterization. Academic Radiology. 2005;12:1512–1520. doi: 10.1016/j.acra.2005.06.009. [DOI] [PubMed] [Google Scholar]