Abstract
We report the cloning and characterization of a tumor-associated carbonic anhydrase (CA) that was identified in a human renal cell carcinoma (RCC) by serological expression screening with autologous antibodies. The cDNA sequence predicts a 354-amino acid polypeptide with a molecular mass of 39,448 Da that has features of a type I membrane protein. The predicted sequence includes a 29-amino acid signal sequence, a 261-amino acid CA domain, an additional short extracellular segment, a 26-amino acid hydrophobic transmembrane domain, and a hydrophilic C-terminal cytoplasmic tail of 29 amino acids that contains two potential phosphorylation sites. The extracellular CA domain shows 30–42% homology with known human CAs, contains all three Zn-binding histidine residues found in active CAs, and contains two potential sites for asparagine glycosylation. When expressed in COS cells, the cDNA produced a 43- to 44-kDa protein in membranes that had around one-sixth the CA activity of membranes from COS cells transfected with the same vector expressing bovine CA IV. We have designated this human protein CA XII. Northern blot analysis of normal tissues demonstrated a 4.5-kb transcript only in kidney and intestine. However, in 10% of patients with RCC, the CA XII transcript was expressed at much higher levels in the RCC than in surrounding normal kidney tissue. The CA XII gene was mapped by using fluorescence in situ hybridization to 15q22. CA XII is the second catalytically active membrane CA reported to be overexpressed in certain cancers. Its relationship to oncogenesis and its potential as a clinically useful tumor marker clearly merit further investigation.
The growing carbonic anhydrase (CA) gene family includes nine enzymatically active CAs and three acatalytic CA-related proteins (CA-RPs). The active CAs catalyze the reversible hydration of CO2 in the reaction CO2 + H2O ⇄ HCO3− + H+ (1–4). The CA isozymes differ in their kinetic properties, their tissue distribution and subcellular localization, and their susceptibility to various inhibitors. They play important roles in diverse physiological processes including respiration, bone resorption, renal acidification, gluconeogenesis, signal transduction, and formation of cerebrospinal fluid and gastric acid (1–5). The CA-RPs are membrane proteins that contain a CA domain, but have amino acid substitutions in one or more of the three Zn-binding histidine residues in the active site of catalytic CAs (6). Although the CA-RPs are acatalytic in terms of CA activity, they show much higher sequence identity between mouse and human than the catalytic CAs, suggesting that each of them has an important function, nonetheless. An association of a CA domain with cancer was provided by the discovery that a complete, acatalytic CA domain forms the ligand-binding domain for each of two members of the receptor protein tyrosine kinase phosphatase (RPTP) family (RPTPβ and RPTPγ), one of which (RPTPγ) has been suggested to be a tumor suppressor gene (7, 8). More recently, a tumor-associated marker protein called MN, which was initially cloned from HeLa cells (9, 10), was found to contain a complete CA domain in the middle of its large extracellular segment and to have CA activity when the recombinant protein was expressed (11). The MN protein, which has since been renamed CA IX, was found to be expressed in tumors of several tissues in which it is not expressed normally (12) and also expressed in normal stomach (13) and proliferative enterocytes of the gut (14). Further evidence that CA IX is related to oncogenesis was provided by the observations that its expression in NIH 3T3 cells led to loss of contact inhibition, a shorter doubling time, a decreased dependence on serum growth factors, and loss of anchorage dependence as evidenced by ability to grow in soft agar (10).
We have discovered still another catalytically active CA associated with human cancers. A transcript was identified in mRNA from a human renal cell cancer (RCC) that turned out to encode a human membrane CA that was overexpressed in the clear cell RCC. In this report, we present the identification, cloning, and characterization of this CA, which we have designated CA XII
MATERIALS AND METHODS
Sera and Tissues.
The study has been approved by the local ethical review board (Ethikkommission der Ärztekammer des Saarlandes). Recombinant DNA work was done with the official permission and according to the rules of the state government of Saarland. Sera and tumor tissues were obtained during routine diagnostic or therapeutic procedures. Sera were stored at −80°C until use. Normal tissues were collected from autopsies of tumor-free patients.
Construction of cDNA Expression Libraries.
The construction of the RCC cDNA expression library has been described elsewhere (15). In brief, a cDNA expression library resulting in 1 × 106 primary clones was established by directionally cloning cDNA derived from the RCC of a 69-year-old woman with an RCC of clear cell type into the EcoRI- and XhoI-digested ZAPII phage (Stratagene).
Immunoscreening of Transfectants.
The immunoscreening for the detection of clones reactive with IgG antibodies in the 1:100-diluted autologous serum was described (15). Briefly, after transfection for primary screening and plaque transfer onto nitrocellulose membranes (Sartorius) and blocking with 5% (wt/vol) low fat milk Tris-buffered saline (Glücksklee, Nestlé, Vevey, Switzerland), nitrocellulose membranes were incubated overnight in 1:300-diluted autologous patient’s serum extensively preabsorbed against Escherichia coli proteins. An alkaline phosphatase-conjugated antibody specific for human IgG (Dianova, Hamburg, Germany) was used to visualize reactive clones.
Sequence and Structure Analysis of Identified Antigens.
