Abstract
Mutations are introduced into rearranged Ig variable genes at a frequency of 10−2 mutations per base pair by an unknown mechanism. Assuming that DNA repair pathways generate or remove mutations, the frequency and pattern of mutation will be different in variable genes from mice defective in repair. Therefore, hypermutation was studied in mice deficient for either the DNA nucleotide excision repair gene Xpa or the mismatch repair gene Pms2. High levels of mutation were found in variable genes from XPA-deficient and PMS2-deficient mice, indicating that neither nucleotide excision repair nor mismatch repair pathways generate hypermutation. However, variable genes from PMS2-deficient mice had significantly more adjacent base substitutions than genes from wild-type or XPA-deficient mice. By using a biochemical assay, we confirmed that tandem mispairs were repaired by wild-type cells but not by Pms2−/− human or murine cells. The data indicate that tandem substitutions are produced by the hypermutation mechanism and then processed by a PMS2-dependent pathway.
During somatic hypermutation of Ig variable (V) genes in B lymphocytes, a large number of nucleotide substitutions are introduced into a small area of the genome for the purpose of generating variant antibodies with high affinity for cognate antigens. Substitutions are distributed at a frequency of 10−2/bp over a 2-kilobase region of DNA that includes the rearranged V gene and noncoding flanking sequences (1–4). Transgenic mice with modified Ig genes have shown that both promoter and enhancer transcription elements are required for hypermutation, perhaps by making the region more accessible to proteins causing mutation (5–8). More than 90% of the mutations are base substitutions, and the rest are deletions and insertions (4, 9). An analysis of the substitutions shows that transitions occur more frequently than transversions, and there are fewer mutations of T than of the other three nucleotides (10). Thus, the substitutions are somewhat asymmetric on each of the two strands (11), but it is not known whether they are preferentially introduced into one strand and/or preferentially removed from one strand. The presence of multiple mutations implies that DNA repair pathways are involved in generating or removing them. We therefore studied the role of nucleotide excision repair and mismatch repair on the frequency and pattern of hypermutation.
A possible role for nucleotide excision repair in somatic hypermutation of V genes has been proposed. Nucleotide excision repair is especially efficient in actively transcribed genes (reviewed in ref. 12), and the generation of base substitutions by the hypermutation process has been broadly correlated with the extent of transcription (5). Furthermore, mutagenesis and transcription are linked in yeast (13). DNA damage in the V region, or transcriptional stalling at pause sites, might be envisaged to initiate nucleotide excision repair events with an occasional error occurring during gap filling (6). Mice lacking the xeroderma pigmentosum group A gene (Xpa) were examined because XPA is one of the first proteins in a plethora of factors that binds to damaged nucleotides. XPA-deficient mice fail to exhibit nucleotide excision repair of thymine dimers and (6–4) photoproducts, and they are susceptible to ultraviolet- and chemical-induced skin tumors (14).
A role for mismatch repair is more likely because mismatched base pairs would be present immediately after hypermutation. In eukaryotic mismatch repair (15–18), mismatched base pairs are recognized by a dimer of the MSH2 protein with either the MSH6 or MSH3 proteins. Subsequent steps involve the action of PMS2, MLH1, and proliferating cell nuclear antigen proteins, which result in excision of the error, followed by resynthesis of the repair patch. The mismatch repair mechanism is strand-specific, although the signal that directs repair to the newly replicated strand in eukaryotes is not known. Mutations could be introduced during resynthesis of the repair patch by an error-prone polymerase, or some mismatches could be removed during repair. Mice lacking the Pms2 gene were examined because PMS2 is essential for mismatch repair. PMS2-deficient mice have no repair as shown by instability of microsatellite repetitive sequences, and they are prone to lymphomas and sarcomas (19).
MATERIALS AND METHODS
Mice.
Xpa−/− and Pms2−/− mice on a C57BL/6 background were produced as previously described (14, 19), and C57BL/6 mice were obtained from Jackson Laboratories. Several mice from each group were given a primary i.p. injection of 50 μg of phenyloxazolone coupled to chicken serum albumin in complete Freund’s adjuvant, boosted 1 month later with 50 μg of antigen in incomplete Freund’s adjuvant, and sacrificed 4 days later. Spleens were removed, and B cells that bound the B220 surface marker and peanut agglutinin were isolated by flow cytometry.
DNA Cloning and Sequencing.
DNA was isolated from about 50,000 cells after proteinase K digestion and phenol/chloroform extraction. To amplify the rearranged VκOx1 gene, the DNA was subjected to 30 rounds of PCR with Pfu polymerase (Stratagene) using a primer specific for the leader sequence on the 5′ side of the gene and a primer specific for the Jκ5 gene segment on the 3′ side. Part of the reaction (1/25th) then was amplified for another 30 rounds using nested primers with restriction sites for cloning the 466-bp product into M13 bacteriophage. The error rate for Pfu polymerase is 1.3 × 10−6 mutations per nucleotide per duplication, which would produce 0.03 mutations per 466-bp clone after 60 rounds of amplification. This amount is 200-fold less than the number found in VκOx1 genes that were generated by somatic hypermutation. M13 plaques were screened for inserts by hybridization to a VκOx1 probe, and positive clones were sequenced.
Statistical Analyses.
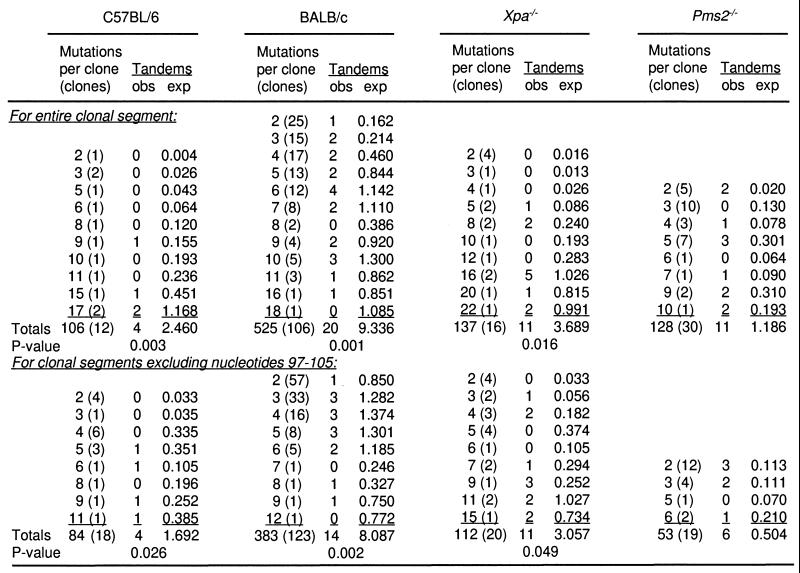
The distribution of types of substitutions between strains was compared by using the χ2 homogeneity test. Computer simulations and selected exact calculations indicate that the expected number of tandem mutations when n mutations are randomly distributed in a sequence of k consecutive nucleotides is equal to n(n−1)/k. To determine the expected mutations in the entire clonal segment listed in the first part of Fig. 4, for C57BL/6, Xpa−/−, and Pms2−/− clones, k was 466; whereas for BALB/c clones, k varied in length from 276 to 466. Exclusion of nucleotides 97–105 in the second part of Fig. 4 results in division of the 466-nt fragment shown in Fig. 1 into two segments, and the expected number of tandem mutations was calculated as n1(n1−1)/k1 + n2(n2−1)/k2. For C57BL/6, Xpa−/−, and Pms2−/− clones, the initial segment included nucleotides −190 through 96 (k1 = 286) and the final segment included nucleotides 106 through 276 (k2 = 171). For BALB/c clones, k1 ranged in length from 96 to 286 nucleotides and k2 had 171 nucleotides.
Figure 4.
Frequency of tandem mutations. Data for BALB/c mice was obtained from hybridomas and transgenes taken from the literature (8, 22, 24, 26–28). Expected numbers of tandem pairs were calculated for each total number of mutations. Two-sided P-values are based on exact Poisson calculations comparing the ratio of total observed tandem mutations to total expected tandem mutations in Pms2−/− mice to the corresponding ratios in the other three strains of mice (29).
Figure 1.
Nucleotide sequence of 5′ flanking and coding region of VκOx1 (21, 22). Dots are placed at 10-base intervals. The flanking region is labeled with negative numbers, and the coding region is labeled with positive numbers. Amino acid codons 34 and 36 in the first complementarity-determining region are underlined.
Mismatch Repair Assay.