Clones reactive with high-titer IgG antibodies were subcloned and submitted to in vivo excision (18) of pBluescript-phagemides (19). The nucleotide sequence of cDNA inserts was determined by using a Sequenase 2.0 kit (United States Biochemical). Initial sequencing was performed according to the manufacturer’s instructions using vector-specific reverse and universal primers. Subsequent sequencing was carried out with internal oligonucleotides. Sequence alignments were performed with dnasis (Pharmacia Biotech) and blast (20) software and on EMBL (21), GenBank (22), PROSITE (23), and TMpred (24) databases.
Cloning of Full-Length Transcript by Rapid Amplification of cDNA Ends.
The unknown 5′ end of the partial primary clone was cloned by using a modified rapid amplification of cDNA ends (25) protocol and adapter ligated cDNA synthesized from the original tissue mRNA. The products obtained after two subsequent rounds of PCR by using adapter-specific and HOM-3.1.3 specific oligonucleotides (3.1.3-RCI 5′-TGG TCC CAA GAC AAG GAG GCA CCC AGC-3′, 3.1.3-RC2 5′-ATG TTT GCA GAT TGA GCT ACA GAG AAC-3′) were separated on agarose gels. DNA bands were excised and cloned into the TA cloning vector (Invitrogen).
Northern Blot Analysis.
Northern blot analyses were performed with RNA extracted from tumors and normal tissues using guanidinium thiocyanate (26). RNA integrity was checked by electrophoresis in formalin/4-morpholinepropanesulfonic acid gels. Gels containing 10 μg of RNA per lane were blotted onto nylon membranes (Hybond N, Amersham). After prehybridization, the membranes were incubated with the specific CA XII cDNA probe overnight at 65°C in Express Hyb solution (CLONTECH). A nick-translated 32P-labeled 754-bp fragment amplified from the 5′ end (sense 5′-CGC GAA GAT GCC CCG GCG CAG CCT-3′; antisense 5′-CTG CTC CTG GGA AAT TTC CAC GGG GTT-3′) was used as a probe. The membranes were then washed at progressively higher stringency, with the final wash in 1× standard saline citrate (SSC; 1[times SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7) and 0.2% SDS at 65°C. Autoradiography was conducted at −70°C using Kodak X-Omat-AR film and intensifying screen. The size of the full-length mRNA was approximated by comparison with the mobilities of 28S and 16S ribosomal RNA. After exposure the filters were stripped and rehybridized with a glyceraldehyde-3-phosphate dehydrogenase probe.
Reverse Transcription–PCR.
Total cellular RNA from a panel of normal tissues was primed with a (dT)18 oligonucleotide and reverse transcribed with Superscript RT (GIBCO). The CA XII transcript was amplified by using sequence-specific primers comprising an 806-bp 3′ fragment (sense 5′-GGA CAA ATG GGG ACA GGA AGG ATC AAG-3′; antisense 5′-GAG GAC ATT TCA TGC TGA CAA AAT GAG-3′).
Cloning of CA XII from Normal Kidney Tissues.
First strand cDNA was synthesized from 10 μg total RNA from normal kidney tissue by using a (dT)18 oligonucleotide and Superscript reverse transcriptase (GIBCO). Amplification of CA XII cDNA was performed by using the transcript-specific oligonucleotides comprising the entire ORF and a large part of the 3′ untranslated region (sense 5′-CGC GAA GAT GCC CCG GCG CAG CCT-3′; antisense 5′-GAG GAC ATT TCA TGC TGA CAA AAT GAG-3′). PCR was performed for 35 cycles with Taq polymerase (Goldstar, Eurogentec, Brussels) with an annealing temperature of 68°C. The resulting 2,492-bp product was cloned into TA cloning vector (Invitrogen) and was sequenced using plasmid- and transcript-specific oligonucleotides. The sequence was aligned and compared with the tumor-derived full-length transcript using dnasis software.
Screening of Allogenic Sera on Identified Specific Phage Clones.
Phage assay with allogenic sera has been described (15). Positive phages found to be reactive with the patient’s serum were plaque purified, mixed with nonrecombinant phages as an internal negative control, and tested with allogenic sera from other patients and healthy controls extensively preabsorbed against E. coli antibodies.
Fluorescence in Situ Hybridization.
Metaphase spreads were prepared from peripheral blood of normal healthy donors by standard procedures (27). A fragment representing full-length CA XII was labeled by nick translation with Bio-16-dUTP according to the manufacturer’s instructions (BRL). Labeled DNA (2 ng) was precipitated together with 20 μg of human placenta DNA as competitor and dissolved in 5 μl of hybridization buffer (50% deionized formamide/2× SSC/50 mM sodium phosphate, pH 7.0/10% dextran sulfate). Hybridization conditions and detection were as described (28).
Production of Histidine-Tagged CA XII for Immunization and Production of Polyclonal Rabbit Antiserum.