Cell-free extracts were prepared from the following lines: HeLa-S3, wild-type human cervical carcinoma; GM0131, wild-type human fibroblasts; HEC-1-A, Pms2−/− human endometrial carcinoma; and C18, Pms2−/− murine embryonic fibroblasts (19). DNA substrates containing single and tandem double mismatches were prepared (20) and contained a nick in the minus strand at position −264, where position +1 is the first transcribed base of the lacZα gene. The G⋅G mispair was at position 88, and the AA⋅⋅CC and GG⋅⋅TT tandem mispairs were at positions 88 and 89, respectively. For in vitro repair of mismatches, 25-μl reactions contained 30 mM Hepes (pH 7.8); 7 mM ATP; 200 μM each of CTP, GTP, and UTP; 100 μM each of dATP, dGTP, dTTP, and dCTP; 40 mM creatine phosphate; 100 μg/ml creatine kinase; 15 mM sodium phosphate (pH 7.5); 1 fmol of heteroduplex DNA substrate; and 50 μg each of cell-free extract protein. Reactions were incubated for 15 min and then processed as described (20).
RESULTS
Hypermutation in Vκ Genes from XPA- and PMS2-Deficient Mice.
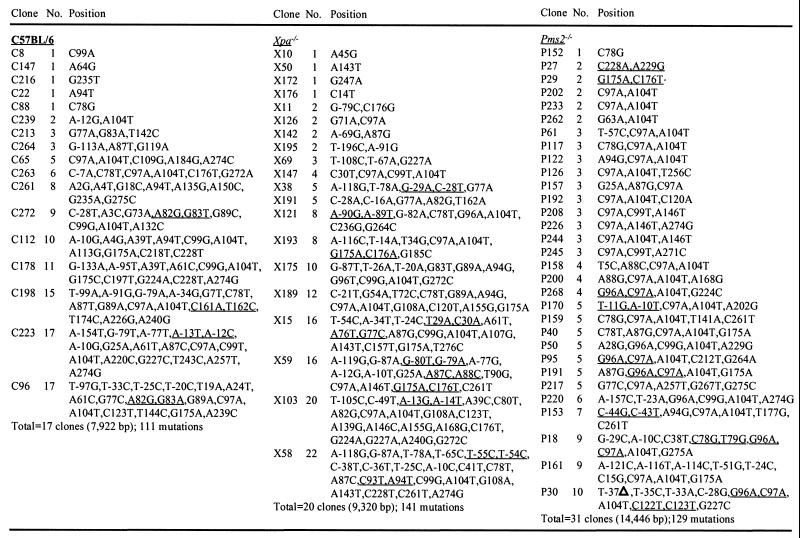
C57BL/6, Xpa−/−, and Pms2−/− mice were immunized with oxazolone coupled to a protein, which elicits a well-characterized immune response (21). After primary and secondary immunizations, the mice were sacrificed, and splenic B cells expressing a receptor for peanut agglutinin, which usually is found on cells residing in germinal centers, were enriched by flow cytometry. DNA was prepared, and oligonucleotides were used to amplify the JκOx1 gene segment rearranged to the Jκ5 gene segment, which encodes a κ light chain that is common in antibodies that bind oxazolone. A 466-bp product, which includes 190 bp of 5′ flanking DNA and 276 bp of coding DNA, was cloned from PCR libraries. The fragment, shown in Fig. 1, was sequenced to unequivocally identify the VκOx1 gene from other closely related Vκ4 family genes (23) and to locate mutations. Approximately half of the clones from all three groups had mutations. For clones with at least one mutation, the overall frequency of mutation was 1.4% mutations per bp for C57BL/6 clones, 1.5% for Xpa−/− clones, and 0.9% for Pms2−/− clones (Fig. 2).
Figure 2.
Number and position of mutations in VκOx1 genes from wild-type and repair-deficient mice. Nucleotide position is numbered according to Fig. 1. No., number of mutations per clone; tandem pairs are underlined; ▵, deletion. The clones are unique in that they have either different V-J joins or different mutations.