5′ truncated CA XII cDNA was amplified with proofreading Pfu DNA polymerase (Stratagene) by using RNA from patient’s RCC and oligonucleotides from each end of the ORF (sense 5′-GGT TCC AAG TGG ACT TAT TTT GGT CCT-3′; antisense 5′-GGG ACC TCA AGC GTG GGC CTC AGT CTC-3′). The product was gel purified and ligated in frame to SmaI-digested, dephosphorylated and gel-purified pQE32 vector (Qiagen, Chatsworth, CA) to produce a fusion protein bearing an N-terminal 6-histidine tag. The construct was transformed into E. coli strain SG13009 (pREP4) and selected on kanamycin/ampicillin-containing plates. Individual colonies were picked and the fusion protein was expressed by induction with 2 mM isopropyl thiogalactoside and purified over Ni-nitrilotriacetate columns. A clone expressing a protein of the expected length was used for large-scale production after sequence verification. Cells were harvested 5 hr after induction, lysed in buffer A (8 M urea/100 mM Na2PO4/10 mM Tris⋅HCl, pH 8.0/0.01% Triton X-100) overnight. Debris was spun down and supernatant was loaded onto preequilibrated Ni-nitrilotriacetate resin, washed with 2 vol of buffer A (pH 8.0) and 10 vol of buffer A (pH 6.3), and eluted with 250 mM imidazole in buffer A. Polyclonal rabbit antisera were obtained from a custom antibody service (Eurogentec) after four immunizations with 100 μg of purified histidine-tagged fusion protein. Sera before and after immunization were tested by Western blot analysis for reactivity with the fusion protein.
Expression of CA XII cDNA in COS Cells.
The 1,292-bp 5′ fragment produced by HindIII, R1 digestion of a full-length clone was subcloned into the eukaryotic expression vector pCXN at the blunt-ended XhoI cloning site (29). COS-7 cells were transfected by the DEAE dextran method (30). After 72 hr, cells were scraped from the 60-mm dishes, washed, sedimented, and sonicated (twice for 20 sec each) in 25 mM Tris⋅H2SO4, pH 7.5 + 1 mM each of benzamidine, o-phenanthroline, and phenylmethylsulfonyl fluoride. The crude extract (200 μg of protein) was analyzed for CA activity as described (31, 32) in the presence and absence of 1 μM acetazolamide. For Western blot analysis, 20 μg of cell extract protein was resolved by SDS/PAGE (33) and the expressed protein was identified with polyclonal antibody to CA XII as first antibody, peroxidase-conjugated goat anti-rabbit IgG (Sigma) as second antibody, and the enhanced chemiluminescence reagent (Amersham). For deglycosylation, an equivalent amount of extract protein (20 μg) was treated with 100 milliunits of endoglycosidase (PNGase F) before SDS/PAGE and Western blot analysis (34).
RESULTS
Isolation of a Partial cDNA Clone and Full-Length Cloning of Human CA XII.
We screened 1.8 × 106 recombinant phage from a λ cDNA expression library prepared from mRNA isolated from a clear cell type RCC by using autologous serum from the RCC patient. Seven clones representing five different transcripts were identified. When these clones were used to probe Northern blots, one clone (HOM-RCC-3.1.3) was found to hybridize with a 4.5-kb transcript that appeared to be overexpressed in the RCC compared with the surrounding normal tissue (15, 17). Sequence analysis of this 1,670-bp clone identified a short coding sequence without an obvious translation start site. The 5′ end of this partial transcript was cloned by the rapid amplification of cDNA ends technique (25) in a 1,032-bp fragment. Alignment and connection of the two clones yielded a cDNA encoding 7 bp of 5′ untranslated sequence preceding a classical initiation codon (35) followed by a 1,062-bp ORF and 1,584 bp of 3′ untranslated sequence.
Nucleotide Sequence, Deduced Amino Acid Sequence, and Alignment of CA XII with Other Human CAs.
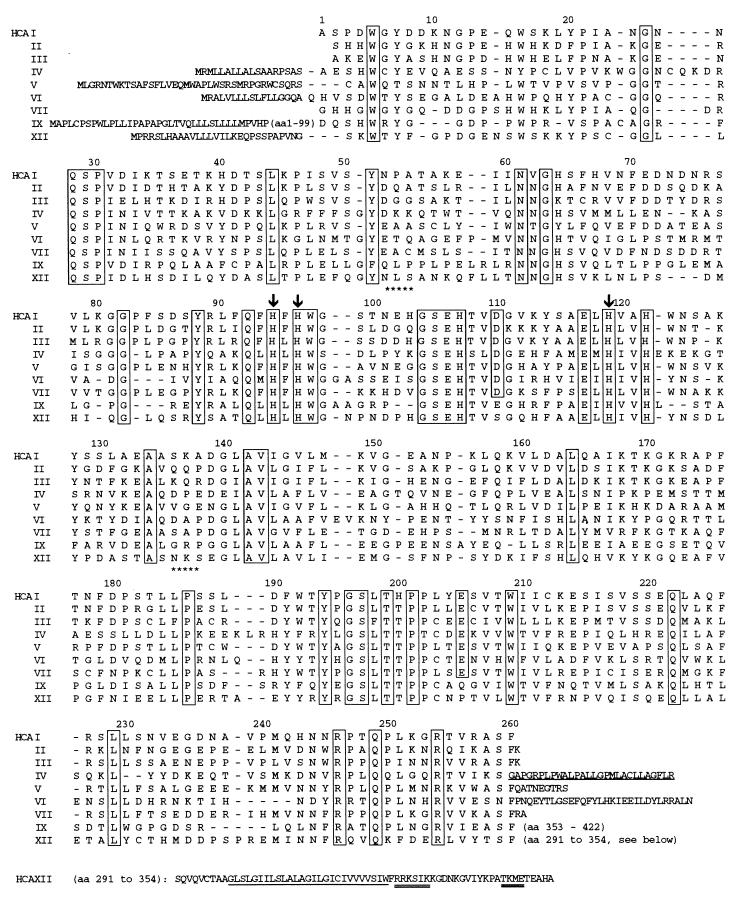
The ORF predicts a polypeptide sequence of 354 amino acids with a predicted mass of 39,448 Da that has features of a type I membrane protein. Database searches showed overall homologies to conserved regions of human CAs and CA-RPs of 30 to 42%. In accordance with the convention of naming CAs and CA-RPs in the order of their discovery, we designated this protein CA XII. Fig. 1 presents the predicted amino acid sequence of CA XII aligned below the sequences of human CAs I–VII and IX. The N-terminal extension of 29 amino acids fulfills the criteria for a signal sequence preceding the predicted cleavage between Gly-1 and Ser-1 (36, 37). A 261-amino acid CA-homology domain follows that contains most of the amino acid residues conserved among the active CAs (1). These are indicated by the boxes. The numbering above the aligned sequences indicates the amino acid number of the residues in CA I. The CA XII sequence contains all three Zn-binding histidine residues (His-94, His-96, and His-119) that are present in the active sites of catalytically active CAs (1, 38). Note also that His-64 is conserved. His-64 has been shown to contribute to the efficiency of high-activity CAs by serving as a proton shuttle between the Zn-bound water molecule and surrounding buffer molecules (39–41). The extracellular domain contains two potential sites for asparagine glycosylation and four cysteine residues, two of which are near positions of two cysteines in CA IV and CA VI that form an important disulfide bond that stabilizes the structure of each of these two CAs (42). Hydropathy plots indicate a 26-amino acid hydrophobic segment (underlined in Fig. 1) that presumably serves as a transmembrane domain, that is followed by a 29-amino acid hydrophilic cytoplasmic C terminus that contains two potential sites for phosphorylation.
Figure 1.
Amino acid sequences of CA XII aligned below with sequences of other catalytic human CAs. Numbers above the aligned sequences correspond to numbering of amino acids in CA I. The conserved residues are boxed. Arrows above His-94, His-96, and His-119 indicate Zn-binding, active site residues. Stars below NLS and NKS indicate asparagine glycosylation sites. The N-terminal extensions represent the signal sequences of membrane CAs (IV, IX, and XII) and mitochondrial CA V. Cysteines at positions 23 and 203 (CA I numbering) are conserved between CA VI and CA XII. The C-terminal extension of CA XII (amino acid residues 291–354) includes the 26-amino acid hydrophobic domain (underlined) and potential sites for phosphorylation by casein kinase II, protein kinase C, and cAMP-dependent kinase (double underline). The underlined hydrophobic C-terminal extension of CA IV is cleaved off in glycosyl-phosphatidylinositol anchoring and is not found in the mature, glycosyl-phosphatidylinositol-anchored enzyme.
Expression of CA XII Immunoreactivity and Catalytic Activity in COS Cells.
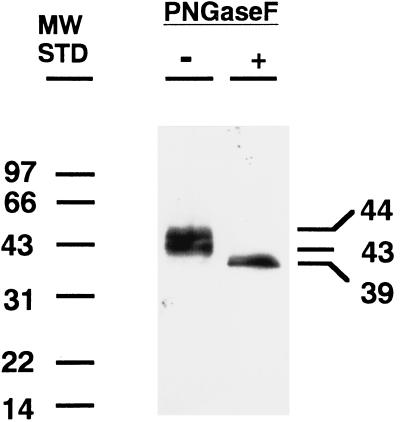
To characterize the protein expressed from the CA XII cDNA, we subcloned the coding sequence into a eukaryotic expression vector and measured the immunoreactivity and catalytic activity of the expressed protein in extracts of transfected COS cells. Fig. 2 shows the Western blot of untreated and PNGase F-treated extracts. A 43- to 44-kDa doublet was identified in the CA XII transfected COS cells by the polyclonal antibody to CA XII that collapsed to a single band at ≈39 kDa after PNGase F treatment, consistent with removal of two oligosaccharide chains. Table 1 presents results of measurements of CA activity in the extracts of COS cells infected with vectors expressing CA XII, CA IV, and vector only. The extract of CA XII transfected cells had ≈16% the activity of the extract from cells transfected with bovine CA IV, which is one of the highest activity isozymes. Both CA XII and CA IV activities were sensitive to inhibition by acetazolamide. All of the immunoreactive CA XII sedimented with membranes when the extract was centrifuged at 50,000 × g for 30 min (unpublished observations). Although purification of CA XII will be required to compare its kinetic properties with those of other CAs, it is clear from the above results that expressed recombinant CA XII has appreciable catalytic activity.
Figure 2.
Western blot of proteins in extracts of COS cells transfected with pCXN vector expressing CA XII that were incubated with or without endoglycosidase PNGase F. A 43- to 44-kDa doublet was identified with polyclonal antibody to CA XII that was reduced to ≈39 kDa by treatment with 100 milliunits of endoglycosidase PNGase F.
Table 1.