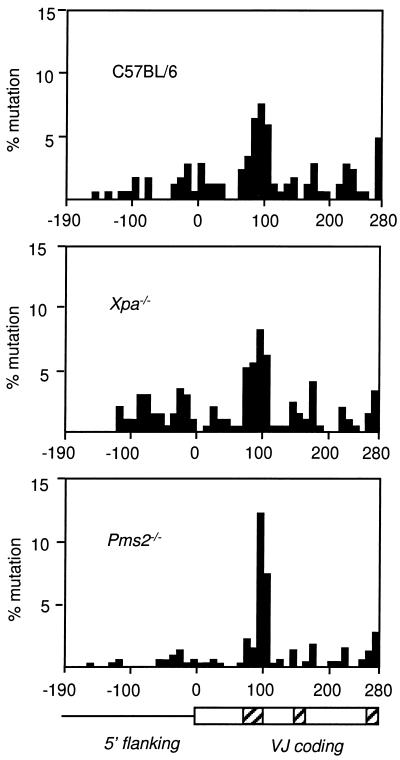
The distribution of mutations in the 5′ flanking and coding regions is diagrammed in Fig. 3. As expected, VκOx1 genes from all three strains had an accumulation of substitutions in the first complementarity determining region. It has been shown that variant amino acids in codons 34 and 36 bind oxazolone with higher affinity than the original amino acids, and B cells expressing antibodies with these mutations are preferentially selected (24, 25). The distribution was more skewed in V genes from Pms2−/− mice, which may be the result of a lower frequency of mutation followed by selection of those B cells with occasional substitutions in residues 34 and 36. When mutations at the codons that encode amino acids 34 (nucleotides 97–99) and 36 (nucleotides 103–105) were excluded, the frequency of mutation in Pms2−/− clones was lower at 0.5% (77 mutations per 14,260 bp), compared with C57BL/6 clones at 1.2% (92 mutations per 7,820 bp) and Xpa−/− clones at 1.4% (125 mutations per 9,200 bp).
Figure 3.
Mutation frequency of 10-nt increments. A diagram of the gene with flanking (line) and coding (box) regions is shown at the bottom. The hatched rectangles within the coding region denote the three complementarity-determining areas of the antibody gene up to the J gene segment. The abscissa is numbered according to Fig. 1, and the frequency of mutation per 10-nt increments is calculated as the number of mutations per increment, divided by 10, divided by the number of clones, multiplied by 100.
Tandem Substitutions in V Genes from Pms2−/− Mice.
We next compared the spectra of mutation in clones from the three strains of mice. As shown in Table 1, the types of substitutions in C57BL/6, Xpa−/−, and Pms2−/− clones did not differ significantly from each other (P > 0.4). However, a significantly higher than expected frequency of tandem substitutions, or two mutations in a row, was found in Pms2−/− clones. Tandem substitutions occurred in all of the mouse strains, as recorded in Fig. 2, and were located throughout the 5′ flanking and coding regions. In Pms2−/− mice, five examples of identical tandem pairs of GC to AA spanning codons 33 and 34 (nucleotides 96 and 97) were observed in clones P268, P95, P191, P18, and P30. The repeated occurrence of these tandem mutations was not an artifact that arose during PCR amplification of splenic DNA for the following reasons: (i) the clones were derived from two separate PCR amplifications; (ii) they had different V-J junctions at residue 95, indicating they came from independent precursor B cells; and (iii) they had unique single substitutions that were not shared by other clones. Thus, this tandem pair was generated independently in several genes, and then B cells expressing antibodies with these mutations were selected for the A in the first position of residue 34, which changes the amino acid to encode an antibody with high affinity for oxazolone.
Table 1.
Types of substitutions
| Substitution | C57BL/6 92 mut % | Xpa−/−123 mut % | Pms2−/−76 mut % |
|---|---|---|---|
| A to G | 19 | 19 | 17 |
| A to T | 16 | 8 | 8 |
| A to C | 11 | 6 | 8 |
| T to C | 9 | 8 | 7 |
| T to A | 2 | 7 | 4 |
| T to G | 1 | 2 | 5 |
| C to T | 10 | 17 | 12 |
| C to A | 2 | 3 | 3 |
| C to G | 2 | 2 | 9 |
| G to A | 17 | 19 | 19 |
| G to T | 4 | 3 | 1 |
| G to C | 7 | 6 | 7 |
Mutations in nucleotides 97–99 and 103–105 were excluded from the analysis because of strong immunological selection for mutations in these codons.
The tandem substitutions in clones from C57BL/6 and Xpa−/− mice frequently were found in V genes with a large number of mutations, where there is a higher probability for adjacent mutations to occur by chance. In contrast, the tandem pairs in Pms2−/− clones often were seen in genes with few mutations; for example, the only two mutations in clones P27 and P29 were next to each other. To determine whether the apparent differences between Pms2−/− and the other strains were significant, the expected frequency of tandem mutations was calculated by assuming a random distribution of mutations based on the length of sequence and the number of mutations per clone, and compared with the observed frequency. Data from VκOx1 genes reported in the literature from hybridomas and transgenes from BALB/c mice also were included in the analysis, which is summarized in Fig. 4. Because all of the strains tended to have more tandem mutations than would be expected by chance, the excess was compared between the strains. An analysis of mutations in the entire clonal segment of 466 nucleotides showed that V genes from C57BL/6, BALB/c, and Xpa−/− strains did not differ from each other (P > 0.4), whereas V genes from Pms2−/− mice were significantly more susceptible to tandem mutations compared with the other strains, with respective P values of 0.003, 0.001, and 0.016. To eliminate any bias for selection of mutations in codons 34 and 36, the analysis also was performed on data that excluded mutations in nucleotides 97–105. As seen in the second half of Fig. 4, Pms2−/− V genes still had significantly more tandem mutations than the other strains, with P values of 0.026, 0.002, and 0.049.