Expression of CA activity
| Construct | CA activity, enzyme units/mg cell protein
|
|
|---|---|---|
| − acetazolamide | + acetazolamide (1 μM) | |
| pCXN (vector only) | 0.25* | 0.0 |
| pCXN-HCAXII-5 | 2.25 | 0.3 |
| pCXN-HCAXII-7 | 2.15 | 0.2 |
| pCXN-bovine CA IV | 12.3 | 0.25 |
*The low activity seen in vector-only transfections is acetazolamide-sensitive and attributable to a low level of CA II expression in COS cells (A. Waheed, personal communication). Inhibition of CA activity was 87% and 91% for CA XII clones 5 and 7, respectively, and 98% for CA IV.
Tissue Expression Studies by Reverse Transcription–PCR and Northern Analysis.
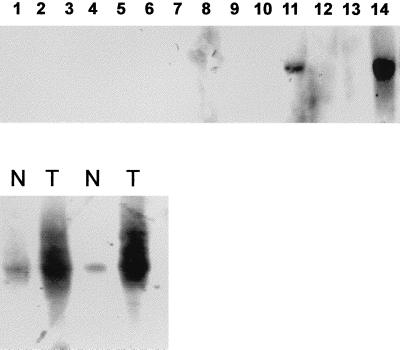
Reverse transcription–PCR with CA XII-specific primers detected this transcript in RNA from many normal tissues (lung, liver, colon, stomach, skeletal muscle, skin, kidney, spleen, tonsil, lymph node, peripheral blood lymphocytes, activated peripheral blood lymphocytes, bladder, breast, uterus, ovary, brain, prostate, and skin) as well as in a panel of different human neoplasms (RCC, colorectal cancer, lung cancer, and breast cancer). However, on Northern blots of RNA from normal tissues, the ≈4.5-kb transcript was detectable only in RNA from kidney, colon, and phorbol 12-myristate 13-acetate-activated peripheral blood lymphocytes (Fig. 3). Analysis of RNA from RCC tumor tissues demonstrated that CA XII is overexpressed (as compared with the corresponding normal renal tissue) in 10% of renal carcinomas of clear cell type as was the case in the tumor of the RCC patient from whose cDNA library the CA XII cDNA was cloned (15–17).
Figure 3.
(Upper) Lanes: Northern blots of mRNAs from normal human tissues and from pairs of specimens derived from RCC (T) and adjacent normal renal tissue (N). Normal tissues were lanes 1, testis; 2, stomach; 3, lung; 4, muscle; 5, liver; 6, spleen; 7, bladder; 8, prostate; 9, breast; 10, lung; 11, colon; 12, peripheral blood lymphocytes; 13, liver; and 14, kidney. Significant signals were seen only in mRNA from kidney and colon. (Lower) Lanes: normal kidney (N) adjacent to mRNA from kidney tumor (RCC) (T).
Chromosomal Localization.
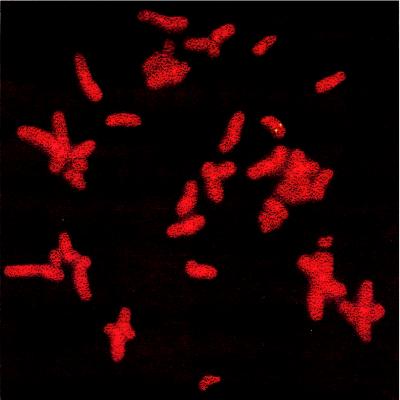
Fluorescence in situ hybridization was performed to localize the gene for human CA XII. Several independent hybridizations revealed a strong signal on the long arm of chromosome 15, namely at 15q22 (Fig. 4). No hybridization signal was seen on chromosomes 1, 8, 16, or 17 where other CAs have been localized (1).
Figure 4.
Chromosome localization of CA XII. Fluorescence in situ hybridization with full-length CA XII showed signal on chromosome 15q22.
CA XII Is Not Mutated in RCC.
To exclude mutations of the tumor-derived CA XII transcript as a reason for immunogenicity in the autologous patient, we cloned the normal counterpart cDNA from normal human kidney by reverse transcription–PCR amplification of the entire transcript. Sequencing revealed no differences from the tumor-derived cDNA, ruling out mutational alterations as a mechanism for the initiation of the humoral immune response in the patient.
Incidence of Anti-CA XII Antibodies in Human Sera.
Other human sera were tested in the above described phage assay for antibodies to CA XII. Four of 30 RCC patients and 1 of 30 normal controls showed antibodies against the CA XII phage clone, whereas none of 11 astrocytoma patients and none of 17 patients suffering from Hodgkin’s disease had any serum response.
DISCUSSION
Two other active CAs are membrane proteins like CA XII. CA IV, the first membrane isozyme described, is a glycosyl-phosphatidylinositol-anchored protein expressed on the apical surface of epithelial cells in colon (43), kidney (44), male genitourinary tract (45, 46), and on the plasma face of endothelial cells of several capillary beds, including the pulmonary microvasculature (47). CA IV facilitates the reversible hydration of CO2 at sites where CO2 and HCO3− flux across membranes needs to be very rapid. The second membrane CA described is CA IX, which, like CA XII, was discovered as a tumor-associated CA and has recently been found to be expressed in many RCCs of clear cell type (48). The CA domain is in the middle of the multidomain CA IX that has been reported to have transforming potential and to be overexpressed in tumors of tissues in which it is not normally expressed (10, 12, 48). N-terminal to the CA domain is a highly acidic (44%) proteoglycan-related domain (59 residues) that shows 37% sequence identity to the keratan sulfate attachment domain of the human cartilage aggregation proteoglycan, aggrecan (49). Whether the CA activity in CA IX is required for its transforming potential, or the CA domain might function in a ligand-binding role analogous to the acatalytic domain in RPTPβ and RPTPγ (7, 8, 50), remains to be established.
The evidence that CA XII is expressed in human cancers comes from several independent sources. While this work was in progress, we became aware of U.S. patent number 5,589,579, in which the inventors R. M. Torczynski and A. P. Bollon claim the DNA sequence and protein it encodes to be novel and specific for human lung cancer cells (51). The patent summarized Northern blot data showing expression in normal pancreas and kidney, and 50-fold more expression in the lung carcinoma cell line A549. The fact that overexpression of CA XII was seen in 10% of RCC of clear cell type provides further evidence that overexpression of CA XII is cancer-related, even if not limited to lung cancers. Although we also found low levels of expression detectable by PCR in many other tissues, the appropriate transcript on Northern blots was only seen in RNA from kidney, intestine, and phorbol myristate acetate-activated peripheral blood lymphocytes.
After this work was completed, we learned of a nearly identical sequence entered in GenBank by S. V. Ivanov, I. Kuzmin, M. H. Wei, S. Pack, L. Geil, E. Stanbridge, and M.I. Lerman entitled “A new family member, CA 12, of α-carbonic anhydrases is downregulated by the VHL gene” (GenBank accession no. AF037335). This sequence contains 108 additional base pairs of 5′ untranslated region. Regulation of CA XII expression by the VHL tumor suppressor gene could explain why we found CA XII overexpressed in RCCs, because inactivation of both copies of the VHL gene is reported to be an early event in development of clear-cell carcinomas (52).
The CA XII cDNA reported here was detected by an IgG antibody in the serum of the cancer patient (15–17). Serum antibodies to CA I and CA II, both rather widely expressed cytoplasmic CAs, have been described in humans with different autoimmune diseases like Sjögren syndrome, immune cholangiopathies, scleroderma, and systemic lupus erythematosus (20, 53–56). However, serum antibodies to CAs in tumor patients have not been investigated. In preliminary studies, we found 4 of 30 RCC patients to have high-titer antibodies against CA XII. As has been reported for autoantibodies to other CAs (54, 56, 57), we also detected antibodies to CA XII in 1 of 30 apparently healthy controls tested. The significance of this finding is not yet clear.
Antibodies to a protein in cancer patients might result from a response to a new epitope created by mutational alteration of the protein, such as in mutant p53 or mutant ras (58, 59). However, we established by sequencing that the CA XII cDNA in the RCC of our patient was identical to the cDNA derived from normal kidney. Thus, other mechanisms must be operative in this patient. It is known that overexpression itself may initiate immune responses to structurally normal proteins, as has been reported for HER-2/neu (60). Possibly, overexpression of the CA XII protein in the RCC of our patients led to the immune responses. An important question to be resolved by extending these studies is whether the autoantibodies to CA XII will be useful markers in screening (or following progression in) patients with RCC and other cancers.
The finding of a second membrane CA associated with human cancer raises another interesting question: Is overexpression of these membrane CAs simply a byproduct of malignant transformation, or do the membrane CAs actually contribute to malignant transformation and/or tumor progression? Conceivably, CA IX and CA XII (both of which are expressed in RCCs) could contribute a ligand-binding domain that is involved in transformation. Alternatively, the transformation might be enhanced by the presence of CA activity at the membrane of highly proliferating cells. If the CA activity at the membrane itself played a role in transformation, it would raise the possibility that isozyme-specific CA inhibitors might be developed that would diminish or abrogate the transforming potential of the membrane CAs.
Acknowledgments
We thank Dr. Lloyd Old for continuing motivating discussion. This work was supported by the Deutsche Forschungsgemeinschaft and by National Institutes of Health Grant DK40163.
ABBREVIATIONS
- RCC
renal cell cancer
- CA
carbonic anhydrase
- CA-RP
carbonic anhydrase-related protein
- RPTP
receptor protein tyrosine kinase phosphatase
- PNGase
endoglycosidase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF051882).