Repair of Tandem Mismatches by Cell Extracts.
Given that Pms2−/− mice had a significant increase in the number of tandem mutations, a biochemical assay was used to measure the ability of extracts from wild-type and DNA repair-deficient cell lines to repair heteroduplexes containing single base or tandem mispairs. M13 DNA substrates were made that contained a covalently closed (+) strand and a (−) strand with a nick located several hundred bp away from a single G⋅G mismatch or a tandem mismatch, either AA⋅⋅CC or GG⋅⋅TT. The two strands encode either blue or white (colorless) M13 plaque colors, depending on the sequence and position of the mismatch. If an unrepaired heteroduplex is introduced into an Escherichia coli strain deficient in methyl-directed heteroduplex repair, plaques will have a mixed color phenotype on selective plates, caused by expression of both strands of the heteroduplex. However, repair occurring during incubation of the substrate in a repair-proficient cell extract will reduce the percentage of mixed plaques. Repair catalyzed by the DNA mismatch repair system also will increase the ratio of the (+) strand phenotype relative to that of the (−) strand phenotype, because the nick directs repair to the (−) strand (30).
As expected based on previous reports (18, 31), extracts of HeLa cells efficiently repair a G⋅G mismatch in a strand-specific manner (Table 2). This same extract also repaired two different tandem mismatches. The increased ratio of (+) to (−) strand plaque phenotypes indicates that repair of the tandem mispairs was preferentially directed to the (−) strand containing the nick. Extracts of a second human cell line, GM0131, gave similar results with all three mispairs. Extracts of the PMS2-defective human endometrial carcinoma cell line HEC-1-A failed to repair the G⋅G mismatch (31), as did extracts of fibroblasts derived from a Pms2−/− mouse. Interestingly, the PMS2-deficient human and murine cell extracts failed to repair both tandem mismatches (Table 2).
Table 2.
Mismatch repair activity in cell extracts
| Cell extract | Mismatch (+):(−) | Plaque phenotype, %
|
Repair efficiency, % | (+) Strand to (−) strand
|
|||
|---|---|---|---|---|---|---|---|
| Mixed | (+) | (−) | Ratio | Change in ratio | |||
| None | G⋅G | 51 | 14 | 35 | — | 0.4 | — |
| AA⋅⋅CC | 53 | 9 | 38 | — | 0.24 | — | |
| GG⋅⋅TT | 38 | 36 | 26 | — | 1.4 | — | |
| HeLa, human, wild type | |||||||
| (4) | G⋅G | 19 | 65 | 16 | 63 | 4.0 | 10.0 |
| (2) | AA⋅⋅CC | 21 | 34 | 44 | 60 | 0.77 | 3.2 |
| (2) | GG⋅⋅TT | 15 | 72 | 13 | 61 | 5.5 | 3.9 |
| GM0131, human, wild type | |||||||
| (1) | G⋅G | 28 | 42 | 30 | 45 | 1.4 | 3.5 |
| (1) | AA⋅⋅CC | 24 | 39 | 36 | 55 | 1.1 | 4.6 |
| (1) | GG⋅⋅TT | 12 | 72 | 16 | 68 | 4.5 | 3.2 |
| HEC-1-A, human, Pms2−/− | |||||||
| (2) | G⋅G | 56 | 11 | 33 | 0 | 0.33 | 0.8 |
| (1) | AA⋅⋅CC | 55 | 10 | 35 | 0 | 0.29 | 1.2 |
| (1) | GG⋅⋅TT | 39 | 38 | 23 | 0 | 1.6 | 1.1 |
| C18, murine, Pms2−/− | |||||||
| (4) | G⋅G | 50 | 12 | 38 | 2 | 0.32 | 0.8 |
| (3) | AA⋅⋅CC | 50 | 13 | 37 | 6 | 0.35 | 1.4 |
| (2) | GG⋅⋅TT | 39 | 34 | 27 | 1 | 1.3 | 0.9 |
Numbers in parentheses are independent determinations.