References
- 1.Sly W S, Hu P. Annu Rev Biochem. 1995;64:375–401. doi: 10.1146/annurev.bi.64.070195.002111. [DOI] [PubMed] [Google Scholar]
- 2.Tashian R E. Adv Genet. 1992;30:321–356. doi: 10.1016/s0065-2660(08)60323-5. [DOI] [PubMed] [Google Scholar]
- 3.Wistrand P J. Ann N Y Acad Sci. 1984;429:195–206. doi: 10.1111/j.1749-6632.1984.tb12333.x. [DOI] [PubMed] [Google Scholar]
- 4.Wistrand P J, Knuuttila K G. Kidney Int. 1989;35:851–859. doi: 10.1038/ki.1989.63. [DOI] [PubMed] [Google Scholar]
- 5.Tashian R E. BioEssays. 1989;10:186–192. doi: 10.1002/bies.950100603. [DOI] [PubMed] [Google Scholar]
- 6.Hewett-Emmett D, Tashian R E. Mol Phylogenet Evol. 1996;5:50–77. doi: 10.1006/mpev.1996.0006. [DOI] [PubMed] [Google Scholar]
- 7.Barnea G, Silvennoinen O, Shaanan B, Honegger A M, Canoll P D, D’Eustachio P, Morse B, Levy J B, LaForgia S, Huebner K, et al. Mol Cell Biol. 1993;13:1497–1506. doi: 10.1128/mcb.13.3.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wary K K, Lou Z W, Buchberg A M, Siracusa L D, Druck T, LaForgia S, Huebner K. Cancer Res. 1993;53:1498–1502. [PubMed] [Google Scholar]
- 9.Liao S Y, Brewer C, Zavada J, Pastorek J, Pastorekova S, Manetta A, Berman M L, DiSaia P J, Stanbridge E J. Am J Pathol. 1991;145:598–609. [PMC free article] [PubMed] [Google Scholar]
- 10.Pastorek J, Pastorekova S, Callebaut I, Mornon J P, Zelnik V, Opavsky R, Zatovicova M, Liao S, Portetelle D, Stanbridge E J. Oncogene. 1994;9:2877–2888. [PubMed] [Google Scholar]
- 11.Opavsky R, Pastorekova S, Zelnik V, Gibadulinova A, Stanbridge E J, Zavada J, Kettmann R, Pastorek J. Genomics. 1996;33:480–487. doi: 10.1006/geno.1996.0223. [DOI] [PubMed] [Google Scholar]
- 12.Zavada J, Zavadova Z, Pastorekova S, Ciampor F, Pastorek J, Zelnik V. Int J Cancer. 1993;54:268–274. doi: 10.1002/ijc.2910540218. [DOI] [PubMed] [Google Scholar]
- 13.Pastorekova S, Parkkila S, Parkilla A-K, Opavsky R, Zelnik V, Saarnio J, Pastorek J. Gastroenterology. 1997;112:398–408. doi: 10.1053/gast.1997.v112.pm9024293. [DOI] [PubMed] [Google Scholar]
- 14.Saarnio J, Parkkila S, Parkkila A-K, Waheed A, Casey M C, Zhou X Y, Pastorekova S, Pastorek J, Karttunen T, Haukipuro K, et al. J Histochem Cytochem. 1998;46:497–504. doi: 10.1177/002215549804600409. [DOI] [PubMed] [Google Scholar]
- 15.Sahin U, Tuereci O, Schmitt H, Cochlovius B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I, Pfreundschuh M. Proc Natl Acad Sci USA. 1995;92:11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sahin U, Tuereci O, Pfreundschuh M. Curr Opin Immunol. 1997;9:709–716. doi: 10.1016/s0952-7915(97)80053-2. [DOI] [PubMed] [Google Scholar]
- 17.Tuereci O, Sahin U, Pfreundschuh M. Mol Med Today. 1997;3:342–349. doi: 10.1016/s1357-4310(97)01081-2. [DOI] [PubMed] [Google Scholar]
- 18.Short J M, Fernandez J M, Sorge J A, Huse W D. Nucleic Acids Res. 1988;16:7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alting-Mees M A, Sorge J A, Short J M. Methods Enzymol. 1992;216:483–495. doi: 10.1016/0076-6879(92)16044-k. [DOI] [PubMed] [Google Scholar]
- 20.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1992;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Emmert D B, Stoehr P J, Stoesser G, Cameron G N. Nucleic Acids Res. 1994;22:3445–3449. doi: 10.1093/nar/22.17.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benson D A, Boguski M, Lipman D J, Ostell J. Nucleic Acids Res. 1994;22:3441–3444. doi: 10.1093/nar/22.17.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bairoch A, Bucher P. Nucleic Acids Res. 1994;22:3583–3589. [PMC free article] [PubMed] [Google Scholar]
- 24.Hofmann K, Stoffel W. Biol Chem Hoppe Seyler. 1993;347:166–168. [Google Scholar]
- 25.Frohman M A, Dush M K, Martin G R. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 27.Arahi D T, Sparkes R S. Cytogenetics. 1963;2:57–61. doi: 10.1159/000129767. [DOI] [PubMed] [Google Scholar]
- 28.Gottert E, Klein V, Piontek K, Overmyer K, Zang K D, Meese E. Hum Genet. 1993;92:623–626. doi: 10.1007/BF00420950. [DOI] [PubMed] [Google Scholar]
- 29.Niwa H, Yamamura K, Miyazaki J. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 30.Tamai S, Waheed A, Cody L B, Sly W S. Proc Natl Acad Sci USA. 1996;93:13647–13652. doi: 10.1073/pnas.93.24.13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maren T H. J Pharmacol Exp Ther. 1960;130:26–29. [PubMed] [Google Scholar]