DISCUSSION
The role of nucleotide excision repair in hypermutation previously was studied in repair-deficient human cells (32) and cell lines (33), and hypermutation was observed. However, these studies were carried out with cells mutant in the Xpb and Xpd genes, which not only are nucleotide excision repair proteins but are also components of the basal RNA polymerase II transcription apparatus. Because both Xpb and Xpd are essential genes necessary for life, a definitive experiment to completely eliminate their function is not possible. Mice with a homozygous defect in the Xpc gene (34) also had hypermutation, but XPC is not required for transcription-coupled nucleotide excision repair. In contrast, total disruption of XPA function is both compatible with viability and eliminates all nucleotide excision repair in mammalian cells, whether transcription-coupled or not. The results presented here and in a recent report by Jacobs et al. (35) indicate that mice with a completely inactivating mutation in the Xpa gene (14) still respond to antigenic challenge with mutated antibodies. This finding conclusively demonstrates that somatic hypermutation of Ig genes does not require nucleotide excision repair.
A connection of hypermutation to other DNA repair pathways has not been explored. Because the generation of somatic mutations in V genes may involve the formation of base⋅base mismatches, it is logical to ask whether mismatch repair affects the hypermutation process. A variety of observations in yeast, mice, and human cells indicate that the PMS2 protein is essential for DNA mismatch repair (15–17, 19, 31, 36), including our finding that extracts of cells derived from PMS2−/− mice do not repair a base⋅base mismatch (Table 2). The results in Figs. 2 and 3 demonstrate that PMS2-deficient mice produce mutated antibodies when challenged with antigen. Thus, the Pms2 gene product is not essential for the hypermutation process. The data also rule out a previous model where direct and indirect palindromic sequences would template mutations during repair of mismatched bases that are formed by misalignment of the sequences (11).
The mutation frequency in the VκOx1 gene was somewhat lower in PMS2-deficient mice than in wild-type mice, comparing all mutations (0.9% vs. 1.4%) or only those thought to be under no selective pressure (0.5% vs. 1.2%). This 2-fold difference could be caused by several possibilities. First, PMS2 may actually help convert mismatches into mutations via excision of the parental nucleotide in the mismatch and replacement by using the mutant strand as a template. A precedence for this concept already exists in E. coli based on repair by MutT and MutY proteins (37). If the PMS2 gene product does indeed participate in converting mismatches into mutations, this function would be distinct from its known role in correcting replication errors. Second, and perhaps more likely, the decrease in frequency may be because of the genetic background of Pms2−/− mice and the experimental conditions. Many factors can affect the ability of B cells to undergo mutation, such as transcription of the rearranged V gene, germinal center formation, quality of antigen stimulation, T cell participation, quality of the cell sort, and selection for B cells expressing the variant protein. If any one of these is altered, the frequency of mutation will be lower.
A recent paper by Cascalho et al. (38) reports a 6- to 22-fold decrease in the level of mutation in V genes from mice deficient for PMS2, whereas we saw only a 2-fold decrease. This discrepancy may be caused by different genetic backgrounds and experimental protocols. Cascalho and coworkers studied quasi-monoclonal mice bred to Pms2−/− mice, did not immunize the mice, examined peripheral blood lymphocytes, and sorted the cells for the presence of an idiotype. In this study, we used Pms2−/− mice, deliberately immunized the mice, isolated spleen cells, and sorted the cells for expression of B220 and a receptor for peanut agglutinin. In determining the frequency of mutation, a critical factor is whether to include the unmutated sequences in the calculations as Cascalho et al. (38) did. As nonmutated clones likely come from cells that were not activated to mutate, a more accurate analysis would be to determine the frequency for clones that have at least one mutation, in which case the decreases in the Cascalho et al. study (38) become less dramatic. Furthermore, it is unlikely that the conventional mismatch repair pathway generates mutations, because B cells from mice defective for other mismatch repair proteins have equal frequencies of hypermutation compared with B cells from wild-type mice (35, 39).
The results in Table 2 demonstrate that extracts of wild-type human cells, but not extracts of PMS2-deficient human or murine cells, are capable of repairing tandem mismatches by preferentially removing nucleotides from the nicked strand. We demonstrate here that the mammalian DNA mismatch repair system can recognize and repair tandem mismatches and that the Pms2 gene is required for repair of tandem mispairs. Tandem base substitution errors are rare in studies where undamaged DNA is replicated by either the DNA polymerases or the mammalian replication apparatus (40). However, the results in Fig. 4 demonstrate that tandem errors frequently are produced during mutation of the VκOx1 gene. They may be preferentially removed in wild-type mice because they cause a greater distortion of the DNA helix than single mispairs. Some tandem mutations occur in clones that have no other mutations, implying that they are produced during a single event. Tandem substitutions may be introduced during error-prone polymerization (41); for example, DNA polymerase β frequently generates tandem misincorporations during filling of a 5-nt gap (42). Clusters (3) and blocks (43) of mutation have been observed before in V genes, implying that tandem mutations are a common feature of hypermutation.