- 32.Sundaram V, Rumbolo P, Grubb J, Strisciuglio P, Sly W S. Am J Hum Genet. 1986;38:125–136. [PMC free article] [PubMed] [Google Scholar]
- 33.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Waheed A, Zhu X L, Sly W S, Wetzel P, Gros G. Arch Biochem Biophys. 1992;294:550–556. doi: 10.1016/0003-9861(92)90724-b. [DOI] [PubMed] [Google Scholar]
- 35.Kozak M. Mamm Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 36.von Heijne G. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 37.Perlman D, Halvorson H O. J Mol Biol. 1983;167:391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- 38.Hewett-Emmett D, Tashian R E. In: Carbonic Anhydrases: Cellular Physiology and Molecular Genetics. Dodgson S J, Tashian R E, Gros G, Carter N D, editors. New York: Plenum; 1991. pp. 15–32. [Google Scholar]
- 39.Tu C K, Silverman D N, Forsman C, Jonsson B H, Lindskog S. Biochemistry. 1989;28:7913–7918. doi: 10.1021/bi00445a054. [DOI] [PubMed] [Google Scholar]
- 40.Engstrand C, Forsman C, Liang Z W, Lindskog S. Biochim Biophys Acta. 1992;1122:321–326. doi: 10.1016/0167-4838(92)90412-7. [DOI] [PubMed] [Google Scholar]
- 41.Ren X, Lindskog S. Biochim Biophys Acta. 1992;1120:81–86. doi: 10.1016/0167-4838(92)90427-f. [DOI] [PubMed] [Google Scholar]
- 42.Whitney P L, Briggle T V. J Biol Chem. 1982;257:12056–12059. [PubMed] [Google Scholar]
- 43.Fleming R E, Parkkila S, Parkkila A-K, Rajaniemi H, Waheed A, Sly W S. J Clin Invest. 1995;96:2907–2913. doi: 10.1172/JCI118362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown D, Zhu X L, Sly W S. Proc Natl Acad Sci USA. 1990;87:7457–7461. doi: 10.1073/pnas.87.19.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parkkila S, Parkkila A-K, Kaunisto K, Waheed A, Sly W S, Rajaniemi H. J Histochem Cytochem. 1993;41:751–757. doi: 10.1177/41.5.8468457. [DOI] [PubMed] [Google Scholar]
- 46.Kaunisto K, Parkkila S, Parkkila A-K, Waheed A, Sly W S, Rajaniemi H. Biol Reprod. 1995;52:1350–1357. doi: 10.1095/biolreprod52.6.1350. [DOI] [PubMed] [Google Scholar]
- 47.Fleming R E, Crouch E C, Ruzicka C A, Sly W S. Am J Physiol. 1993;265:L627–L635. doi: 10.1152/ajplung.1993.265.6.L627. [DOI] [PubMed] [Google Scholar]
- 48.McKiernan J M, Buttyan R, Bander N H, Stifelman M D, Katz A E, Chen M-W, Olsson C A, Sawczuk I S. Cancer Res. 1997;57:2362–2365. [PubMed] [Google Scholar]
- 49.Doege K J, Sasaki M, Kimura T, Yamada Y. J Biol Chem. 1991;266:894–902. [PubMed] [Google Scholar]
- 50.Peles E, Natir M, Campbell P L, Sakurai T, Martinez R, Lev S, Clary D O, Shilling J, Barnea G, Plowman G D, et al. Cell. 1995;82:251–260. doi: 10.1016/0092-8674(95)90312-7. [DOI] [PubMed] [Google Scholar]
- 51.Torczynski, R. M. & Bollon, A. P. (1996) U.S. Patent 5,589,579.
- 52.Linehan W M, Klausner R. In: The Genetic Basis of Human Cancer. Vogelstein B, Kinzler K W, editors. New York: McGraw-Hill; 1998. pp. 455–473. [Google Scholar]
- 53.Gordon, S. C., Quattrociocchi-Longe, T. M., Khan, B. A., Kodali, V. P., Chen, J., Silverman, A. L. & Kiechle, F. L. Gastroenterology 108, 1802–1809. [DOI] [PubMed]
- 54.Kiechle F L, Quattrochiocchi-Longe T M, Brinton D A. Am J Clin Pathol. 1994;101:611–615. doi: 10.1093/ajcp/101.5.611. [DOI] [PubMed] [Google Scholar]
- 55.Inagaki Y, Jinno-Yoshida Y, Hamasaki Y, Ueki H. J Dermatol Sci. 1991;2:147–154. doi: 10.1016/0923-1811(91)90060-b. [DOI] [PubMed] [Google Scholar]
- 56.Itoh Y, Reichlin M. Arthritis Rheum. 1992;35:73–82. doi: 10.1002/art.1780350112. [DOI] [PubMed] [Google Scholar]
- 57.Kino-Ohsaki J, Nishimori I, Morita M, Okazaki K, Yamamoto Y, Onishi S, Hollingworth M A. Gastroenterology. 1996;110:1579–1586. doi: 10.1053/gast.1996.v110.pm8613065. [DOI] [PubMed] [Google Scholar]
- 58.Canevari S, Pupa S M, Menard S. Ann Oncol. 1996;7:227–232. doi: 10.1093/oxfordjournals.annonc.a010564. [DOI] [PubMed] [Google Scholar]
- 59.Schlichtholz B, Legros Y, Gillet D, Gaillard C, Marty M, Lane D, Calvo F, Soussi T. Cancer Res. 1992;52:6380–6384. [PubMed] [Google Scholar]
- 60.Disis M L, Calenoff E, McLaughlin G, Murphy A E, Chen W, Groner B, Jeschke M, Lydon N, McGlynn E, Livingston R B, et al. Cancer Res. 1993;54:16–20. [PubMed] [Google Scholar]