Acknowledgments
We thank Michael Neuberger for the generous gift of antigen; Francis Chrest for assistance in flow cytometry; David Orren for advice; and Richard Wood for many useful discussions and comments on the manuscript. The work was partially supported by National Institutes of Health Grants GM45413 and GM32741 to R.M.L.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviation: XPA, xeroderma pigmentosum group A.
A commentary on this article begins on page 6576.
References
- 1.Pech M, Hochtl J, Schnell H, Zachau H G. Nature (London) 1981;291:668–670. doi: 10.1038/291668a0. [DOI] [PubMed] [Google Scholar]
- 2.Kim S, Davis M, Sinn E, Patten P, Hood L. Cell. 1981;27:573–581. doi: 10.1016/0092-8674(81)90399-8. [DOI] [PubMed] [Google Scholar]
- 3.Gearhart P J, Bogenhagen D F. Proc Natl Acad Sci USA. 1983;80:3439–3443. doi: 10.1073/pnas.80.11.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lebecque S G, Gearhart P J. J Exp Med. 1990;172:1717–1727. doi: 10.1084/jem.172.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goyenechea B, Klix N, Yelamos J, Williams G T, Riddell A, Neuberger M S, Milstein C. EMBO J. 1997;16:3987–3994. doi: 10.1093/emboj/16.13.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters A, Storb U. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- 7.Tumas-Brundage K, Manser T. J Exp Med. 1997;185:239–250. doi: 10.1084/jem.185.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Betz A G, Milstein C, Gonzalez-Fernandez A, Pannell R, Larson T, Neuberger M S. Cell. 1994;77:239–248. doi: 10.1016/0092-8674(94)90316-6. [DOI] [PubMed] [Google Scholar]
- 9.Wilson P C, de Bouteiller O, Liu Y-L, Potter K, Banchereau J, Capra J D, Pascual V. J Exp Med. 1998;187:59–70. doi: 10.1084/jem.187.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith D S, Creadon G, Jena P K, Portanova J P, Kotzin B L, Wysocki L J. J Immunol. 1996;156:2642–2652. [PubMed] [Google Scholar]
- 11.Golding G B, Gearhart P J, Glickman B W. Genetics. 1987;115:169–176. doi: 10.1093/genetics/115.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedberg E C. Annu Rev Biochem. 1996;65:15–42. doi: 10.1146/annurev.bi.65.070196.000311. [DOI] [PubMed] [Google Scholar]
- 13.Korogodin V I, Korogodina V L, Fajszi C, Chepurnoy A, Mikhova-Tsenova N, Simonyan N V. Yeast. 1991;7:105–117. doi: 10.1002/yea.320070204. [DOI] [PubMed] [Google Scholar]
- 14.Nakane H, Takeuchi S, Yuba S, Saijo M, Nakatsu Y, Murai H, Nakatsuru Y, Ishikawa T, Hirota S, Kitamura Y, et al. Nature (London) 1995;377:165–168. doi: 10.1038/377165a0. [DOI] [PubMed] [Google Scholar]
- 15.Modrich P, Lahue R. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 16.Kolodner R. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 17.Umar A, Kunkel T A. Eur J Biochem. 1996;238:297–307. doi: 10.1111/j.1432-1033.1996.0297z.x. [DOI] [PubMed] [Google Scholar]
- 18.Umar A, Buermeyer A B, Simon J A, Thomas D C, Clark A B, Liskay R M, Kunkel T A. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 19.Baker S M, Bronner C E, Zhang L, Plug A W, Robatzek M, Warren G, Elliott E A, Yu J, Ashley T, Arnheim N, et al. Cell. 1995;82:309–319. doi: 10.1016/0092-8674(95)90318-6. [DOI] [PubMed] [Google Scholar]
- 20.Thomas D C, Umar A, Kunkel T A. In: Methods: A Companion to Methods in Enzymology. Friedberg E C, editor. New York: Academic; 1995. pp. 187–197. [Google Scholar]
- 21.Kaartinen M, Griffiths G M, Markham A F, Milstein C. Nature (London) 1983;304:320–324. doi: 10.1038/304320a0. [DOI] [PubMed] [Google Scholar]
- 22.Rada C, Gonzalez-Fernandez A, Jarvis J M, Milstein C. Eur J Immunol. 1994;24:1453–1457. doi: 10.1002/eji.1830240632. [DOI] [PubMed] [Google Scholar]
- 23.Milstein C, Even J, Jarvis J M, Gonzalez-Fernandez A, Gherardi E. Eur J Immunol. 1992;22:1627–1634. doi: 10.1002/eji.1830220642. [DOI] [PubMed] [Google Scholar]
- 24.Griffiths G M, Berek C, Kaartinen M, Milstein C. Nature (London) 1984;312:271–275. doi: 10.1038/312271a0. [DOI] [PubMed] [Google Scholar]
- 25.Alzari P M, Spinelli S, Mariuzza R A, Boulot G, Poljak R J, Jarvis J M, Milstein C. EMBO J. 1990;9:3807–3814. doi: 10.1002/j.1460-2075.1990.tb07598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez-Fernandez A, Milstein C. Proc Natl Acad Sci USA. 1993;90:9862–9866. doi: 10.1073/pnas.90.21.9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berek C, Jarvis J M, Milstein C. Eur J Immunol. 1987;17:1121–1129. doi: 10.1002/eji.1830170808. [DOI] [PubMed] [Google Scholar]
- 28.Lozano F, Rada C, Jarvis J M, Milstein C. Nature (London) 1993;363:271–273. doi: 10.1038/363271a0. [DOI] [PubMed] [Google Scholar]
- 29.Brownlee K A. Statistical Theory and Methodology in Science and Engineering. New York: Wiley; 1965. pp. 183–185. [Google Scholar]
- 30.Thomas D C, Roberts J D, Kunkel T A. J Biol Chem. 1991;266:3744–3751. [PubMed] [Google Scholar]
- 31.Risinger J I, Umar A, Barrett J C, Kunkel T A. J Biol Chem. 1995;270:18183–18186. doi: 10.1074/jbc.270.31.18183. [DOI] [PubMed] [Google Scholar]
- 32.Wagner S D, Elvin J G, Norris P, McGregor J M, Neuberger M S. Int Immunol. 1996;8:701–705. doi: 10.1093/intimm/8.5.701. [DOI] [PubMed] [Google Scholar]
- 33.Kim N, Kage K, Matsuda F, Lefranc M-P, Storb U. J Exp Med. 1997;186:413–419. doi: 10.1084/jem.186.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen H M, Cheo D L, Friedberg E, Storb U. Mol Immunol. 1997;34:527–533. doi: 10.1016/s0161-5890(97)00064-3. [DOI] [PubMed] [Google Scholar]
- 35.Jacobs, H., Fukita, Y., van der Horst, G. T. J., de Boer, J., Weeda, G., Essers, J., de Wind, N., Engelward, B. P., Samson, L., Verbeek, S., et al. (1998) J. Exp. Med., in press. [DOI] [PMC free article] [PubMed]
- 36.Li G M, Modrich P. Proc Natl Acad Sci USA. 1995;92:1950–1954. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vidmar J J, Cupples C G. Can J Microbiol. 1993;39:892–894. doi: 10.1139/m93-133. [DOI] [PubMed] [Google Scholar]
- 38.Cascalho M, Wong J, Steinberg C, Wabl M. Science. 1998;279:1207–1210. doi: 10.1126/science.279.5354.1207. [DOI] [PubMed] [Google Scholar]
- 39.Phung, Q. H., Winter, D. B., Cranston, A., Tarone, R. E., Bohr, V. A., Fishel, R. & Gearhart, P. J. (1998) J. Exp. Med., in press. [DOI] [PMC free article] [PubMed]
- 40.Roberts J D, Kunkel T A. In: DNA Replication in Eukaryotic Cells. DePamphilis M L, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 217–247. [Google Scholar]
- 41.Seidman M M, Bredberg A, Seetharam S, Kraemer K H. Proc Natl Acad Sci USA. 1987;84:4944–4948. doi: 10.1073/pnas.84.14.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beard W A, Osheroff W P, Prasad R, Sawaya M R, Jaju M, Wood T G, Kraut J, Kunkel T A, Wilson A H. J Biol Chem. 1996;271:12141–12144. doi: 10.1074/jbc.271.21.12141. [DOI] [PubMed] [Google Scholar]
- 43.Klein R, Jaenichen R, Zachau H G. Eur J Immunol. 1993;23:3248–3271. doi: 10.1002/eji.1830231231. [DOI] [PubMed] [Google Scholar]